ABSTRACT
As a recycling center, lysosomes are filled with numerous acid hydrolase enzymes that break down waste materials and invading pathogens. Recently, lysosomal cell death has been defined as “lysosomal membrane permeabilization and the consequent leakage of lysosome contents into cytosol.” Here, we show that the neuraminidase (NA) of H5N1 influenza A virus markedly deglycosylates and degrades lysosome-associated membrane proteins (LAMPs; the most abundant membrane proteins of lysosome), which induces lysosomal rupture, and finally leads to cell death of alveolar epithelial carcinoma A549 cells and human tracheal epithelial cells. The NA inhibitors peramivir and zanamivir could effectively block the deglycosylation of LAMPs, inhibit the virus cell entry, and prevent cell death induced by the H5N1 influenza virus. The NA of seasonal H1N1 virus, however, does not share these characteristics. Our findings not only reveal a novel role of NA in the early stage of the H5N1 influenza virus life cycle but also elucidate the molecular mechanism of lysosomal rupture crucial for influenza virus induced cell death.
IMPORTANCE The integrity of lysosomes is vital for maintaining cell homeostasis, cellular defense and clearance of invading pathogens. This study shows that the H5N1 influenza virus could induce lysosomal rupture through deglycosylating lysosome-associated membrane proteins (LAMPs) mediated by the neuraminidase activity of NA protein. NA inhibitors such as peramivir and zanamivir could inhibit the deglycosylation of LAMPs and protect lysosomes, which also further interferes with the H5N1 influenza virus infection at early stage of life cycle. This work is significant because it presents new concepts for NA's function, as well as for influenza inhibitors' mechanism of action, and could partially explain the high mortality and high viral load after H5N1 virus infection in human beings and why NA inhibitors have more potent therapeutic effects for lethal avian influenza virus infections at early stage.
INTRODUCTION
Lysosomes are membrane-bound organelles that are found in the cytoplasm of most cells and contain various hydrolytic enzymes that are usually active at an acidic pH (<5) (1). Lysosomes are known primarily to degrade macromolecules or infected pathogens from the endocytic, autophagic, and phagocytic pathways, which are essential for innate immunity recognition, antigen presentation, and pathogen elimination (2). Lysosome-associated membrane protein 1 (LAMP1) and LAMP2 constitute ca. 50% of the proteins in the lysosome membrane and function to maintain the structural integrity of lysosomal compartment to prevent hydrolytic enzyme release (3). Most hydrolytic enzymes and membrane proteins in the lysosome are highly glycosylated to prevent themselves from being digested in the hostile environment of the lysosome (4). Furthermore, lysosomal rupture is reported to induce cell death through the release of hydrolytic lysosomal enzymes, since partial release of enzymes could lead to apoptosis, whereas total lysosomal rupture induces necrosis (5). Previous reports have shown that HIV, adenovirus, and poliovirus could cause lysosomal rupture; however, the underlying molecular mechanism remains unclear (6).
Influenza A virus infections have caused several pandemics in the last century (7–9). Hemagglutinin (HA) and neuraminidase (NA) are the two main glycoproteins on the influenza viral envelope. As reported previously, HA mediates virus entry into the host cell by interacting with sialic acid, whereas NA cleaves sialic acid to release new viral particles at the end of the viral life cycle (7, 10).
The NA inhibitors are currently most effective clinical drugs for influenza pandemics (11). NA has been reported to play a crucial role in influenza virus pathogenesis (12). By performing assorted influenza virus experiments between high pathogenic avian influenza (PAI) virus H5N1 and low-pathogenicity influenza virus H1N1, quite a few manuscripts have revealed that NA was an important virus segment protein contributing to the virus pathogenesis both in vivo (mice and chickens) and in vitro (MDCK cells) (13, 14), and the sialidase activity of NA expressed in the infected cells was also detected in the lysosome (14). These findings suggest that, in addition to its role in late-stage viral release from the host cell, NA may also play a role in the lysosome with influenza virus infection.
We show here that, in addition to its classical mechanism of releasing assembled virus particles from the cell membrane at the late stage of the viral life cycle, the NA of influenza virus may directly bind to LAMPs of lysosomes, reduce the glycosylation of LAMPs, disrupt lysosome integrity, and increase the cell death rate. NA inhibitors could effectively protect the lysosomal integrity upon H5N1 influenza virus infection and prevent from cell death.
MATERIALS AND METHODS
Influenza viruses.
The seasonal influenza viruses H1N1 (A/New Caledonia/20/1999 [H1N1]) isolated from human in 1999 and avian influenza virus H5N1 (A/Jilin/9/2004 [H5N1]) isolated from chickens in 2004 were used in the present study. Experiments with live influenza viruses were performed in biosafety level 3 facilities according to governmental and institutional guidelines. The viruses were propagated, and titers were determined as described previously (15). Briefly, the viruses were propagated by inoculation into 10- to 11-day-old, specific-pathogen-free embryonated chicken eggs, and titers were determined using the Reed-Muench method with MDCK cells. The titers are expressed as the 50% tissue culture infective dose (TCID50)/ml of supernatant. Unless otherwise stated, a multiplicity of infection (MOI) of 4 was used for the H1N1 or H5N1 influenza virus examined in the present study.
Antibodies and reagents.
Primary antibodies against LAMP1 (ab24170) (for immunofluorescence), cathepsin K (ab19027), M6PR (ab124767), and cathepsin L (ab58991) were purchased from Abcam (Cambridge, MA). Cleaved cathepsin D (2284) and LAMP1 (3243) (for immunoblotting) were purchased from Cell Signaling Technology (Danvers, MA). Antibodies against the flag tag (F7425) and β-actin (clone AC-15, A5441) were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies against LAMP2 (clone H4B4, sc-18822) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The NA (IT-003-021M2) antibody was purchased from Immune Technology (Foster City, CA), the NP (C01321M) antibody was purchased from Millipore (Billerica, MA), the TFEB (H00007942-M01) antibody was purchased from Abnova (Taipei, Taiwan), and horseradish peroxidase (HRP)-conjugated secondary antibodies and luminal reagents for immunoblotting were purchased from Santa Cruz Biotechnology.
Dulbecco modified essential medium (DMEM), fetal calf serum, and antibiotics were obtained from Gibco/Life Technologies (Carlsbad, CA). F-12/Ham's nutrient medium and the Halt protease inhibitor cocktail were obtained from Thermo Fisher Scientific (Waltham, MA), and MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt] was procured from Promega Corp. (Madison, WI). Acridine orange was acquired from Merck (Billerica, MA). The anti-α-tubulin AF 488 kit, a peroxisome labeling kit for fixed cells, LysoTracker Red DND-99, and ProLong Gold Antifade Mountant with DAPI (4′,6′-diamidino-2-phenylindole) were purchased from Molecular Probes/Life Technologies (Eugene, OR). The NA inhibitors peramivir and zanamivir were purchased from Santa Cruz Biotechnology.
Cell culture.
The human lung adenocarcinoma A549 cell line was purchased from the American Type Culture Collection (ATCC; Rockville, MA) and cultured in F-12/Ham's medium (HyClone); HEK293T (human embryonic kidney cell line), HeLa, MRC-5, and H1650 cells were purchased from the Peking Union Medical College Cell Culture Center (Beijing, China). HEK293T and H1650 cells were propagated in DMEM, and MRC-5 cells were propagated in Eagle minimum essential medium supplemented with nonessential amino acids. All of these cells were cultured in the indicated medium supplemented with 10% fetal bovine serum (Gibco), 100 U of penicillin/ml, and 100 U of streptomycin/ml at 37°C with 5% CO2. For the primary cell, the human tracheal epithelial cells (HTEpiC; ScienCell Research Laboratories, Carlsbad, CA) were cultured in the bronchial epithelial cell medium according to the protocol provided by the manufacturer.
Plasmids.
To determine which viral protein mediates the deglycosylation of LAMPs, the avian influenza virus genes polymerase complex PB1 (GenBank accession no. AAT73504.1), polymerase complex PB2 (GenBank accession no. AAT73545.1), polymerase complex PA (GenBank accession no. AAT70214.1), nucleocapsid protein (NP; GenBank accession no. ABC66772.1), hemagglutinin (GenBank accession no. AAV34704), neuraminidase (GenBank accession no. AAS89006), matrix protein 1 (M1; GenBank accession no. Q6J8D2.1), matrix protein 2 (M2; GenBank accession no. AAT70566.1), nonstructural protein 1 (NS1; GenBank accession no. AAT73445.1), and nonstructural protein 2 (NS2; GenBank accession no. AAT73391.1) were optimized with human high-frequency codons as described before (16) for cDNA synthesis, and then each of these gene was fused with a flag tag at the C terminus and cloned into Peak13 vector (provided by B. Seed, Harvard Medical School, Boston, MA). The full-length cDNAs encoding human LAMP1, LAMP2A, and avian influenza virus NA (Jilin) A/Jilin/9/2004 (H5N1) were amplified by using PCR from a cDNA library of A549 cells or cells infected with H5N1 influenza virus. All of these gene products were verified by DNA sequencing (Invitrogen, Beijing, China).
Plasmid transient transfection.
293T cells were transfected at 50 to 70% confluence with mixture of Lipofectamine 2000 transfection reagent (Invitrogen) and each plasmid according to the manufacturer's instructions. The transient transfections of A549 cells were performed with X-treme Gene HP reagents (Roche Applied Science, Indianapolis, IN) according to the protocol provided by the manufacturer.
Immunoblot analysis.
Immunoblot analysis was performed as described previously (17). Cells were collected and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 0.5% deoxycholate sodium, 1 mM Na3VO4, 1 mM NaF, and protease inhibitor cocktails), the total protein concentration of each sample was determined with a BCA protein assay kit (TianGen, Beijing, China), and the samples were boiled with 2× loading buffer after equalization of the protein concentration. Samples were resolved by 10 or 12% SDS-polyacrylamide gel electrophoresis (PAGE), and proteins were transferred onto a nitrocellulose filter membrane. Membranes were blocked in 2% albumin and then incubated with the appropriate primary antibodies overnight and then with HRP-conjugated secondary antibodies at room temperature for 1 h. Binding of secondary antibodies was detected using the Kodak film exposure detection system, and the film was scanned and analyzed. For detection of the second primary antibody, the nitrocellulose membrane was stripped with stripping buffer (1% SDS, 25 mM glycine [pH 2.0]) and then incubated with another primary antibody.
LAMP2-Fc and Fc protein purification and degradation detection.
The methods for constructing the cell lines expressing the LAMP2-Fc and control human IgG Fc fusion proteins have been previously described (17). Briefly, LAMP2-Fc and Fc were expressed by the HEK293T/LAMP2-Fc and HEK293T/Fc stable-expression cell lines, respectively. LAMP2-Fc is a recombinant protein composed of the soluble extracellular region (amino acids 1 to 376) of the human LAMP2A protein fused to the IgG Fc at its C terminus. The fusion protein was purified with an AKTA FPLC system (GE Healthcare, Piscataway, NJ), and the protein concentration was measured using the BCA assay kit. Equal amounts of LAMP2-Fc or Fc protein were conjugated to new protein A-agarose beads (Santa Cruz) and washed several times with sterile H2O. Samples were then rinsed with 20 mM acetate buffer (pH 4.0 or 5.0), 20 mM phosphate buffer (pH 6.0), or phosphate-buffered saline (PBS; pH 7.4) with an equal amount of LAMP2-Fc fusion or control Fc protein. Next, 5 μl (105 TCID50) of each virus or vehicle was added to each group and stored at 37°C for 8 h. Finally, an equal volume of 2× loading buffer was added to the samples, which were then boiled for 5 min. Samples were separated using SDS-PAGE (10% gel) and probed with an appropriate antibody.
Real-time quantitative PCR (q-PCR) analysis.
Cells were lysed with TRIzol reagent (Invitrogen), and total RNA was isolated according to the standard protocol provided by the manufacture. cDNA was synthesized from 1.5 μg of total RNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems/Life Technology). PCR amplification assays were performed with the FastStart Universal SYBR green Master mix and Rox (Roche) on a LightCycler 480 PCR system. The relative gene expression levels were calculated using the CT value and normalized to the expression of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reference gene. The specific primers that were used were as follows: GAPDH forward, 5′-GGTGGTCTCCTCTGACTTCAACA-3′; GAPDH reverse, 5′-GTTGCTGTAGCCAAATTCGTTGT-3′; M1 forward, 5′-CTCTCTATCATCCCGTCAG-3′; M1 reverse, 5′-GTCTTGTCTTTAGCCATTCC-3′; M2 forward, 5′-ATTGTGGATTCTTGATCGTC-3′; and M2 reverse, 5′-TGACAAAATGACCATCGTC-3′.
siRNA transfections.
All small interfering RNAs (siRNAs) used in the present study were designed and synthesized by Ribobio (Guangzhou, China). Before siRNA transfection, A549 cells were seeded at 3 × 105 cells/well in 12-well plates. After 24 h, the cells were transfected with Lipofectamine RNAiMAX reagent (Invitrogen) and a 50 nM concentration of the indicated siRNA diluted in Opti-MEM (Invitrogen) according to the manufacturer's guidance. Next, 48 h after siRNA transfection, the downstream experiments were performed. The siRNA sequences for LAMP1 were 5′-CAAUGCGAGCUCCAAAGAAdTdT-3′ and 3′-dTdT GUUACGCUCGAGGUUUCUU-5′, and the siRNA sequences for LAMP2 were 5′-GCGGUCUUAUGCAUUGGAAdTdT-3′ and 3′-dTdT-CGCCAGAAUACGUAACCUU-5′.
Neuraminidase activity assay.
The method for neuraminidase activity assay in different pH buffers was as described before (18). In brief, 5 μl of H1N1 or H5N1 influenza A virus suspension (i.e., a 50% tissue culture infective dose of 105) or the vehicle AF in different pH buffers (the same as described above) were incubated with 5 μl of 4 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid sodium salt hydrate (4-MUα-Neu5Ac; Sigma-Aldrich) at 37°C for 30 min. After stopping the reaction by adding 1 ml of 100 mM carbonate buffer (pH 10.7), the released 4-methylumbelliferone was measured with a fluorescence microplate reader (Synergy 2; BioTek Instruments, Inc., Winooski, VT) with a excitation at 355 nm and an emission at 460 nm. Relative fluorescence units (RFU) were used to show the NA activity.
Confocal microscopy.
A549 cells were grown on coverslips in 24-well plates; at the indicated time points after virus infection, the culture supernatant was removed, and the coverslips were washed three times with PBS. The cells were then fixed with 4% paraformaldehyde for 10 min, penetrated with 0.2% Triton X-100 for 5 min, and blocked with 10% goat serum for additional 30 min. The coverslips were then incubated with the indicated primary antibody at room temperature for 1 h and then with Alexa Fluor 488- or Alexa Fluor 568-labeled secondary antibody (Molecular Probes). The subcellular localization of each target protein was observed using confocal laser scanning microscopy (FV-1000; Olympus, Tokyo, Japan), and the images were analyzed using Olympus Fluoview 3.0 software. For the NP-positive nucleus analysis, at least 64 pictures were continuously obtained by the confocal microscope automatically, and the images were assessed with ImageJ software (National Institutes of Health, Bethesda, MD).
To label cellular lysosomes, the cells were incubated with 300 nM LysoTracker Red DND 99 (Molecular Probes) in fresh medium for 30 min at 37°C, and the cells were then extensively washed with PBS and fixed with 4% paraformaldehyde for 15 min. Lastly, the coverslips were covered with the ProLong Gold antifade mounting medium with DAPI regent. We calculated the colocalization efficiency directly using Fluoview FV10-ASW 3.0 (Olympus) software according to the standard Pearson correlation coefficient (r) method as recommended by the manufacturer.
Cell viability assays.
A549 cells were infected with virus at an MOI of 4 or an equal volume of vehicle for the indicated time points. Cell viability was then determined by the MTS assay (Promega). In the rescue experiments, peramivir (30 μg/ml) or zanamivir (30 μg/ml) was added 1 h before viral administration or at the indicated time points. In the siRNA knockdown group, A549 cells were transfected with siRNA as described before, and the MTS assay was performed 48 h after virus infection.
Fluorescence-activated cell sorting analysis (FACS).
At the indicated time points after experimental treatment, the cell medium was removed, and the cells were refed with medium containing 75 nM LysoTracker Red DND-99, followed by incubation at 37°C in 5% CO2 for 30 min. The cells were then washed three times with PBS, harvested with trypsin-EDTA, and washed with 1 ml of PBS. Finally, the stained cells were fixed with 4% paraformaldehyde and analyzed by FACSAria II cytometer (BD, San Jose, CA) with an excitation at 561 nm. Fluorescence-activated cell sorting (FACS) data analysis was performed with Flow Jo software (Tree Star, Inc., Ashland, OR).
Immunoprecipitation.
293T cells were transfected with GFP2, NS1 (H5N1)-flag, or NA (H5N1, Jilin)-flag expression plasmids in six-well plates. At 30 h posttransfection, whole-cell extracts were obtained using RIPA lysis buffer and centrifuged at 15,000 rpm for 15 min at 4°C. Then, 1 mg of total protein from each sample was incubated with the flag or isotype control antibody at 4°C overnight on a rotator. Next, 60 μl of 50% protein A-agarose beads (Santa Cruz) was added to the samples, and the mixture was incubated for an additional 2 h at 4°C. The beads were then washed extensively five times with TBS buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl) and boiled for 5 min in 50 μl of 2× loading buffer. Immunoblotting analysis of the samples was then performed as described previously.
Statistics.
All data are shown as means ± standard errors of the mean (SEM). Measurements at single time points were analyzed by analysis of variance and, if they demonstrated significance, were further analyzed by a two-tailed t test. All statistical tests were conducted using Prism 5.0 (GraphPad Software, San Diego, CA). P < 0.05 indicates statistical significance, and P < 0.01 indicates high significance.
RESULTS
Lysosomes were ruptured after influenza virus infection.
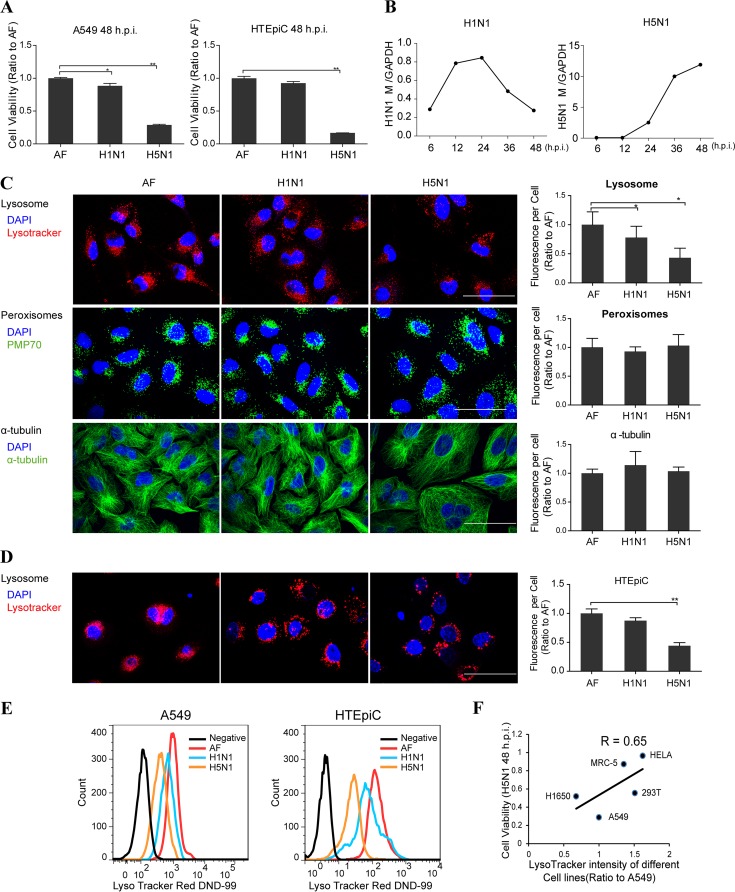
H5N1 influenza virus could induce cell death in various cell lines (19, 20) (Fig. 1A) and extremely high viral load in infected cells (Fig. 1B) or patients (21). To identify potential function of organelles involved in the cell death induced by H5N1 influenza virus, we used confocal microscopy to examine the amounts and structure of several cellular organelles, including the lysosome, peroxisome, and cytoskeleton tubulin. In A549 cells at 24 h postinfection with live seasonal H1N1 and H5N1 viruses, the number of lysosomes (labeled with LysoTracker) in cells decreased significantly when infected with the H5N1 virus but decreased slightly when infected with the H1N1 virus compared to those in cells of vehicle controls (Fig. 1C). However, no significant differences were observed with the peroxisome and tubulin present in each group (Fig. 1C). The lysosomes in primary cells human tracheal epithelial cells (HTEpiC) also decreased significantly after H5N1 virus infection (Fig. 1D). Using fluorescence-activated cell sorting (FACS) analysis, we also observed a significant decrease in the overall number of lysosomes per cell in A549 cells and HTEpiC infected with the H5N1 virus and a moderate decrease when they were infected with the H1N1 virus (Fig. 1E). Next, we infected A549, HeLa, HEK293T, H1650, and MRC-5 cells with the H5N1 virus and found that cell survivals at 48 h postinfection were positively correlated to the cellular lysosomal numbers before infection (Fig. 1F), suggesting lysosome may have protective function in influenza virus infections.
FIG 1.
Lysosomes were decreased significantly upon H5N1 influenza virus infection. (A) MTS assay of cell viability of A549 cells and primary HTEpiC treated with vehicle or H1N1 or H5N1 virus at 48 h postinfection. (B) q-PCR analysis of influenza viral gene M in A549 cells infected with H1N1 or H5N1 virus. (C) A549 cells were infected with either seasonal H1N1 or H5N1 influenza virus or treated with the vehicle as a control. At 24 h postinfection, the cells were stained by LysoTracker or immunolabeled for peroxisomal membrane protein PMP70 and α-tubulin, respectively. Images were obtained using confocal microscopy, and the fluorescence signal of each cell shown on the right was estimated by examining more than 100 cells. (D) Confocal microscopy determination of changes of lysosomes in primary HTEpiC that were infected with either seasonal H1N1 or H5N1 influenza virus or treated with the vehicle allantoic fluid (AF) as a control at 24 h postinfection (h.p.i.). (E) FACS analysis of A549 cells and HTEpiC after staining with LysoTracker at 24 h after seasonal H1N1 or H5N1 infection or treatment with vehicle, with unstained cells as the negative control. (F) Pearson correlation coefficients between cell viability at 48 h postinfection with H5N1 virus and the relative lysosome numbers measured by FACS using LysoTracker before infection in A549, HeLa, MRC-5, H1650, and HEK293T cells. All scale bars indicate 50 μm, and data represent means ± the SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
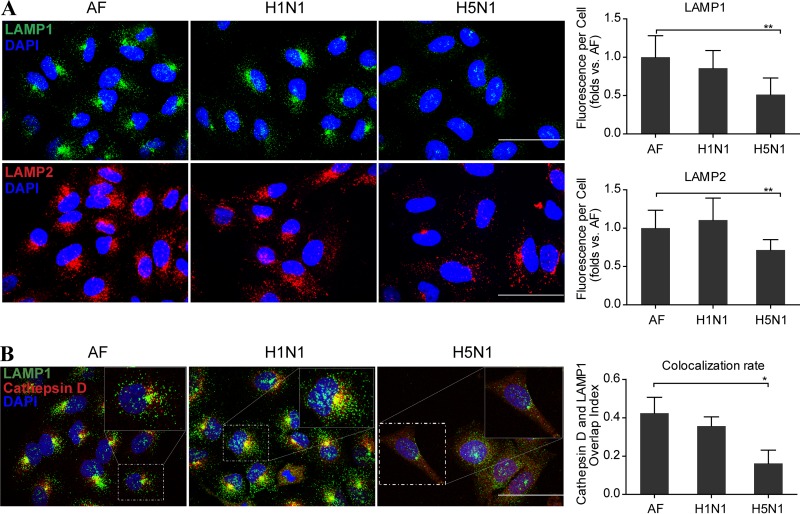
Using immunofluorescent assay, we also examined the changes of lysosomal membrane protein LAMP1, LAMP2, and hydrolytic enzyme cathepsin D and found that LAMP1 and LAMP2 were significantly decreased 24 h after H5N1 virus infection (Fig. 2A); moreover, lysosome hydrolytic enzyme cathepsin D changed to diffuse throughout the cytosol and the colocalization of cathepsin D and LAMP1 was also significantly decreased after H5N1 virus infection (Fig. 2B). Collectively, these results suggest that the lysosome integrity is disrupted upon H5N1 virus infection.
FIG 2.
Lysosomes were ruptured at a late stage of H5N1 influenza virus infection. (A) Representative images of LAMP1 (green) and LAMP2 (red) in A549 cells infected with the seasonal H1N1 virus or H5N1 virus or treated with AF at 24 h postinfection. The fluorescence signal of each cell analyzed from 50 to 100 cells is shown on the right. (B) Confocal microscopy of the cathepsin D and LAMP1 after immunolabeling in A549 cells infected with seasonal H1N1 virus or H5N1 virus or treated with vehicle at 24 h postinfection. The colocalization rate of these two proteins was analyzed for 50 to 100 cells using Fluoview software. All scale bars indicate 50 μm, and the data represent means ± the SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
LAMP1 and LAMP2 were deglycosylated after influenza virus infection.
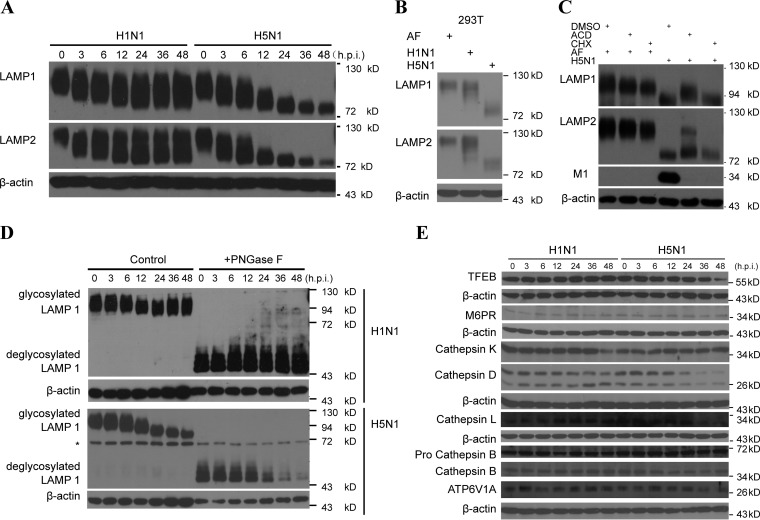
Since the lysosomal membrane glycoproteins LAMP1 and LAMP2 are important for maintaining the structural integrity of the lysosomal compartment (22), we investigated changes of these proteins in A549 cells infected with the H1N1 and H5N1 virus. We observed the significant deglycosylation of LAMP1 and LAMP2 upon live H5N1 virus infection in A549 cells quickly after the virus infection (Fig. 3A) and HEK293T cells (Fig. 3B). Treatment with the mRNA transcriptional inhibitor actinomycin D or the protein biosynthesis inhibitor cycloheximide showed similar patterns of deglycosylation of LAMP1 and LAMP2 as the control upon the H5N1 influenza virus infection (Fig. 3C), which indicated that the viral proteins themselves could induce the LAMP1 and LAMP2 deglycosylation directly. The digestion assay with the glycosidase PNGase F suggested that protein deglycosylation is involved in the decrease of LAMPs after H5N1 influenza virus infection (Fig. 3D).
FIG 3.
LAMP1 and LAMP2 were significantly deglycosylated after H5N1 influenza virus infection. (A) Immunoblot analysis of LAMP1 and LAMP2 in A549 cells infected with seasonal H1N1 or H5N1 virus at the indicated times (in hours) postinfection. β-Actin was used as a control. (B) Immunoblot analysis of LAMP1 and LAMP2 in 293T cells infected with H1N1 or H5N1 virus or treated with AF at 48 h postinfection. β-Actin was used as a loading control. (C) Immunoblot analysis of LAMP1 and LAMP2 in A549 cells infected with seasonal H1N1 or H5N1 virus in the presence of actinomycin D (ACD) or cycloheximide (CHX) at 24 h postinfection. (D) Immunoblot analysis of highly glycosylated LAMP1 and those that were digested with the glycosidase PNGase F in A549 cells infected with H1N1 or H5N1 virus at the indicated time points. The asterisk (*) indicates an unspecific band. (E) Immunoblot analysis of TFEB, M6PR, cathepsin B, cathepsin D, cathepsin K, cathepsin L, ATPase, H+ transporting, lysosomal 70-kDa, and V1 subunit A (ATP6V1A) in A549 cells at the indicated time points in hours postinfection (h.p.i.) with H1N1 or H5N1 virus.
We also studied two genes that regulate lysosome functions: transcription factor EB (TFEB), which is a master activator of lysosomal biogenesis (23), and mannose-6-phosphate receptor (M6PR), which traffics hydrolytic enzymes transported to the lysosome (2). The protein levels of TFEB and M6PR were not affected significantly before 36 h after H5N1 influenza virus infection (Fig. 3E). Next, we examined levels of some important hydrolytic enzymes: cathepsin B, cathepsin D, cathepsin K, and cathepsin L. Compared to H1N1 virus infection, the active cathepsin B, mature cathepsin D and cathepsin L proteins were slightly downregulated upon H5N1 virus infection at late stage, but the cathepsin K remained unchanged (Fig. 3E). In addition, the expression level of ATP6V1A (ATPase, H+ transporting, lysosomal 70 kDa, V1 subunit A), which is a component of the lysosomal proton vacuolar-type H+-ATPase (Fig. 3E), also remained unchanged upon influenza virus infection. These results indicate that, under conditions of lysosomal rupture, released lysosomal components, such as hydrolytic enzymes, might contribute to the cell death (6).
Influenza virus NA induced lysosome rupture.
To elucidate the molecular mechanism of lysosomal rupture induced by the H5N1 virus, we transfected all coding genes PB1, PB2, PA, NP, HA, NA, M1, M2, NS1, and NS2 (for the abbreviation definitions, see Materials and Methods) of the H5N1 virus, into HEK293T cells. We showed that the deglycosylation of LAMP1 and LAMP2 was induced in HEK293T cells upon expression of the NA gene (Fig. 4A). This result was also observed in A549 cells (Fig. 4B). Similarly, after digestion by the glycosidase PNGase F, LAMPs were deglycosylated significantly (Fig. 4C). In addition, the decrease of the lysosomal numbers per cell (Fig. 4D) and cell survival rate (Fig. 4E) was also confirmed using FACS analysis of cells transfected with the NA gene of the H5N1 virus. Furthermore, the NA protein of H5N1 virus could also induce more seriously LAMP1 and LAMP2 deglycosylation compared to the same amount of NA protein from H1N1 virus in A549 cells (Fig. 4F).
FIG 4.
Influenza virus NA could bind to the LAMPs directly and induce the deglycosylation of LAMPs. (A) Plasmids encoding H5N1 influenza viral proteins (PB1, PB2, PA, NP, HA, NA, M1, M2, NS1, and NS2) (for the abbreviation definitions, see Materials and Methods) were separately transfected in 293T cells, and LAMP1 and LAMP2 were detected by immunoblot analysis. The expression of each viral protein was detected using flag antibody. (B) Immunoblot analysis of LAMP1 or LAMP2 in A549 cells transfected with control vector or the NA-flag plasmid. (C) Immunoblot analysis of glycosylated LAMP1 and those digested with glycosidase PNGase F in 293T cells transfected with plasmids encoding NA of H5N1 or vector. An asterisk (*) indicates an unspecific band. (D) FACS analysis of lysosomes stained with LysoTracker in 293T cells transfected with NA of the H5N1 virus or vector. (E) MTS assay of cell viability of A549 cells transfected with control vector or NA(H5N1) gene at 48 h posttransfection. (F) Immunoblot analysis of LAMP1 and LAMP2 in A549 cells treated with NA protein of H1N1 or H5N1 virus which was expressed and purified from 293T cells at the indicated amounts. β-Actin was used as a control. (G) Confocal microscopy analysis showing the interaction between viral NA and the lysosomal membrane marker LAMP1 in A549 cells at 3 h after H1N1 or H5N1 virus infection or treated with the vehicle as a control. Scale bars, 50 μm. (H) 293T cells were transfected with vector, NS1-flag, or NA-flag encoding plasmids, and an immunoprecipitation (IP) assay was performed with anti-flag or control IgG. Cell lysates (input) and immunoprecipitated complexes were analyzed by immunoblotting with anti-LAMP1, anti-LAMP2, anti-NS1-flag, anti-NA-flag, or anti-β-actin antibodies. (I) MTS assay of the cell viability of A549 cells, which were transfected with nontarget control siRNA (NC) or LAMP1- or LAMP2-specific siRNA and then treated with vehicle, H1N1 virus, or H5N1 virus. The knockdown efficiency is shown on the right. (J) q-PCR analysis of influenza viral M gene in A549 cells after transfection of LAMP1, LAMP2, or nontarget control (NC) siRNA and infected with H5N1 virus. (K) MTS assay of the viability of A549 cells transfected with vector control, LAMP1, or LAMP2 and then infected with H5N1 virus. The overexpression efficiency is shown on the right. All scale bars indicate 50 μm, and data represent means ± the SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
We further observed the colocalization of LAMP1 and NA of influenza virus as early as 3 h after live influenza virus infection in A549 cells under confocal microscopy (Fig. 4G). We also demonstrated the colocalization of NA-GFP and LAMP1-RFP or LAMP2-RFP after transfections in A549 cells (data not shown). Moreover, the NA of H5N1 was shown binding to LAMP1 and LAMP2 by coimmunoprecipitation assays (Fig. 4H). Furthermore, the cell death and viral loads were increased when LAMP1 and LAMP2 were knocked down in cells before viral infection (Fig. 4I and J), and the survival rate of alveolar epithelial cells was increased upon H5N1 virus infection when LAMP1 and LAMP2 were overexpressed in A549 cells (Fig. 4K). Taken together, these data suggest that the NA of H5N1 virus may bind to LAMP1 and LAMP2, deglycosylate them, disrupt lysosomal integrity, and possibly induce cell death.
NA activity mediated LAMP deglycosylation.
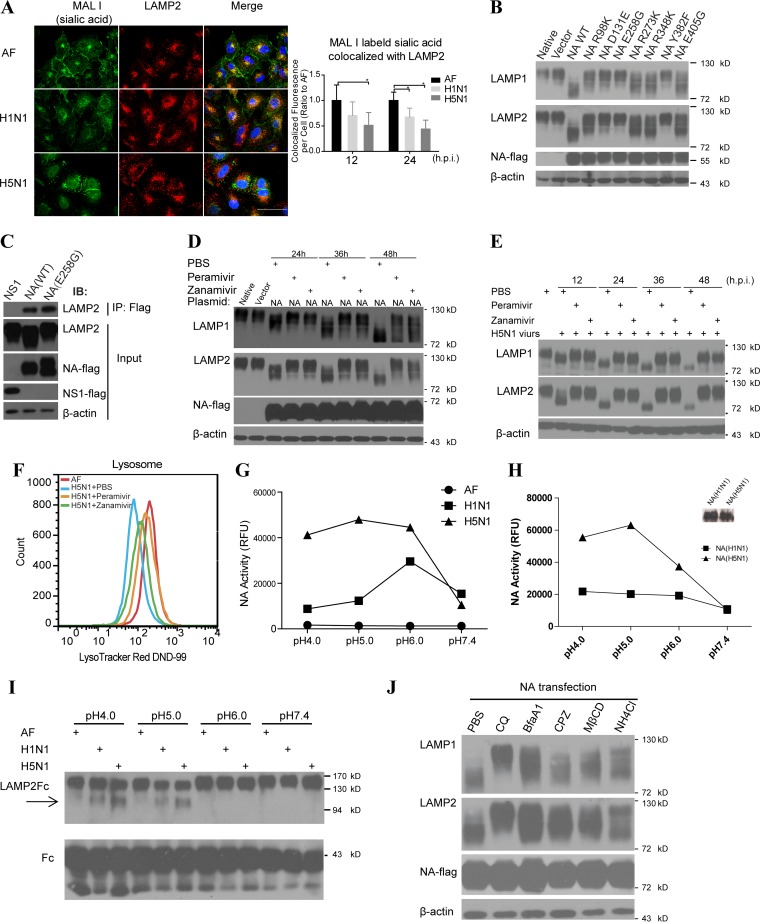
Sialic acids are usually located on the surface of glycoprotein and are essential for maintaining the integrity of glycoprotein especially in the hazard acid lysosomes (24); moreover, the sialic acids are also the targets of NA of influenza virus (25), so the interaction between NA and LAMPs might cause the decrease of sialic acid on the lysosomes which could contribute to the deglycosylation of LAMP1 and LAMP2. We labeled sialic acid (linked [α2,3]) with Maackia Amurensis Lectin I (MAL I) and lysosome marker LAMP1, the fluorescence intensity of sialic acid colocated with LAMP2 were observed significantly decreased after H5N1 virus infection (Fig. 5A), as well as Sambucus Nigra Lectin-labeled sialic acid (α2,6; data not shown). Examination of the structure of influenza virus NA has revealed conservative amino acids in its catalytic pocket (26). Since these sites are crucial for the neuraminidase activity, we mutagenized each of the conserved catalytic amino acids and assessed the impact on interaction with the LAMPs. Similar with previous reports that some mutations could significantly affect the NA activity (27), we found that mutations of R98K, D131E, E258G, and Y382F in the NA catalytic pocket site could markedly prevent deglycosylation of the LAMPs (Fig. 5B) but did not significantly affect the interaction between mutant NA and LAMP2 (Fig. 5C), indicating that the catalytic activity of NA plays a critical role in NA-induced LAMPs deglycosylation.
FIG 5.
Influenza virus NA induced the LAMP deglycosylation depending on the its NA activity and low-pH environment. (A) Immunofluorescent colocalization of MAL I-labeled sialic acid and lysosomal membrane marker LAMP2 in A549 cells infected with H1N1 or H5N1 virus or treated with vehicle as a control. The sialic acid fluorescence signal colocalized with LAMP2 per cell (on the right) was estimated by examining 50 to 100 cells. Scale bars, 50 μm. (B) Immunoblot analysis of LAMP1 and LAMP2 deglycosylation in 293T cells transfected with plasmids encoding vector, wild-type (WT) NA, or mutant NAs using NA-flag as an overexpression control and β-actin as a loading control. (C) 293T cells were transfected with NS1-flag, wild-type NA-flag, or mutant NA (E258G) encoding plasmids, respectively, and the immunoprecipitation (IP) assay was performed with anti-flag antibody. Cell lysates (input) and immunoprecipitated complexes were analyzed by immunoblotting with anti-LAMP2 and anti-flag antibodies. (D) 293T cells were transfected with control plasmid or NA(H5N1)-encoding plasmids. After 6 h, peramivir (30 μg/ml), zanamivir (30 μg/ml), or PBS was separately added into the media of these cultured cells. The deglycosylation of LAMP1 and LAMP2 was analyzed by immunoblotting, and flag was used for NA detection and β-actin as a loading control. (E) Immunoblot analysis of LAMP1 and LAMP2 deglycosylation in A549 cells infected with H5N1 virus and treated with PBS, peramivir (30 μg/ml), or zanamivir (30 μg/ml), respectively, at the indicated time points. (F) FACS analysis of the lysosome numbers in A549 cells treated with AF and in cells infected with H5N1 virus and treated with peramivir (30 μg/ml), zanamivir (30 μg/ml), or PBS. (G) Assay of NA activity of H1N1 or H5N1 influenza virus or vehicle control AF in different buffers (pH 4.0, 5.0, 6.0, or 7.4) at 37°C. RFU, relative fluorescence units. (H) Activity assay evaluating 0.1 μg of H1N1 or H5N1 influenza virus NA protein in different buffers (pH 4.0, 5.0, 6.0, or 7.4) at 37°C. The amounts of NA protein were also determined by immunoblot analysis (shown on the right). (I) The extracellular region of LAMP2 fused to the human IgG-Fc fragment (LAMP2-Fc) and the control protein human IgG-Fc (Fc) were conjugated to protein A-beads. After incubation with vehicle or H1N1 or H5N1 virus in different buffers (pH 4.0, 5.0, 6.0, or 7.4) at 37°C for 6 h, protein changes were detected by immunoblotting with anti-LAMP2 antibody or Fc antibody as a control. (J) 293T cells were transfected with NA(H5N1) gene and treated with control PBS, chloroquine (CQ), bafilomycin A1 (BfaA1), chlorpromazine (CPZ), methyl-β-cyclodextrin (MβCD), or NH4Cl, and then, 48 h later, LAMP1 and LAMP2 were detected by immunoblotting with NA-flag as an overexpression control and β-actin as a loading control. All data represent means ± the SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
Currently, the most widely clinical used effective drugs against influenza are NA catalytic activity blockers, such as peramivir and zanamivir. We showed that the treatment of 293T cells with either of these drugs could effectively inhibit LAMPs deglycosylation (Fig. 5D) and protect the LAMPs after H5N1 influenza virus infection in A549 cells (Fig. 5E). We then examined the numbers of lysosomes per cell in cells infected with the live H5N1 virus and found that treatment with the NA inhibitors could ameliorate lysosomal rupture (Fig. 5F). Collectively, these data demonstrate that the NA inhibitors peramivir and zanamivir could reduce the deglycosylation of LAMPs and protect from lysosomal rupture induced by influenza virus infection.
Higher NA catalytic activity of the H5N1 virus at low pH.
We have shown that NA of H5N1 virus could bind directly to LAMPs, degrade LAMPs, and markedly induce lysosomal rupture, while NA of seasonal H1N1 virus may have much less effect. Previous studies have shown that the NA catalytic activity of different strains may be different (14, 28). We examined the NA catalytic activity of both avian H5N1 influenza virus and seasonal H1N1 influenza virus, and the NA catalytic activity of H5N1 virus was much higher than NA catalytic activity of H1N1 under acidic pH conditions, which are more common in lysosomes, although their activities are similar at pH 7.4 (Fig. 5G). We also demonstrated a higher activity of the NA protein from H5N1 influenza virus than NA from H1N1 influenza virus expressed in the 293T cells (Fig. 5H). Moreover, we also observed here that the NA of the live H5N1 virus prefers to cleave glycosylated LAMPs at low pH (Fig. 5I). Furthermore, treatment with the lysosomotropic pH-elevating agents chloroquine, bafilomycin A1, and NH4Cl separately could alleviate the LAMPs deglycosylation after NA transfection in 293T cells (Fig. 5J). These results suggest that the NA catalytic activity of avian H5N1 influenza virus may have markedly higher impact on inducing lysosomal rupture than that of seasonal H1N1 influenza virus.
Effects of NA at the early stage of influenza virus life cycle.
Our studies have demonstrated that avian H5N1 influenza virus NA protein induces lysosomal rupture, which may play an important role in early stage of virus life cycle, in addition to the late stage of virus life cycle. To investigate the role of NA in the early stage of virus life cycle, we need to design experiments excluding effects of NA at the late stage of the virus life cycle. Previous studies have shown that the time from influenza virus cell entry to new influenza virion production in cells is on average 6 h (29, 30). We therefore investigated the effects of NA in cells within 6 h after virus infection. Using NA inhibitors at different time points, we found the cell survival was rescued more significantly at the early stage of the first virus life cycle (Fig. 6A), suggesting that influenza virus NA may play an important role at the early stage of the virus life cycle. In order to analyze the effects of NA inhibitors on the infection of influenza virus at early stage of life cycle, we measured the viral NP-positive nuclei at 4 h after virus infection (Fig. 6B) and found that the percentage of NP-positive nuclei in H1N1 or H5N1 influenza virus-infected cells was significantly decreased by the NA inhibitors, respectively. Also, the viral load was also reduced at the early stage of viral life cycle when the cells were treated with NA inhibitors before infection (Fig. 6C). These results suggest NA activity of H1N1 or H5N1 influenza virus may indeed have effects at the early stage of virus life cycle.
FIG 6.
Neuraminidase inhibitors could decrease influenza virus cell entry. (A) A549 cells were treated with PBS or the NA inhibitors peramivir (30 μg/ml) or zanamivir (30 μg/ml) 1 h before or at the indicated times after H5N1 virus infection, and an MTS assay was performed to determine the viability of these cells at 48 h postinfection. (B) A549 cells were treated with PBS or the NA inhibitors peramivir (30 μg/ml) and zanamivir (30 μg/ml) 1 h before H1N1 or H5N1 virus infection and 4 h postinfection; the NP-positive nucleus was analyzed by confocal microscopy, and the percentages of NP-positive nuclei were determined using ImageJ software. (C and D) q-PCR analysis of the influenza viral gene M at the indicated time points in A549 cells that infected with H1N1 virus (C) or H5N1 virus (D) and treated with PBS, peramivir (30 μg/ml), or zanamivir (30 μg/ml) 1 h before the infection. All data represent the means ± the SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
DISCUSSION
In summary, our data indicate that influenza A virus NA, especially H5N1 virus NA, may bind LAMP1 and LAMP2 proteins, promote the deglycosylation of LAMPs, induce lysosomal rupture (which may lead to block virus clearance), increase virus load, and enhance cell death (21). NA inhibitors such as peramivir and zanamivir can effectively inhibit the deglycosylation of the lysosomal structural proteins LAMP1 and LAMP2, protect lysosomal integrity, and increase cell survival.
Our study revealed a novel mechanism of NA, namely, the targeting of LAMPs to induce lysosomal rupture at an early stage of viral infection, in addition to its classic mechanism of releasing assembled virus particles from the cell membrane at the late stage of the viral cycle. We also demonstrated a new mechanism for NA inhibitors, which could protect the integrity of the lysosome, decrease the growth rate of influenza virus-infected cells at an early stage and the high viral load after influenza virus infection, and partially explain why NA inhibitors have potent therapeutic effects for lethal avian influenza virus infections at early stage (31, 32).
As reported before, lysosome cell death could occur after lysosomal membrane permeabilization and the consequent leakage of the lysosomal hydrolases such as cathepsin D into the cytosol (namely, lysosome rupture), and there are numerous stimuli that could lead to lysosome cell death (33). Here, we reported that the lysosome rupture could be induced by the deglycosylation of LAMP1 and LAMP2 after influenza virus infection. Since LAMPs constitute up to 50% of lysosome membrane proteins and are heavily glycosylated, the integrity of LAMPs should be essential for the lysosome function by preventing hydrolases release into cytoplasm (34). Mice lacking both LAMP-1 and LAMP-2 are embryonic lethal (35), suggesting that lysosome functions are critical maintaining mouse survival.
The deficiency of LAMPs may result from the impairment of lysosome functions (36), leading to the inhibition of virus degradation and clearance (37). Influenza virus was reported to enter cells through the endocytic pathway, and endosome maturation occurs through sequential interactions and fusion with lysosomes, which are then degraded and cleared the influenza viruses (38). Therefore, lysosome rupture may enhance virus replication in the infected cells, in addition to releasing hydrolase enzymes into the cytosol.
It is interesting that recombinant H5N1 viruses carrying NA mutations that are resistant to NA inhibitors were reported to be more virulent and were associated with significantly higher virus titers in ferret lungs and livers than wild-type (WT) virus, and the molecular basis for this phenomenon remains unknown (39, 40). A possible interpretation is that these NA mutant recombinant viruses had significantly higher NA enzymatic activity than those of WT NA and that the mutant NA accelerates the deglycosylation of LAMPs and induces more serious lysosome rupture.
We have observed more severe LAMPs deglycosylation caused by H5N1 virus NA than by H1N1 virus NA, whereas a higher enzyme activity for H5N1 influenza virus NA was detected than that for H1N1 influenza virus NA in low-pH conditions. There are more severe lysosome ruptures induced by H5N1 virus NA, and more cell death resulted from H5N1 virus infection. Alveolar epithelial cell death interferes with respiratory blood-oxygen exchange in humans (41), and most avian influenza patients developed acute respiratory distress syndrome (42, 43). Our study may provide a novel molecular mechanism to show why H5N1 is so lethal.
ACKNOWLEDGMENTS
We are especially grateful to W. Dai for assistance with the electron microscopy and J. Jia, Department of Biological Physics, Chinese Academy of Sciences, for assistance with the FACS.
This study was supported by the Ministry of Science and Technology of China (2015CB553400/6), the National Natural Science Foundation of China (81230002 and 81490531), the Ministry of Health (201302017), and the 111 Project (B08007).
C. Jiang conceived the project. X. Ju, Y. Yan, Q. Liu, N. Li, M. Sheng, L. Zhang, Y. Zhao, and Y. Zhang contributed to the experiments and analysis of the data. X. Ju, M. Sheng, P. Yang, and K. Liu performed the cell culture and protein purification. J. Du, Y. Zhong, and H. Zhou assisted with the immunoblotting and q-PCR. Z. Liang and F. Huang assisted with the statistics and bioinformatics. Z. Zou, X. Li, H. Lu, and M. Tian assisted with the biosafety level 3 experiments. D. Li and J. Zhang provided key ideas. C. Jiang and N. Jin designed the experiments and analyzed the data. C. Jiang, X. Ju, Y. Yan, Z. Liang, and N. Jin wrote the manuscript.
REFERENCES
- 1.Eskelinen EL. 2006. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med 27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 2.McGourty K, Thurston TL, Matthews SA, Pinaud L, Mota LJ, Holden DW. 2012. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science 338:963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneede A, Schmidt CK, Holtta-Vuori M, Heeren J, Willenborg M, Blanz J, Domanskyy M, Breiden B, Brodesser S, Landgrebe J, Sandhoff K, Ikonen E, Saftig P, Eskelinen EL. 2011. Role for LAMP-2 in endosomal cholesterol transport. J Cell Mol Med 15:280–295. doi: 10.1111/j.1582-4934.2009.00973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saftig P, Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10:623–635. doi: 10.1038/nrn2709, . [DOI] [PubMed] [Google Scholar]
- 5.Appelqvist H, Waster P, Kagedal K, Ollinger K. 2013. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol 5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- 6.Aits S, Jaattela M. 2013. Lysosomal cell death at a glance. J Cell Sci 126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 7.Horimoto T, Kawaoka Y. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 8.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, Ju X, Liang Z, Liu Q, Zhao Y, Guo F, Bai T, Han Z, Zhu J, Zhou H, Huang F, Li C, Lu H, Li N, Li D, Jin N, Penninger JM, Jiang C. 2014. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun 5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Jiang C. 2009. Avian influenza H5N1: an update on molecular pathogenesis. Sci China C Life Sci 52:459–463. doi: 10.1007/s11427-009-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Jiang C. 2009. Influenza A virus H5N1 entry into host cells is through clathrin-dependent endocytosis. Sci China C Life Sci 52:464–469. doi: 10.1007/s11427-009-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng E, Ye D, Li J, Zhang D, Wang J, Zhao F, Hilgenfeld R, Zheng M, Jiang H, Liu H. 2012. Recent advances in neuraminidase inhibitor development as anti-influenza drugs. ChemMedChem 7:1527–1536. doi: 10.1002/cmdc.201200155. [DOI] [PubMed] [Google Scholar]
- 12.Hulse DJ, Webster RG, Russell RJ, Perez DR. 2004. Molecular determinants within the surface proteins involved in the pathogenicity of H5N1 influenza viruses in chickens. J Virol 78:9954–9964. doi: 10.1128/JVI.78.18.9954-9964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Yu Z, Hu Y, Tu J, Zou W, Peng Y, Zhu J, Li Y, Zhang A, Ye Z, Chen H, Jin M. 2009. The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One 4:e6277. doi: 10.1371/journal.pone.0006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Takahashi T, Guo CT, Hidari KI, Miyamoto D, Goto H, Kawaoka Y, Suzuki Y. 2005. Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. J Virol 79:11705–11715. doi: 10.1128/JVI.79.18.11705-11715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Li C, Shu Y, Ju X, Zou Z, Wang H, Rao S, Guo F, Liu H, Nan W, Zhao Y, Yan Y, Tang J, Zhao C, Yang P, Liu K, Wang S, Lu H, Li X, Tan L, Gao R, Song J, Gao X, Tian X, Qin Y, Xu KF, Li D, Jin N, Jiang C. 2012. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal 5:ra16. doi: 10.1126/scisignal.2001931. [DOI] [PubMed] [Google Scholar]
- 16.Haas J, Park EC, Seed B. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol 6:315–324. doi: 10.1016/S0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. 2008. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res 136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi T, Suzuki Y, Nishinaka D, Kawase N, Kobayashi Y, Hidari KI, Miyamoto D, Guo CT, Shortridge KF, Suzuki T. 2001. Duck and human pandemic influenza A viruses retain sialidase activity under low pH conditions. J Biochem 130:279–283. doi: 10.1093/oxfordjournals.jbchem.a002983. [DOI] [PubMed] [Google Scholar]
- 19.Thanunchai M, Kanrai P, Wiboon-Ut S, Puthavathana P, Hongeng S, Thitithanyanont A. 2013. Tropism of avian influenza A (H5N1) virus to mesenchymal stem cells and CD34+ hematopoietic stem cells. PLoS One 8:e81805. doi: 10.1371/journal.pone.0081805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uiprasertkul M, Kitphati R, Puthavathana P, Kriwong R, Kongchanagul A, Ungchusak K, Angkasekwinai S, Chokephaibulkit K, Srisook K, Vanprapar N, Auewarakul P. 2007. Apoptosis and pathogenesis of avian influenza A (H5N1) virus in humans. Emerg Infect Dis 13:708–712. doi: 10.3201/eid1305.060572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawada R, Lowe JB, Fukuda M. 1993. E-selectin-dependent adhesion efficiency of colonic carcinoma cells is increased by genetic manipulation of their cell surface lysosomal membrane glycoprotein-1 expression levels. J Biol Chem 268:12675–12681. [PubMed] [Google Scholar]
- 23.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. 2009. A gene network regulating lysosomal biogenesis and function. Science 325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 24.Yogalingam G, Bonten EJ, van de Vlekkert D, Hu H, Moshiach S, Connell SA, d'Azzo A. 2008. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev Cell 15:74–86. doi: 10.1016/j.devcel.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matrosovich M, Herrler G, Klenk HD. 2015. Sialic acid receptors of viruses. Top Curr Chem 367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 27.Ghate AA, Air GM. 1998. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. Eur J Biochem 258:320–331. doi: 10.1046/j.1432-1327.1998.2580320.x. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi T, Suzuki T, Hidari KI, Miyamoto D, Suzuki Y. 2003. A molecular mechanism for the low-pH stability of sialidase activity of influenza A virus N2 neuraminidases. FEBS Lett 543:71–75. doi: 10.1016/S0014-5793(03)00403-4. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson KG WR, Hay AJ. 1998. Textbook of influenza. Blackwell Science, Oxford, England. [Google Scholar]
- 30.Lamb RA KR. 2001. Orthomyxoviridae: the viruses and their replication, p 1487–1531. In Knipe DM, Howley PM (ed), Fields virology, 4th ed Lippincott, Philadelphia, PA. [Google Scholar]
- 31.Heinonen S, Silvennoinen H, Lehtinen P, Vainionpaa R, Vahlberg T, Ziegler T, Ikonen N, Puhakka T, Heikkinen T. 2010. Early oseltamivir treatment of influenza in children 1–3 years of age: a randomized controlled trial. Clin Infect Dis 51:887–894. doi: 10.1086/656408. [DOI] [PubMed] [Google Scholar]
- 32.Cao B, Li XW, Mao Y, Wang J, Lu HZ, Chen YS, Liang ZA, Liang L, Zhang SJ, Zhang B, Gu L, Lu LH, Wang DY, Wang C. 2009. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N Engl J Med 361:2507–2517. doi: 10.1056/NEJMoa0906612. [DOI] [PubMed] [Google Scholar]
- 33.Boya P, Kroemer G. 2008. Lysosomal membrane permeabilization in cell death. Oncogene 27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 34.Schwake M, Schroder B, Saftig P. 2013. Lysosomal membrane proteins and their central role in physiology. Traffic 14:739–748. doi: 10.1111/tra.12056. [DOI] [PubMed] [Google Scholar]
- 35.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, von Figura K, Knecht E, Saftig P. 2004. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. 2007. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saftig P, Beertsen W, Eskelinen EL. 2008. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy 4:510–512. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 38.Grove J, Marsh M. 2011. The cell biology of receptor-mediated virus entry. J Cell Biol 195:1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilyushina NA, Seiler JP, Rehg JE, Webster RG, Govorkova EA. 2010. Effect of neuraminidase inhibitor-resistant mutations on pathogenicity of clade 2.2 A/Turkey/15/06 (H5N1) influenza virus in ferrets. PLoS Pathog 6:e1000933. doi: 10.1371/journal.ppat.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen HL, Ilyushina NA, Salomon R, Hoffmann E, Webster RG, Govorkova EA. 2007. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J Virol 81:12418–12426. doi: 10.1128/JVI.01067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan MC, Chan RW, Yu WC, Ho CC, Chui WH, Lo CK, Yuen KM, Guan YI, Nicholls JM, Peiris JS. 2009. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir Res 10:102. doi: 10.1186/1465-9921-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu T, Qiao J, Zhao L, Wang G, He G, Li K, Tian Y, Gao M, Wang J, Wang H, Dong C. 2006. Acute respiratory distress syndrome induced by avian influenza A (H5N1) virus in mice. Am J Respir Crit Care Med 174:1011–1017. doi: 10.1164/rccm.200511-1751OC. [DOI] [PubMed] [Google Scholar]
- 43.Tang X, He H, Sun B, Wan J, Ban C, Zhang C, Wang S, Xia J, Li J, Liu Y, Cao B, Tong Z. 2014. ARDS associated with pneumonia caused by avian influenza A H7N9 virus treated with extracorporeal membrane oxygenation. Clin Respir J 9:380–384. doi: 10.1111/crj.12140. [DOI] [PubMed] [Google Scholar]