Abstract
Reactive chlorine species (RCS), such as hypochlorous acid (i.e., bleach), are antimicrobial oxidants produced by the innate immune system. Like many redox-regulated transcription factors, the Escherichia coli repressor NemR responds to RCS by using the reversible oxidation of highly conserved cysteines to alter its DNA-binding affinity. However, earlier work showed that RCS response in NemR does not depend on any commonly known oxidative cysteine modifications. We have now determined the crystal structure of NemR, showing that the regulatory cysteine, Cys106, is in close proximity to a highly conserved lysine (Lys175). We used crystallographic, biochemical, and mass spectrometric analyses to analyze the role of this lysine residue in RCS sensing. Based on our results, we hypothesize that RCS treatment of NemR results in the formation of a reversible Cys106-Lys175 sulfenamide bond. This is, to our knowledge, the first description of a protein whose function is regulated by a cysteine–lysine sulfenamide thiol switch, constituting a novel addition to the biological repertoire of functional redox switches. Antioxid. Redox Signal. 23, 747–754.
Introduction
Reactive chlorine species (RCS), including hypochlorous acid (HOCl) and reactive chloramines such as N-chlorotaurine (NCT), are powerful antimicrobial oxidants produced by neutrophils during inflammation (2). To survive RCS exposure, Escherichia coli mounts a complex and still incompletely understood stress response that is coordinated by several different transcriptional regulators (2). These are distinct from the transcription factors that sense reactive oxygen species (ROS) and include the repressor NemR (2, 3).
NemR integrates the E. coli stress responses to both RCS and toxic electrophiles (3, 6, 8). NemR controls expression of three proteins: itself, NemA, and GloA. NemA is an oxidoreductase able to reduce electrophiles, including glyoxal (GO), methylglyoxal (MGO), oxidized quinones, and N-ethylmaleimide (6, 8). GloA plays a key role in GO and MGO detoxification (6). We previously demonstrated that MGO accumulates in RCS-stressed E. coli as part of a pathway facilitating production of polyphosphate, a biopolymer that prevents the aggregation of oxidized proteins (4). It is therefore not surprising that both NemA and GloA are necessary for HOCl survival in E. coli (3).
The response of NemR to both RCS (3) and electrophiles (6, 8) involves reversible redox modifications of distinct cysteine residues. NemR has six cysteines located at positions 21, 98, 106, 116, 149, and 153. Surprisingly, while Cys106 is sufficient for the response to RCS (3), the response to electrophiles requires Cys21 and Cys116 instead (6). This makes NemR a very unusual sensor that uses distinct redox cysteine modifications to sense different stress conditions. In response to electrophiles, NemR forms intermolecular disulfide bonds involving Cys21 and Cys116, resulting in cross-linked dimers that are unable to bind DNA to repress transcription (6). In our previous work (3), we showed that oxidation of Cys106 by RCS also prevents DNA binding. We were unable, however, to identify any of the common cysteine redox modifications in RCS-treated NemR and excluded formation of disulfide bonds or sulfenic acid (−SOH). Nevertheless, we found that the oxidative modification of Cys106 was reversible by the thiol-reducing agent, dithiothreitol (DTT), indicating that the regulatory modification was neither a sulfinic (−SO2H) nor a sulfonic acid (−SO3H) (3).
Innovation.
During inflammation, the innate immune system attacks bacteria by generating reactive chlorine species (RCS), including the powerful oxidant, hypochlorous acid. The Escherichia coli transcription factor NemR is able to sense RCS to regulate expression of stress response genes. We present data supporting a model in which NemR does so via formation of a reversible sulfenamide bond between conserved cysteine and lysine residues. These results have not only identified a previously undescribed regulatory protein modification but also make an important contribution to the molecular understanding of how bacteria sense and respond to inflammatory oxidants.
The objective of this study was therefore to identify the regulatory modification of Cys106 in RCS-oxidized NemR. We employed crystallographic, biochemical, and mass spectrometry methods and present evidence that RCS treatment likely leads to the formation of a reversible sulfenamide bond between Cys106 and the nearby Lys175. This is, to our knowledge, the first example of a cysteine–lysine bond occurring in a natural protein. This result makes a significant contribution to the molecular understanding of how bacteria sense and respond to RCS.
Results
The crystal structure of NemR shows lysine 175 in close proximity to cysteine 106
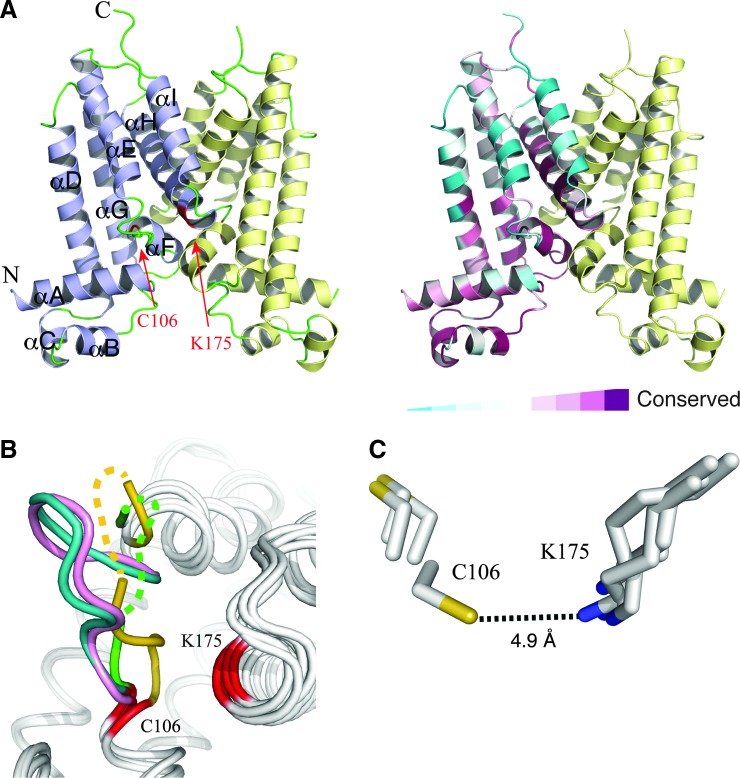
We determined the crystal structure of the single cysteine NemRC106 only variant of E. coli NemR by molecular replacement using the Pseudomonas aeruginosa PA2196 structure as a search model (5). NemR is a dimer in solution and, as expected, crystallizes as such. The subunit structure of NemR contains an all-α fold, with nine α-helices (αA-αI) of varying lengths (Fig. 1A). The C-terminal two α-helices (αH-αI) are largely responsible for dimer formation. Two short helices (αB-αC) near the N-terminus form the canonical helix-turn-helix DNA-binding motif and have been shown previously to be responsible for DNA binding (5). E. coli NemR displays a high degree of structural similarity to three bacterial NemR homologs whose structures are known [P. aeruginosa PA2196 (5); Acinetobacter baylyi ACIAD2740 (PDB 3KNW); and Rhodobacter sphaeroides RSP_1435 (PDB 3BRU)]. Alignment of E. coli NemR with these structures showed a root mean square deviation of 1.42 Å (A. baylyi), 2.41 Å (P. aeruginosa), and 3.18 Å (R. sphaeroides) (the Cα atom positions).
FIG. 1.
The crystal structure of Escherichia coli NemRC106 only. (A) Shown is a schematic representation of NemR in two color schemes. Left panel: the two subunits of NemRC106 only dimer are colored in blue and yellow, respectively. Helices within one subunit are labeled αA–αI. Cys106 and Lys175 are colored red. Right panel: residues in one subunit are colored based on their sequence conservation. (B) Structural flexibility of the loop between helices αE and αF (EF loop). The four NemRC106 only subunit structures within the crystallographic asymmetric unit are superimposed and shown as white coils. The EF loops in the four subunits are colored in blue, pink, green, and orange, respectively. Cys106 and Lys175 are colored in red. Part of the loop structure in two subunits (green and orange) is not defined due to poor electron density. (C) Cys106 and Lys175 are in close proximity. Side chains of Cys106 and Lys175 as observed in the asymmetric unit are shown as stick models and colored using the following scheme: carbon (white), nitrogen (blue), and sulfur (yellow).
We have previously shown that Cys106, which is the only cysteine conserved among all NemR homologs, is responsible for E. coli stress responses to RCS (3). It is located at the C-terminal end of a loop between helices αE and αF (the EF loop). Comparison of the four independent NemR subunit structures in the crystallographic asymmetric unit revealed that the EF loop constitutes one of the conformationally most flexible regions within the NemR structure. The conformation of the loop is dramatically different in the four structural copies (Fig. 1B). In two subunits, the electron density for part of the loop was too poor to allow confident model building. We found the side chain of Cys106 to adopt two distinct conformations (Fig. 1C). In one conformation, Cys106 points directly to the C-terminal end of helix αH, bringing the Sγ of Cys106 into very close proximity to the Nζ of Lys175 (distance: 4.9 Å). Sequence analysis revealed that Lys175 is conserved among 85.5% of NemR homologs (183 of 214 genera), and many sequences that lack a lysine at this position contain arginine instead (16 of 214 genera). This high degree of conservation together with the known reactivity of sulfenyl chlorides with lysine and arginine residues to form reversible sulfenamide sulfur–nitrogen cross-links (1) led us to consider the possibility that NemR RCS sensing works through the formation of a reversible sulfenamide bond.
Fluorescence and mass spectrometric analyses of RCS-treated NemR are consistent with the formation of a Cys106–Lys175 sulfenamide bond
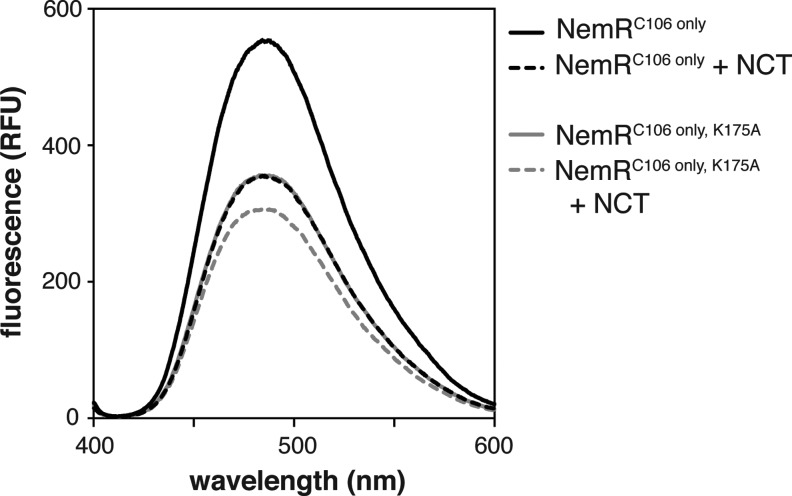
To test whether RCS treatment causes a loss in free amino content, which would be expected if Lys175 did indeed undergo sulfenamide formation, we conducted fluorescence spectroscopy with fluorescamine, a fluorophore known to react with accessible primary amino groups. We found that oxidation of NemRC106 only with NCT substantially reduced the free amino content of NemR, consistent with the oxidation of one or more amino groups (Fig. 2, compare black solid and black dashed lines). Interestingly, substitution of Lys175 with Ala (NemRC106 only, K175A) yielded in the same decrease of fluorescamine binding as observed in NCT-treated NemRC106 only (Fig. 2, solid gray line), suggesting that NCT treatment does indeed involve Lys175. NCT-mediated oxidation of NemRC106 only, K175A resulted in only a small additional change in free amino content (Fig. 2, dashed gray line). These results strongly supported the idea that oxidation of NemR leads to the reversible modification of the free amine group in Lys175.
FIG. 2.
Oxidation reduces the free amine content of NemRC106 only. Oxidized and reduced NemR variants (1 ml, 50 μg ml−1) were mixed with fluorescamine (0.334 μl, 3 mg ml−1 in acetone) and fluorescence was measured, indicating the proportion of accessible lysine side chains in each sample.
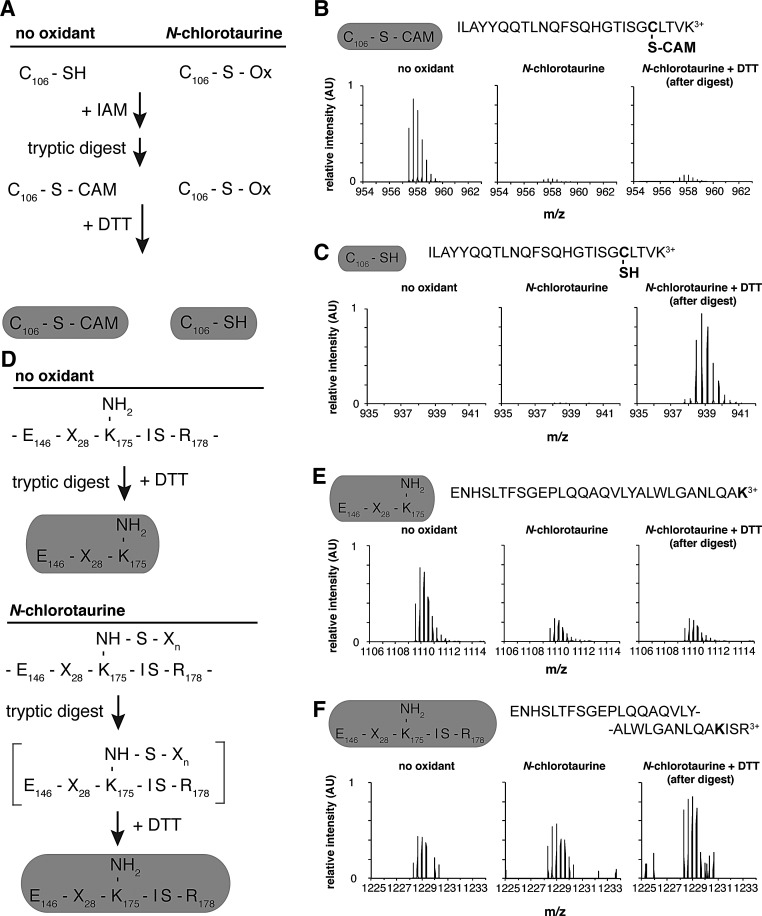
To directly test whether oxidation of NemRC106 only results in sulfenamide formation, we conducted mass spectrometry and compared the molecular mass of reduced and NCT-oxidized NemRC106 only. We found the expected mass of 22,418 Da for reduced NemRC106 only. In contrast, oxidation with NCT resulted in two species with masses of 22,416 (i.e., −2 Da) and 22,450 Da (i.e., +32 Da). These masses are consistent with a mixture of NemR species containing either sulfenamide (−2 Da) or irreversible sulfinic acid (+32 Da) modifications and fit to previous observations that NCT treatment of NemRC106 only is only partially reversible (3). Subsequent liquid chromatography/mass spectrometry (LC-MS) analysis of tryptically digested NemRC106 only further supported our finding. To distinguish between reduced and reversibly oxidized thiol modifications, we prepared reduced and NCT-treated NemRC106 only and incubated the proteins with iodoacetamide (IAM) to irreversibly alkylate all reduced cysteine residues (Fig. 3A). Whereas the Cys106-containing peptide in reduced NemRC106 only showed a mass fully consistent with alkylation, NCT treatment caused the complete disappearance of the alkylated peptide (Fig. 3B). Treatment of the digested peptides with DTT caused the reappearance of the reduced peptide (Fig. 3C), indicating that Cys106 was reversibly oxidized. Very similar results were observed with the Lys175-containing peptide (Fig. 3D), which decreased in abundance upon oxidation of NemR as well (Fig. 3E). DTT incubation of the tryptic digest led to the reappearance of the lysine-containing peptide (Fig. 3F). However, the newly appeared peptide was slightly longer, missing the cleavage immediately after Lys175. These results suggested that modification of Lys175 affects its accessibility to trypsin, and were fully consistent with the formation of a Cys106-Lys175 sulfenamide in oxidized NemR.
FIG. 3.
Mass spectra of oxidized NemR are consistent with the formation of a cysteine 106–lysine 175 sulfenamide bond. (A) To investigate modifications of Cys106 and Lys175 upon NCT oxidation, we blocked any unmodified cysteine 106 with IAM in reduced and oxidized NemRC106 only and examined tryptic peptides both before and after DTT reduction. (B) Normalized LC-MS spectra revealed a reduction of the 957.48 Da IAM-modified Cys106-containing peptide (+3 charged) in NCT-treated NemRC106 only. (C) Conversely, the 938.47 Da Cys106-containing peptide (+3 charged) without IAM modification could be released by DTT reduction from a digest of IAM-treated NCT-oxidized NemRC106 only. (D) Predicted results of tryptic digestion of the Lys175-containing region of NemRC106 only with and without NCT oxidation. DTT reduction should have no effect on an unmodified lysine, while it should reduce a sulfenamide bond. (E) NCT treatment of NemRC106 only makes the potential cleavage site, Lys175, less accessible to trypsin and decreases the abundance of the 1109.57 Da Lys175-containing peptide (+3 charged). (F) DTT reduction of a tryptic digest of IAM-treated NCT-oxidized NemRC106 only increases the amount of the 1228.32 Da Lys175-containing peptide (+3 charged) without tryptic cleavage at Lys175. The predicted Cys106-Lys175 covalently linked peptide was not detected in any sample. However, this peptide is very large (6.1 kDa) and may not be easily detectable with LC-MS. DTT, dithiothreitol; IAM, iodoacetamide; LC-MS, liquid chromatography/mass spectrometry; NCT, N-chlorotaurine.
Disruption of lysine 175 prevents DNA binding
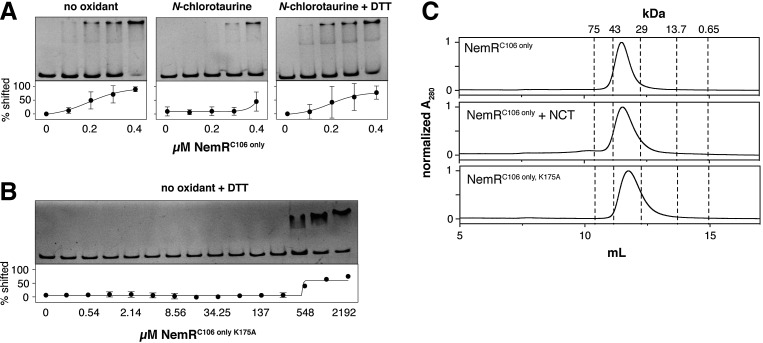
We reasoned that if RCS sensing by NemR did indeed depend on Cys106-Lys175 sulfenamide formation, disruption of Lys175 should result in a decrease in DNA-binding affinity similar to the one observed in oxidized NemR. As previously shown (3), treatment with NCT reduced the affinity of NemRC106 only for a DNA fragment containing the nemR promoter sequence (PnemR) (Fig. 4A). This loss of DNA-binding activity was largely reversible by addition of DTT (Fig. 4A). Strikingly, even in the absence of RCS, NemRC106 only, K175A binding to the promoter required more than 1,000-fold higher protein concentrations compared with NemRC106 only (Fig. 4B). Size exclusion chromatography revealed that neither oxidation of NemRC106 only nor the substitution of Lys175 to Ala affected the oligomerization state of NemR, suggesting that the low DNA-binding affinity observed in NemRC106 only, K175A is not due to major structural changes in the protein, but presumably caused by more subtle changes in the DNA-binding site. Importantly, in the structure of the P. aeruginosa NemR homolog, PA2197, bound to DNA, Lys175 is not in direct contact with the DNA (5).
FIG. 4.
Disruption of lysine 175 prevents NemRC106 only DNA binding, but does not affect its oligomerization state. (A) EMSAs of the binding of NemRC106 only (0, 100, 200, 300, 400 nM) to nemR promoter DNA (PnemR, 10 nM) before and after treatment with NCT. To test the reversibility of oxidative inactivation, NCT-oxidized NemRC106 only was incubated with the thiol reductant, DTT (5 mM). Representative gels are shown along with quantification results (mean±SD, n=3). (B) EMSA of the binding of NemRC106 only, K175A to 10 nM PnemR DNA in the presence of 5 mM DTT (mean±SD, n=3). Concentration of NemR is plotted on a log2 scale. (C) Size exclusion chromatography was used to compare the oligomeric state of reduced and oxidized NemRC106 only and reduced NemRC106 only, K175A. Dotted lines indicate elution volume of protein standards of known molecular weight. EMSA, electrophoretic mobility shift assay.
Discussion
Analysis of the NemR crystal structure revealed a close proximity between the side chains of Cys106 and Lys175 and led us to hypothesize that NemR responds to RCS by forming a reversible sulfenamide bond. Although the distance between the two residues in the current structure (reduced form) is somewhat longer than what is required for efficient bond formation, the inherent structural flexibility within the EF loop makes it possible for RCS-oxidized Cys106 to approach Lys175. In a preliminary study, we have attempted to determine the crystal structure of the RCS-oxidized NemRC106 only. Although the overall architecture of the protein is maintained, the EF loop structures in all four subunits became disordered, suggesting a possible mechanism for how oxidation leads to bond formation between Cys106 and Lys175.
Sulfenamide formation has been previously reported in only a few other proteins, including protein tyrosine phosphatases (PTPs) (9) and the Bacillus subtilis transcription factor, OhrR (2, 7). However, in both of these cases, the oxidized cysteine reacts with a backbone amide of the polypeptide chain causing a cyclic sulfenamide formation. In PTPs, sulfenamide formation is thought to protect the active site cysteine from irreversible overoxidation (9), and it is possible that formation of a Cys106-Lys175 sulfenamide in NemR plays a similar role, especially given the high reactivity of RCS with cysteine residues (2). Based on the conservation of Cys106 and Lys175 among NemR homologs from diverse bacterial genera, we propose that formation of a regulatory sulfenamide bond may represent a broadly distributed redox switch mechanism, although the functions of most NemR homologs have not yet been examined.
It has been known for some time that RCS can oxidize cysteines to sulfenyl chlorides and that these can react with lysine or arginine residues to form covalent sulfur–nitrogen bonds, but this has been considered to be a mechanism by which RCS damages proteins (1). NemR, however, appears to employ this mechanism as a novel thiol switch that uses this chemistry to specifically sense RCS. These findings help explain how NemR distinguishes between RCS and ROS and provide important insights into the molecular mechanism by which bacteria sense and respond to inflammatory oxidants.
Notes
Reagents and growth media
Bacterial growth media were from Difco. Unless otherwise specified, all other chemicals and reagents were from either Sigma-Aldrich or Fisher.
Construction of nemR plasmids
The nemRT61A, T292A, T346A, T445A, T447C, T457A allele from plasmid pNEMR40 (3), encoding the NemRC106 only variant protein (NemRC21S, C98S, C116S, C149S, C153S), was subcloned into the BamHI and HindIII sites of plasmid pSUMO2 (Life Sensors, Inc.) to generate plasmid pSUMO2-NemR. The QuickChange site-directed mutagenesis kit (Stratagene) was used to mutate pSUMO2-NemR with primer 5′ CGA ATC TGC AGG CCG CGA TTT CGC GCA GTT TC 3′. This yielded plasmid pNEMR47, containing the nemRT61A, T292A, T346A, T445A, T447C, T457A, A523G, A524C, A525G allele, encoding NemRC106 only, K175A (NemRC21S, C98S, C116S, C149S, C153S, K175A).
Purification of NemR proteins
NemR variants with N-terminal His6-SUMO tags were overproduced in E. coli BL21(DE3) (Novagen) and purified using a nickel-loaded HiTrap Chelating HP affinity column (GE Healthcare Life Sciences) and an ÄKTA fast protein liquid chromatography system (Amersham Biosciences). The resulting protein was treated with small ubiquitin-like modifier (SUMO) protease (Life Sensors, Inc.) to remove the His6-SUMO tag and purified with an additional passage through a nickel-loaded HiTrap Chelating HP affinity column. The resulting protein was stored at −80°C in 50 mM potassium phosphate, pH 8.0, 400 mM NaCl, 5 mM DTT, 1 mM EDTA, and 10% glycerol.
Crystallization, data collection, and structure determination
For crystallization, NemRC106 only was further purified by ion-exchange chromatography on a Source Q column (GE Healthcare) with a buffer containing 25 mM Tris (pH 8.0), 1 mM DTT, and 0–1 M NaCl gradient. NemRC106 only (25 mg ml−1 in 25 mM Tris (pH 8.0), 1 mM DTT, and 200 mM NaCl) was crystallized in 3.2 M NaCl and 100 mM sodium acetate (pH 4.6) at 22°C using the sitting-drop vapor diffusion method. Crystals were cryoprotected in 3.5 M NaCl, 100 mM sodium acetate (pH 4.6), and 35% sucrose. Diffraction data were collected at Advanced Photon Source LS-CAT 21-ID-F (Argonne National Laboratory). Crystals were cryocooled in liquid nitrogen during data collection. The crystal diffracted to 2.2 Å and belonged to the space group of P212121 with two NemR dimers in the asymmetric unit. All data were processed using HKL2000 (HKL Research). The structures were solved by molecular replacement using the crystal structure of P. aeruginosa NemR (PDB entry: 3RD3) as the initial search model. The PHENIX software suite was used for structure determination and refinement. Model building was done with COOT. Data collection and structure refinement statistics are shown in Table 1.
Table 1.
Crystallographic Data Statistics
| Data collection | |
|---|---|
| Space group | P212121 |
| Unit cell parameters: | |
| a, b, c (Å) | 67.4, 67.3, 213.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Molecules/ASU | 2 dimers |
| Wavelength (Å) | 0.9787 |
| Resolution (Å) | 2.2 |
| Unique reflections | 50363 |
| Redundancya | 7.3 (7.5) |
| Completeness (%) | 99.9 (100.0) |
| Average I/σ (I) | 23.9 (3.0) |
| Rmerge | 0.08 (0.73) |
| Refinement | |
|---|---|
| Resolution range (Å) | 46.5–2.2 |
| Rwork (%) | 20.7 |
| Rfree (%) | 25.5 |
| RMS deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.03 |
| B-factor average (A2) | 60.1 |
| Ramachandran plot | |
| Most favored (%) | 97.3 |
| Allowed (%) | 2.6 |
| Outliers (%) | 0.1 |
| PDB accession code | 4YZE |
Values in parentheses are for the specified high-resolution bin.
Bioinformatics analysis
Custom Python scripts using the Biopython 1.59 (biopython.org) and Enthought Canopy (enthought.com) packages were used to search for and sort NemR homologs from the National Center for Biotechnology Information databases (accessed on March 3, 2015). Using the C-terminal sensing domain of the E. coli MG1655 NemR (residues 59–199) as a search sequence, and setting a BLAST e-value cutoff of 1, we retrieved the sequences of NemR homologs from 214 bacterial genera. Sequence alignments were performed using MUSCLE 3.8 (www.drive5.com/muscle/) and visualized using WebLogo 3.1 (weblogo.threeplusone.com). Mutagenic primers were designed with PrimerX (bioinformatics.org/primerx/).
Oxidation of NemR
Oxidized NemR was prepared using a variation of our previously described method (3). NCT was prepared fresh before each experiment (3). Purified NemR was exchanged into 50 mM sodium phosphate, pH 8, and 150 mM NaCl with Bio-Spin® columns with Bio-Gel® P-30 (Bio-Rad, Inc.) before use. Protein samples were incubated on ice for 30 min in the presence or absence of NCT at a 0.5:1 molar ratio of NCT to NemR monomers, then exchanged into 50 mM sodium phosphate, pH 8, and 150 mM NaCl with or without 10% glycerol using Bio-Spin columns.
DNA-binding assays
Electrophoretic mobility shift assays were performed as previously described (3), except that the buffer, in which NemR and PnemR DNA fragments were incubated before electrophoresis, was 50 mM sodium phosphate, pH 8, 150 mM NaCl, and 10% glycerol. To assess the reversibility of NemR oxidation, 5 mM DTT was added to this incubation.
Size exclusion chromatography
The oligomeric state of NemR was determined using a Superdex 75 10/300 GL size exclusion chromatography column (GE Healthcare Life Sciences) and an ÄKTA fast protein liquid chromatography system (Amersham Biosciences) according to the manufacturer's instructions. The GE Healthcare Gel Filtration Calibration Kit LMW was used to verify the molecular weight of oxidized and reduced NemR.
Fluorescamine assay for accessible amino group content
One milliliter of NemR (50 μg ml−1 in 50 mM sodium phosphate, pH 8, 150 mM NaCl) was mixed with 0.334 μl of fluorescamine (3 mg ml−1 in acetone) (Sigma-Aldrich) and incubated for 15 min in the dark at room temperature. Resulting fluorescent products (λex=388 nm and λem=400–600 nm) were measured with a Hitachi F4500 fluorescence spectrophotometer.
Mass spectrometry analysis of NemR peptides
Protein digestion was performed in Lo-Bind reaction tubes (Eppendorf). Before digestion, 100 μg aliquots of NemR were treated with NCT (as described above) or left untreated as a control. NemR was then precipitated with five volumes of ice-cold acetone at −20°C for 4–16 h. The precipitated protein was centrifuged (13,000 g, 30 min, 4°C) and washed twice carefully by rinsing the pellet with ice-cold acetone. All the acetone was removed and the pellet was redissolved in 6 M urea, 0.5% SDS, 200 mM Tris–HCl pH 8.5, and 10 mM EDTA containing 100 mM iodoacetamide and incubated for 1 h at 37°C. Alkylation was stopped by acetone precipitation as described above. The resulting pellet was washed thoroughly to remove residual iodoacetamide. The pellet was then redissolved in 50 μl of 8 M urea. Five hundred fifty microliters of a 50 mM ammonium bicarbonate solution was added. Two micrograms of Promega sequencing grade-modified trypsin (Promega, Madison, WI) in 10 μl of 50 mM acetic acid was added, and the protein was digested for 12–15 h. One aliquot of NCT-treated NemR was then reduced by addition of DTT to a final concentration of 20 mM and further incubated for 1 h. Digestion or subsequent reduction was stopped by the addition of trifluoroacetic acid (TFA) to a final concentration of 0.1%. The complete sample was then aspirated through an Agilent Bond Elut Omix C18 100 μl tip preconditioned with 50% acetonitrile (ACN; Agilent). The peptides bound to the tip were washed four times with 100 μl 0.1% TFA and eluted with 100 μl 75% ACN 0.1% TFA into a fresh tube. ACN was removed in a vacuum centrifuge, and the peptides were reconstituted to a final volume of 60 μl with 0.2% TFA.
MS analysis was performed as follows: 15 μl of the peptide sample was separated using an HP Ultimate 3000 system (Dionex) at 60°C. A 300 μm×5 mm Acclaim PepMap100 C18 microprecolumn with a 3-μm particle size (Dionex) was used for enrichment and desalting. Loading was performed for 10 min in 95% solvent A (0.1% TFA) and 5% solvent B (0.1% TFA, 50% ACN [v/v]) at a flow rate of 30 μl min−1. Separation was performed on a 75 μm×25 cm Acclaim PepMap 100 C18, 3 μm, 100-Å column (Dionex) with a linear gradient of 95% A, 5% C (0.1% formic acid, 84% ACN) to 60% A, 40% C over 60 min at a flow rate of 0.400 μl min−1. Mass spectra were obtained online by an LTQ Orbitrap Elite Instrument (Thermo Fisher Scientific, Waltham, MA). Fourier transform mass spectrometry was performed in a mass/charge range of 300–2000. The 20 most intense peaks in each MS spectrum were selected for MS/MS fragmentation (collision energy 35 V) in the collision-induced dissociation mode. The exclusion list size was set to 500, with an exclusion duration time of 35.00 s. Peak intensity was normalized across LC-MS runs using the three most intense peaks of NemR peptides, GSFYHYFR (538.75, 2+), GVIALLSQALENGR (720.91, 2+), and EHLLATGEQLSLQR (797.93, 2+).
Abbreviations Used
- DTT
dithiothreitol
- EMSA
electrophoretic mobility shift assay
- GO
glyoxal
- HOCl
hypochlorous acid
- IAM
iodoacetamide
- LC-MS
liquid chromatography/mass spectrometry
- MGO
methylglyoxal
- NCT
N-chlorotaurinet
- NEM
N-ethylmaleimide
- PTP
protein tyrosine phosphatase
- RCS
reactive chlorine species
- ROS
reactive oxygen species
- SUMO
small ubiquitin-like modifier
- TFA
trifluoroacetic acid
Acknowledgments
The authors thank the Indiana University Mass Spectrometry Facility for the analysis of full-length NemR. This work was funded by National Institutes of Health grants, GM065318 (to U.J.) and GM095769 (to Z.X.), as well as by grants of the Deutsche Forschungsgemeinschaft, LE 2905/1-1 (to L.I.L.) and Schw823/3-1 (to U.J.).
References
- 1.Fu X, Mueller DM, and Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: a potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry 41: 1293–1301, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Gray MJ, Wholey WY, and Jakob U. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67: 141–160, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray MJ, Wholey WY, Parker BW, Kim M, and Jakob U. NemR is a bleach-sensing transcription factor. J Biol Chem 288: 13789–13798, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JC, and Jakob U. Polyphosphate is a primordial chaperone. Mol Cell 53: 689–699, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y, Kang Y, and Choe J. Crystal structure of Pseudomonas aeruginosa transcriptional regulator PA2196 bound to its operator DNA. Biochem Biophys Res Commun 440: 317–321, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Lee C, Shin J, and Park C. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88: 395–412, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, Soonsanga S, and Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104: 8743–8748, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umezawa Y, Shimada T, Kori A, Yamada K, and Ishihama A. The uncharacterized transcription factor YdhM is the regulator of the nemA gene, encoding N-ethylmaleimide reductase. J Bacteriol 190: 5890–5897, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, and Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry 46: 709–719, 2007 [DOI] [PubMed] [Google Scholar]