Abstract
In China, a majority of the highly pathogenic porcine reproductive and respiratory syndrome (HP-PRRSV) strains were seeded by the 2006 outbreak. However, the most recently emerged (2013-2014) HP-PRRSV strain has a very different genetic background. It is a NADC30-like PRRSV strain recently introduced from North America that has undergone genetic exchange with the classic HP-PRRSV strains in China. Subsequent isolation and characterization of this variant suggest high pathogenicity, so it merits special attention in control and vaccine strategies.
TEXT
Porcine reproductive and respiratory syndrome (PRRS) is characterized by respiratory distress in nursery swine and reproductive failure in sows and has resulted in huge economic losses to the global swine industry since its first recognition in the United States in 1987 (1). In China, highly pathogenic PRRSV (HP-PRRSV) has been circulating and predominating in the field since the initial outbreak in 2006 and has resulted in the loss of more than one million pigs (2–5). Retrospective studies of that outbreak have shown that the highly pathogenic variant emerged from less pathogenic PRRSV strains in China (6), which were initially introduced from North America in the 1990s. CH-1a was the earliest representative of this group (7). Following the outbreak, more-stringent biosecurity controls and a targeted immunization campaign were undertaken to limit HP-PRRS in China.
However, despite these measures, HP-PRRSV has experienced recurrent population expansions since the initial outbreak. One such reemergence was associated with genetic exchange between two HP-PRRSV viruses circulating in the field (8). Another recent outbreak (2013-2014) is probably in the early stage of emergence. It has occurred in several provinces of China and is characterized by high fever, cough, anorexia, red discoloration of the body, and blue ears. Diseased pigs also have multiple visceral lesions. Their lungs display consolidation, and their lymph nodes are enlarged and hemorrhagic. The rates of morbidity and mortality due to this new HP-PRRSV are very high. An affected farm in Jilin Province had a morbidity rate of 100% and a mortality rate of 76.6% (230/300).
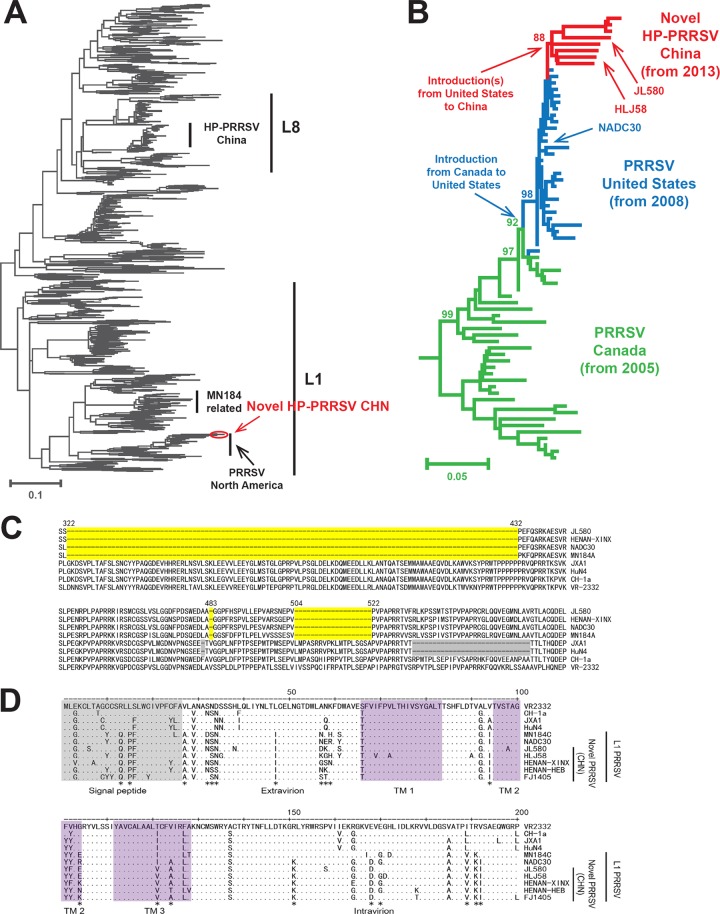
We obtained the representative open reading frame 5 (ORF5) sequences from two farms that experienced the disease, i.e., JL580 and HLJ58 from Jilin Province and Heilongjiang Province, respectively. Phylogenetic analyses using the PhyML version 3.0 software (9) suggested that these viruses are distantly related to the classic HP-PRRSV strains in China, which belong to lineage 8 (Fig. 1A). Instead, they nested deeply within diverse lineage 1, which originated in Canada and is now prevalent in both the United States and Canada (10). Interestingly, the virus is also closely related to a group represented by NADC30, a moderately virulent strain isolated in 2008 in the United States (11). Furthermore, the phylogenetic topology of the diversity surrounding this new HP-PRRSV suggests, with high resolution, a transmission chain from Canada to the United States and then to China, all on a relatively recent time scale (Fig. 1B). This demonstrates the recent expansion of PRRSV over a large geographic area. Within China, the new HP-PRRSV strain has affected not only Jilin and Heilongjiang but also central and southern China, and strains isolated there include HENAN-XINX (accession numbers KF416327 to KF416329), HENAN-HEB (KF416334), HENAN-JIAOZ (KF416333), and FJ1405 (KM453701). The nucleotide sequences of the complete JL580 genome and HLJ58 ORF5 were obtained by the Sanger sequencing approach.
FIG 1.
Genotyping of the HP-PRRSV strain that has newly emerged in China. The phylogenetic positions of all representative type 2 PRRSV strains (A) and the most closely related viruses (B) are shown on a tree based on their ORF5 nucleotide sequences. HP-PRRSV strains newly emerged in China are shown in red, and related viruses from the United States and Canada are shown in blue and green, respectively. Only the important viral groups and strains are labeled to the right of the tree. The genetic relatedness of these strains is also supported by the distribution of the gaps in the nsp2 protein alignment (C). Deletions in JL580 and related virus strains are shaded yellow, and deletions in the classic HP-PRRSV strains are shaded gray. Alignment and comparison of amino acid mutations of GP5 (D). The signal peptide and transmembrane (TM) domains are demarcated and shaded. Important amino acid sequence differences between L1 PRRSV and classical HP-PRRSV are indicated by asterisks.
To further characterize the virus, strain JL580 was isolated on Marc-145 cells and confirmed with an immunofluorescence assay using a monoclonal antibody directed against the PRRSV N protein. Sequence alignment revealed three discontinuous deletions (a total of 131 amino acids) in nonstructural protein 2 (nsp2) that resembled those in the NADC30 and MN184 strains but were not present in the classic Chinese HP-PRRSV strains (Fig. 1C). These deletions can be used as molecular markers to distinguish the JL580-like HP-PRRSV strains from other type 2 PRRSV strains in China. Importantly, in the ORF5 region, JL580 is more closely related to less pathogenic NADC30 than to the classic Chinese HP-PRRSV, and a majority of the altered amino acids lie within the extravirion domain of the protein (Fig. 1D).
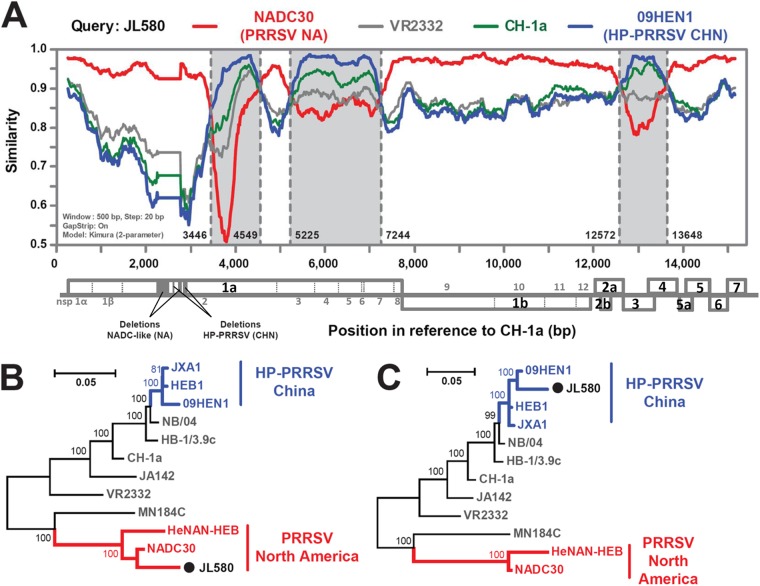
To test for recombination, we performed similarity comparisons within a 500-bp window sliding along the genome alignment (20-bp step size) with SimPlot v3.5.1 (12). The analysis revealed that JL580 is the result of recombination between the NADC30-like viruses and the classic HP-PRRSV strains circulating in China (Fig. 2A). From the similarity plot, we identified six recombination breakpoints: two located in nsp2 (nucleotides [nt] 3446 and 4549) and the others located in nsp3 (nt 5225), nsp7 (nt 7244), ORF2a (nt 12572), and ORF4 (nt 13648). These breakpoints separate the genome into seven regions; four are closely related to PRRSV strains from North America (represented by NADC30) (Fig. 2B), and the remaining three are closely related to classic HP-PRRSV strains in China (Fig. 2C). This suggests that after its introduction from the United States, the virus gained genetic diversity by recombining with local HP-PRRSV strains in China. Interestingly, two other related viruses (i.e., HENAN-XINX and HENAN-HEB) are also possible recombinants, although their recombination patterns differ from that of JL580 (data not shown; the SimPlot results are available upon request). Because these two sequences were not generated in this study, their recombination events cannot be verified here.
FIG 2.
Recombination analysis of strain JL580. (A) Genome scale similarity comparisons of JL580 (query) with NADC30 (red), 09HEN1 (blue), CH-1a (green), and VR2332 (gray). Recombination breakpoints are shown as black dotted lines, with the locations indicated at the bottom. The background color of the major parental regions (parental region A) is white, whereas that of the minor parental regions (parental region B) is gray. Below the similarity plot is a full genome structure, with reference to CH-1a, in which the positions and boundaries of the major ORFs, nsp-encoding genes within ORF1a and ORF2b, and gaps are shown. Phylogenies of parental regions A (B) and B (C) are shown below the similarity plot. The major parental group (NADC30-like viruses) is shown in red; the minor parental group (classic HP-PRRSV in China) is shown in blue.
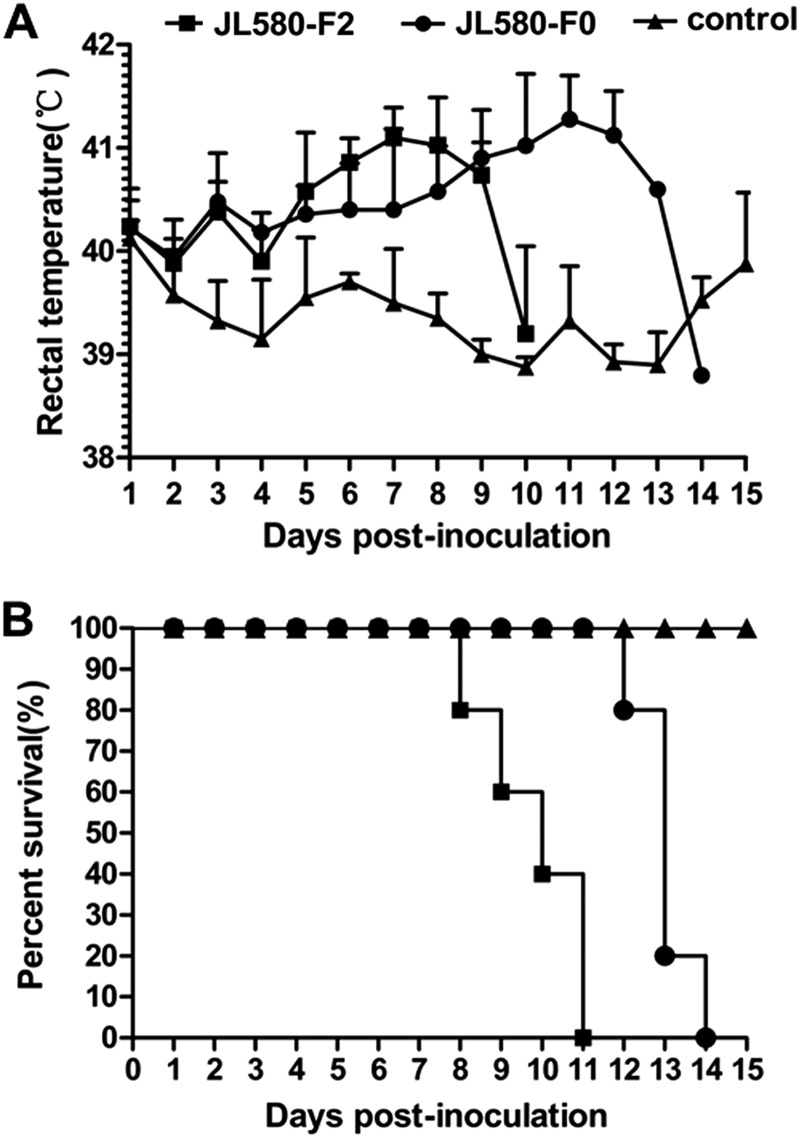
To determine the pathogenicity of JL580, 14 6-week-old PRRSV-free piglets were randomly divided into three groups and maintained in individual biosafety rooms. The piglets in group A (n = 5) were inoculated intramuscularly (1 ml) and intranasally (2 ml) with JL580-F2 (the second passage in Marc-145 cells; 3 × 104.0 50% tissue culture infective doses [TCID50] in 3 ml per pig), respectively, according to the doses and methods previously reported (13). The piglets in group B (n = 5) were infected with JL580 homogenate supernatant (uncontaminated with pseudorabies virus, porcine parvovirus, classical swine fever virus, or porcine circovirus type 2; passed through a 0.45-μm filter; and confirmed by real-time reverse transcription-PCR) at a dose of 3 × 104.0 copies per pig as described for JL580-F2 (14). The group C piglets (n = 4) were mock infected with 3 ml of Dulbecco's modified Eagle's medium. The animals' behavior was observed every day. Blood samples were collected from individual pigs periodically (0, 3, 7, 10, and 14 days postinfection [dpi]).The piglets in groups A and B showed fever from 3 dpi (Fig. 3A) and also displayed obvious clinical signs, including cough, anorexia, and red discoloration of the body and ears. The piglets in groups A and B died at 7 to 10 and 10 to 14 dpi, respectively (Fig. 3B). Serum samples collected at 3, 7, and 10 dpi from all of the infected pigs were positive for PRRSV. The virus was reisolated from the sera of groups A and B, and the ORF5 region was sequenced to confirm that it was the original virus. The piglets in the control group remained negative for PRRSV, with no obvious clinical signs throughout the experiment. In the infected pigs, the lung lesions showed pulmonary consolidation (Fig. 4C) and histopathology revealed interstitial pneumonia (Fig. 4E). The pathogenicity of JL580 is much higher than that of earlier strains NADC30 (11) and CH-1a (15) and is similar to that of the classic HP-PRRSV strains in China (2–4, 16). The pathogenicity of the classic HP-PRRSV was extremely high at the outset of the 2006 HP-PRRSV outbreak, and piglets injected intravenously with this virus died within 6 to 8 days, and those inoculated intranasally died at 10 dpi (2). It is still unclear whether the other related viruses with different recombination patterns (i.e., HENAN-XINX and HENAN-HEB) have similar pathogenicity. It is worth noticing that the piglets infected with F2 died slightly faster than piglets infected with the initial viral homogenate. This may be associated with the difference in the dose of inoculated virus; the number of viral copies in the 3 × 104.0 TCID50 used for group A is much higher than the 3 × 104.0 copies used for group B.
FIG 3.
Rectal temperatures and mortality rates of piglets inoculated with JL580. (A) Rectal temperatures of piglets inoculated with JL580 F0 (homogenate supernatant) or JL580 F2 (second passage of JL580 in Marc-145 cells). Mean temperatures ± standard deviations (error bars) are shown (groups A and B, n = 5; group C, n = 4). A rectal temperature of >40.5°C was defined as fever. (B) Mortality rates of piglets infected with JL580 F0 or F2.
FIG 4.
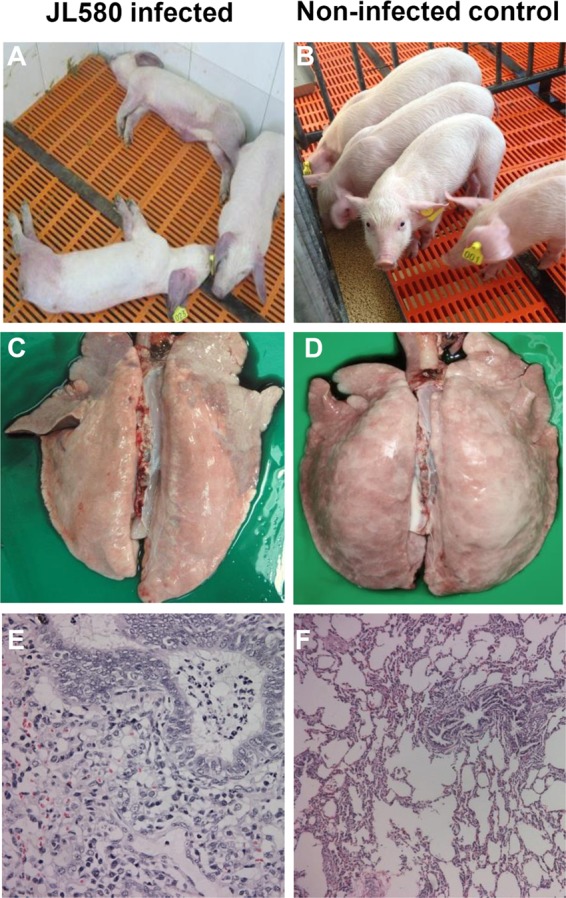
HP-PRRSV syndrome in piglets infected with JL580, including red discoloration of the body and ears (A), consolidation in the lungs (C), and massive lymphomononuclear cell infiltration of the lungs (E), compared with controls (B, D, and F, respectively).
In summary, our study describes and characterizes the importation of a NADC-30-like PRRSV strain from North America that has spread to several provinces in China and recombined with local HP-PRRSV strains. It has (i) high pathogenicity and (ii) a mixed genetic background distinct from that of the local strains, which makes it hard to control with current measures. Therefore, we suggest that surveillance data and vaccine strategies be updated to prevent the disease from spreading further.
Nucleotide sequence accession numbers.
The nucleotide sequences of the complete JL580 genome and HLJ58 ORF5 have been deposited in GenBank (accession no. KR706343 and KR706344, respectively).
ACKNOWLEDGMENTS
We thank Xia Yang for her helpful discussions.
This study was supported by the National Natural Science Foundation of China (31270045).
REFERENCES
- 1.Keffaber KK. 1989. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl 2:1–9. [Google Scholar]
- 2.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. 2007. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis 13:1434–1436. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J 174:577–584. doi: 10.1016/j.tvjl.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Yang H. 2010. Porcine reproductive and respiratory syndrome in China. Virus Res 154:31–37. doi: 10.1016/j.virusres.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 6.An TQ, Tian ZJ, Xiao Y, Li R, Peng JM, Wei TC, Zhang Y, Zhou YJ, Tong GZ. 2010. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis 16:365. doi: 10.3201/eid1602.090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo BQ, Chen ZS, Liu WX, Cui YZ. 1996. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin J Infect Diseases Anim Poult 2:1–5. [Google Scholar]
- 8.Shi M, Holmes EC, Brar MS, Leung FC. 2013. Recombination is associated with an outbreak of novel highly pathogenic porcine reproductive and respiratory syndrome viruses in China. J Virol 87:10904–10907. doi: 10.1128/JVI.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 10.Shi M, Lemey P, Singh Brar M, Suchard MA, Murtaugh MP, Carman S, D'Allaire S, Delisle B, Lambert MÈ Gagnon CA, Ge L, Qu Y, Yoo D, Holmes EC, Leung FC. 2013. The spread of type 2 porcine reproductive and respiratory syndrome virus (PRRSV) in North America: a phylogeographic approach. Virology 447:146–154. doi: 10.1016/j.virol.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Brockmeier SL, Loving CL, Vorwald AC, Kehrli ME Jr, Baker RB, Nicholson TL, Lager KM, Miller LC, Faaberg KS. 2012. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res 169:212–221. doi: 10.1016/j.virusres.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian ZJ, An TQ, Zhou YJ, Peng JM, Hu SP, Wei TC, Jiang YF, Xiao Y, Tong GZ. 2009. An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against HP-PRRS. Vet Microbiol 138:34–40. doi: 10.1016/j.vetmic.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Wei TC, Tian ZJ, Zhou YJ, An TQ, Jiang YF, Xiao Y, Hu SP, Peng JM, Hao XF, Zhang SR, Tong GZ. 2011. Evaluation of the pathogenicity of a highly pathogenic porcine reproductive and respiratory syndrome virus variant in piglets. Agri Sci China 10:1280–1291. doi: 10.1016/S1671-2927(11)60120-X. [DOI] [Google Scholar]
- 15.Xue Q, Zhao YG, Zhou YJ, Qiu HJ, Wang YF, Wu DL, Tian ZJ, Tong GZ. 2004. Immune responses of swine following DNA immunization with plasmids encoding porcine reproductive and respiratory syndrome virus ORFs 5 and 7, and porcine IL-2 and IFNgamma. Vet Immunol Immunopathol 102:291–298. doi: 10.1016/j.vetimm.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, Guo X, Ge X, Yang H. 2009. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol 83:5156–5167. doi: 10.1128/JVI.02678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]