Abstract
Amphibian-like ranaviruses include pathogens of fish, amphibians, and reptiles that have recently evolved from a fish-infecting ancestor. The molecular determinants of host range and virulence in this group are largely unknown, and currently fish infection models are lacking. We show that European sheatfish virus (ESV) can productively infect zebrafish, causing a lethal pathology, and describe a method for the generation of recombinant ESV, establishing a useful model for the study of fish ranavirus infections.
TEXT
Amphibian-like ranaviruses (ALRVs) are a subgroup of closely related viruses within the genus Ranavirus isolated from different amphibian, reptile, and fish host species (1). Epizootic hematopoietic necrosis virus (EHNV), a rainbow trout pathogen belonging to the ALRV found exclusively in Australia, was the first ALRV isolated from fish to be completely sequenced (2). More recently, we obtained the complete genome of a second fish ALRV isolated from European sheatfish (ESV) (3). Both the dot plot analyses as well as a phylogenetic tree, including completely sequenced vertebrate ranaviruses (Fig. 1), confirm that ESV is very closely related to EHNV, with the short branch length among ALRVs supporting its recent evolutionary origin. Most annotated genes are conserved between both fish viruses, although two potentially relevant differences are found. A putative 3β-hydroxysteroid dehydrogenase, whose orthologue in vaccinia virus acts as a virulence factor in vivo (4, 5), is present in all ALRVs but deleted in ESV. Additionally, ESV has acquired a unique set of genes representing a multigene family of unknown function described in EHNV (2). Specifically, while EHNV carries five members of related genes located on consecutive positions on the viral genome, ESV retains the orthologues for only three of these (ESV open reading frames [ORFs] 75R, 76R, and 80L) but has acquired three novel genes belonging to this family that are not found in any other virus and are located at different positions in the genome (ESV ORFs 59L, 110L, and 111L). In poxviruses, the presence of additional copies of viral genes can serve as evolutionary scanning devices for their adaptation to host immune evasion or host range adaptation (6, 7), and multigene families in the related virus family Asfarviridae are known to contribute to the same processes (8, 9).
FIG 1.
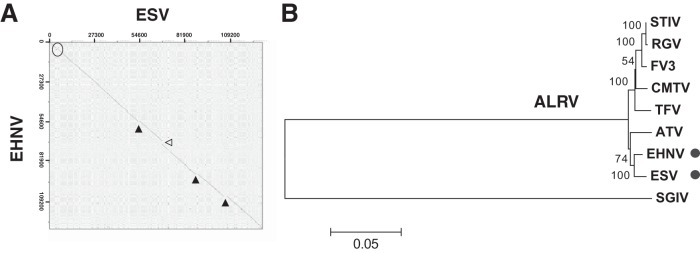
European sheatfish virus (ESV) is a close evolutionary relative of epizootic hematopoietic necrosis virus (EHNV). (A) Dot plot analysis of the complete ESV genome compared to EHNV, indicating sites of sequence insertion (solid triangles), deletion (open triangles), and inversion (circle). (B) Phylogenetic tree calculated by the neighbor-joining method using the concatemeric protein sequence corresponding to the 26 core genes of the indicated ranaviruses. STIV, soft-shelled turtle iridovirus; RGV, Rana grylio virus; FV3, frog virus 3; CMTV, common midwife toad virus; TFV, tiger frog virus; SGIV, Singapore grouper iridovirus. Bootstrap percentages (500 replicates) are indicated next to the branches, and the tree is drawn to scale, with distances given as the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA6. The positions of ESV and EHNV are indicated by dots.
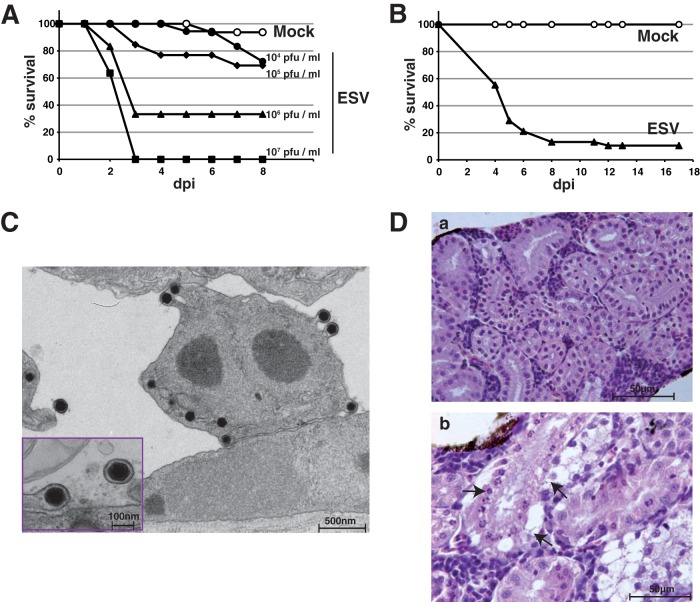
Currently, in vivo studies of fish ranaviruses are restricted to its respective host species, and a genetically tractable model is lacking. The zebrafish (Danio rerio) is regarded as a choice model for the study of pathological disorders, including viral infections (10, 11). Thus, it has been employed to characterize the host response to infections with the human pathogen Chikungunya virus (12) or the pathogenesis of the salmonid rhabdovirus infectious hematopoietic necrosis virus (IHNV) infection (13). The zebrafish also provides a relevant model to study infections with the infectious spleen and kidney iridovirus (ISKNV), a member of the genus Megalocytivirus of fish pathogens included within the Iridoviridae (14). To assess the susceptibility of zebrafish to ESV infection, we immersed larvae for 1 h in a reduced volume of water containing different virus doses (bath immersion) and then transferred them to freshwater tanks. As shown in Fig. 2A, larvae readily succumbed to ESV infection in a dose-dependent matter, with cumulative death rates of up to 100%. Adult fish were similarly susceptible to infection, with cumulative mortality rates of up to 90% within 12 days postinfection (dpi) (Fig. 2B). By using plaque assays on epithelioma papulosum cyprini (EPC) cells, we detected the presence of live virus in whole-body tissue homogenates of dead adult fish obtained at day 3 postinfection (p.i.) as well as in all 4 surviving animals by day 16 after inoculation. Similarly, infectious ESV was recovered from dead larvae between days 2 and 5 p.i. The virus recovered was confirmed to be ESV by PCR analysis using oligonucleotides targeting ESV ORF 114L. Moreover, electron microscopy analyses of ultrathin sections of infected larvae at 2 and 3 dpi showed discrete viral factories, assembling virions and budding viruses at different locations, which were never observed in mock-infected animals (Fig. 2C) that were indistinguishable in size, shape, and composition from ESV particles replicating on cultured EPC cells. By 2 dpi, histopathological examination of adult infected fish showed kidney lesions with chromatin margination and necrosis of the tubular epithelium (Fig. 2D) in all infected animals, as described in ESV-infected catfish (Ictalurus melas) (15). Other typical pathological findings, including disseminated foci of hemorrhage and necrosis of hematopoietic tissue of the kidney and spleen, were not detected in zebrafish. Whether these occur at later stages of the disease remains to be addressed.
FIG 2.
ESV infection causes high mortality rates in larval and adult zebrafish. (A) Groups of AB zebrafish larvae (n > 11) at 6 days postfertilization (dpf) were infected by bath immersion with increasing doses of semipurified ESV obtained by ultracentrifugation of crude viral stocks through a 45% sucrose cushion as indicated, and cumulative mortality was recorded daily up to 8 dpi. Shown is one representative experiment of three. (B) Groups of adult AB zebrafish (mock, n = 28; ESV, n = 38) were infected by bath immersion with a single dose of 2 × 106 PFU/ml of semipurified ESV, and cumulative mortality was recorded daily up to 17 dpi. Shown is one representative experiment of three. (C) Transmission electron micrograph of ultrathin sections of glutaraldehyde-fixed and Epon-embedded ESV-infected larva at 2 dpi. The inset shows a detail of budding viruses from a different section. (D) Hematoxylin and eosin (H&E)-stained section of an adult zebrafish kidney corresponding to mock-infected (a) or ESV-infected (b) animals at 48 h p.i. Arrows indicate nuclear chromatin margination, necrosis, and vacuolation in segments of the renal tubular epithelium. Scale bars are shown in panels C and D.
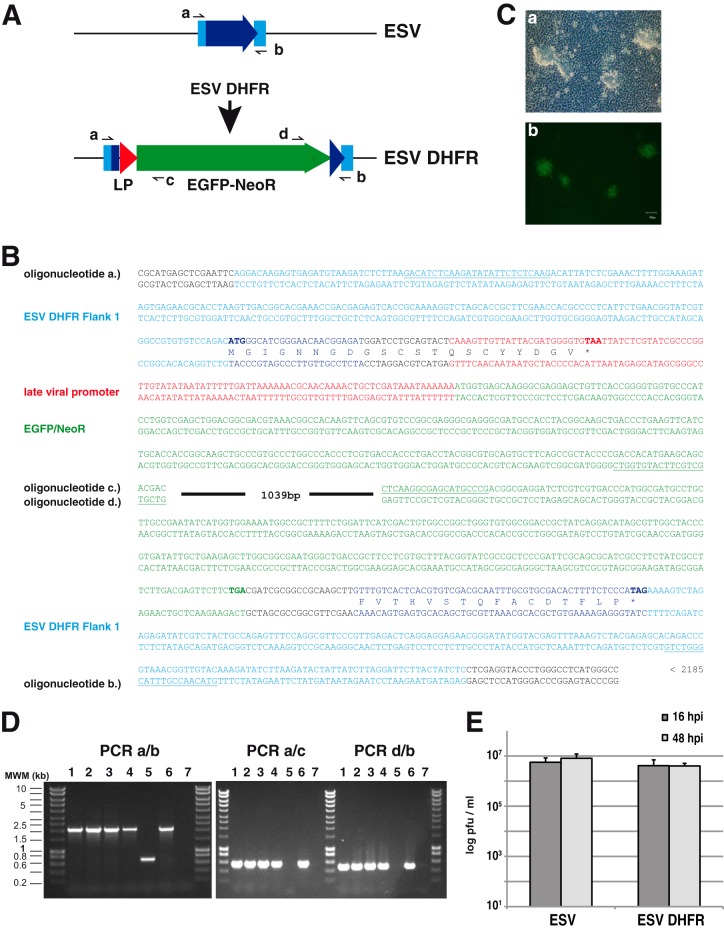
In order to be able to study the contribution of individual viral genes to pathogenesis and other processes, we developed a method to generate recombinant ESV. Homologous recombination in ranaviruses was first demonstrated when a recombinant Bohle iridovirus expressing the β-globin from the cane toad (Bufo marinus) was obtained. In this case, the promoter from ranaviral ICP18 was used to drive a neomycin resistance gene that was used for selection (16). A similar strategy was employed to generate recombinant Ambystoma tigrinum virus (ATV) lacking the homologue of eukaryotic initiation factor-2 (vIF2a) to study its contribution to immune evasion in infected salamanders (17). More recently, a dual selection marker encoding a puromycin resistance-enhanced green fluorescent protein (EGFP) fusion protein was placed under the control of the same viral promoter and used to generate targeted knockouts in frog virus 3 (FV3) (18). However, no recombinant fish ranaviruses have been constructed or tested in vivo. We adapted the methodology to the generation of ESV recombinants by using a distinct selection marker encoding an EGFP/neomycin resistance fusion protein (19) placed under the control of a strong late promoter sequence from the fish ranavirus Singapore grouper iridovirus, which is highly expressed both in cell culture and in vivo (20) (Fig. 3A). We selected the ESV ORF 114 locus for targeted disruption. This gene encodes a putative dihydrofolate reductase (DHFR) that is highly similar to but smaller than the orthologue identified in EHNV, where it was proposed to play a role in host tropism (2). To delete ESV DHFR, a plasmid bearing two approximately 200-bp-long flanking regions corresponding to positions 112506 to 112284 and 111945 to 111726 of the ESV genome as well as the previously mentioned selection cassette was obtained by gene synthesis (Mr. Gene, Germany) (Fig. 3B). EPC cells were transfected with circular plasmid and subsequently infected with parental ESV. The recombinant viruses were obtained by selection in the presence of neomycin and subsequent purification of EGFP-expressing viral plaques (Fig. 3C). Four independent clones of the resulting viruses were selected and amplified, and the correct insertion of the reporter cassette at the expected genomic location as well as absence of unwanted single crossover events were confirmed by PCR (Fig. 3D). Gene expression using the annotated start codon from ORF 114L in the recombinant viruses would result in a 21-amino-acid peptide due to the presence of an early in-frame stop codon, which would only retain the first eight N-terminal residues of the putative DHFR, abrogating its activity (Fig. 3B). Importantly, this system may be adapted for its use in a transient dominant selection procedure (21) by placing both flanking regions together on one side of the selection cassette in the plasmid used for recombination. This would enable the generation of recombinant viruses lacking heterologous DNA sequence, allowing the construction of revertant viruses as controls or of multiple targeted deletion mutants using the same selection procedure.
FIG 3.
Method for the generation of recombinant ESV using homologous recombination. (A) Schematic representation of the genomic structure of the parental ESV and the ESVΔDHFR deletion mutant generated indicating the fragmented locus, the late viral promoter, and the neomycin resistance/EGFP fusion cassette used for selection, as well as the positions of the oligonucleotides used for PCR analyses. (B) Relevant sequences of a fragment of the plasmid used for homologous recombination, representing the genomic sequence of the recombinant virus in this area. The sequence of the viral promoter corresponding to the 92 bp upstream of the initiation codon of Singapore grouper iridovirus ORF 125R is shown in red. The complete sequence of the plasmid is available upon request. The amino acids corresponding to the putative ESV DHFR are shown in blue, and translation frames are indicated. (C) Bright-field (a) and fluorescence microscopy (b) of EPC cells showing developing ESVΔDHFR replication plaques at 24 hpi. (D) PCR analyses using the indicated oligonucleotide pairs of purified DNA derived from four individual recombinant ESV clones (lanes 1 to 4) and the parental ESV (lane 5), along with a positive (plasmid used for the generation of the recombinant virus (lane 6) and negative (no-DNA) control (lane 7). MWM, molecular weight markers. (E) ESV and ESVΔDHFR virus yields at the indicated time points after high-multiplicity infection of EPC cells.
The recombinant virus generated, termed ESVΔDHFR, showed no defect in single-cycle (Fig. 3E) or multiple-cycle (not shown) viral growth assays on fish EPC cells. Both ESV and ESVΔDHFR were able to infect and replicate in the BHK, Vero, and L929 cell lines of mammalian origin when held at 30°C under standard growth conditions (not shown). When zebrafish larvae were infected with increasing doses of the parental ESV or the ESVΔDHFR viruses, no significant differences in the overall mortality rates were observed (Fig. 4A). Similarly, no significant differences in mortality rates were observed in adult zebrafish. Immunostaining using anti-GFP antibodies showed labeling of kidney cells in adult animals infected with the recombinant virus but not with parental ESV at 4 dpi, indicating viral replication and expression of the heterologous gene in vivo (Fig. 4B). Overall, these results show that the putative ESV DHFR activity is not an essential factor for virus replication in cell culture and does not act as a major virulence factor in zebrafish infection. Further experiments on the activity of EHNV and ESV DHFRs will be required to understand their precise roles during infection of their specific hosts.
FIG 4.
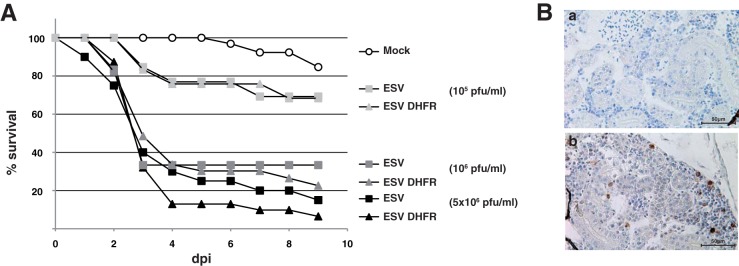
The DHFR gene of ESV is not essential for virulence in infected zebrafish. (A) Cumulative mortality of groups (n >11) of zebrafish (AB strain; www.zfin.org) larvae at 7 days postfertilization (dpf) either mock infected or infected by bath immersion with increasing doses of the parental ESV and the recombinant ESVΔDHFR as indicated. Means from duplicates from one representative experiment of two are shown. (B) Immunohistochemical analysis of paraformaldehyde-fixed specimens of mock-infected (a) and ESVΔDHFR-infected (b) adult zebrafish sacrificed at 4 dpi using anti-GFP antibody, showing specific staining of kidney cells in infected fish. Scale bars are shown.
In summary, we have shown zebrafish to be a convenient model for the study of ranavirus pathogenesis and provided a new platform for the generation of recombinant iridoviruses that might be used to explore the role of additional genes in virus-host interaction as well as for the expression of foreign antigens as novel fish vaccines.
ACKNOWLEDGMENTS
We wish to acknowledge the work of Milagros Guerra, from the electron microscopy unit at Centro de Biología Molecular Severo Ochoa.
This work was supported by grant AGL 2009-08711 from the Spanish Ministerio de Ciencia e Innovación and grant AGL2013-48998-C2-2-R from the Ministerio de Economía y Competitividad. G.A. was supported by the Amarouto Program for senior researchers from the Comunidad Autónoma de Madrid. A.L.B. and V.M. are holders of RyC-2010-06300 and RyC-2010-06516 Ramón y Cajal fellowships from the Spanish Ministerio de Ciencia e Innovación.
REFERENCES
- 1.Chinchar VG, Yu KH, Jancovich JK. 2011. The molecular biology of frog virus 3 and other iridoviruses infecting cold-blooded vertebrates. Viruses 3:1959–1985. doi: 10.3390/v3101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jancovich JK, Brémont M, Touchman JW, Jacobs BL. 2010. Evidence for multiple recent host species shifts among the ranaviruses (family Iridoviridae). J Virol 84:2636–2647. doi: 10.1128/JVI.01991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavian C, López-Bueno A, Fernández Somalo MP, Alcami A, Alejo A. 2012. Complete genome sequence of the European sheatfish virus. J Virol 86:6365–6366. doi: 10.1128/JVI.00618-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore JB, Smith GL. 1992. Steroid hormone synthesis by a vaccinia enzyme: a new type of virus virulence factor. EMBO J 11:3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reading PC, Moore JB, Smith GL. 2003. Steroid hormone synthesis by vaccinia virus suppresses the inflammatory response to infection. J Exp Med 197:1269–1278. doi: 10.1084/jem.20022201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. 2014. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog 10:e1004002. doi: 10.1371/journal.ppat.1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrage TG, Lu Z, Neilan JG, Rock DL, Zsak L. 2004. African swine fever virus multigene family 360 genes affect virus replication and generalization of infection in Ornithodoros porcinus ticks. J Virol 78:2445–2453. doi: 10.1128/JVI.78.5.2445-2453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonso CL, Piccone ME, Zaffuto KM, Neilan J, Kutish GF, Lu Z, Balinsky CA, Gibb TR, Bean TJ, Zsak L, Rock DL. 2004. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J Virol 78:1858–1864. doi: 10.1128/JVI.78.4.1858-1864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, Bitter W. 2004. A star with stripes: zebrafish as an infection model. Trends Microbiol 12:451–457. doi: 10.1016/j.tim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Allen JP, Neely MN. 2010. Trolling for the ideal model host: zebrafish take the bait. Future Microbiol 5:563–569. doi: 10.2217/fmb.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, Wang C-H, Lieschke GJ, Herbomel P, Schwartz O, Levraud J-P. 2013. Real-time whole-body visualization of Chikungunya virus infection and host interferon response in zebrafish. PLoS Pathog 9:e1003619. doi: 10.1371/journal.ppat.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig M, Palha N, Torhy C, Briolat V, Colucci-Guyon E, Brémont M, Herbomel P, Boudinot P, Levraud J-P. 2011. Whole-body analysis of a viral infection: vascular endothelium is a primary target of infectious hematopoietic necrosis virus in zebrafish larvae. PLoS Pathog 7:e1001269. doi: 10.1371/journal.ppat.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Zhang L, Weng S, Huang Z, Lu J, Lan D, Zhong X, Yu X, Xu A, He J. 2008. A zebrafish (Danio rerio) model of infectious spleen and kidney necrosis virus (ISKNV) infection. Virology 376:1–12. doi: 10.1016/j.virol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Pozet F, Morand M, Moussa A, Torhy C, de Kinkelin P. 1992. Isolation and preliminary characterization of a pathogenic icosahedral deoxyribovirus from the catfish Ictalurus melas. Dis Aquat Org 14:35–42. doi: 10.3354/dao014035. [DOI] [Google Scholar]
- 16.Pallister J, Goldie S, Coupar B, Shiell B, Michalski WP, Siddon N, Hyatt A. 2007. Bohle iridovirus as a vector for heterologous gene expression. J Virol Methods 146:419–423. doi: 10.1016/j.jviromet.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Jancovich JK, Jacobs BL. 2011. Innate immune evasion mediated by the Ambystoma tigrinum virus eukaryotic translation initiation factor 2alpha homologue. J Virol 85:5061–5069. doi: 10.1128/JVI.01488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Ward BM, Yu KH, Chinchar VG, Robert J. 2011. Improved knockout methodology reveals that frog virus 3 mutants lacking either the 18K immediate-early gene or the truncated vIF-2alpha gene are defective for replication and growth in vivo. J Virol 85:11131–11138. doi: 10.1128/JVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewals B, Boudry C, Gillet L, Markine-Goriaynoff N, de Leval L, Haig DM, Vanderplasschen A. 2006. Cloning of the genome of Alcelaphine herpesvirus 1 as an infectious and pathogenic bacterial artificial chromosome. J Gen Virol 87:509–517. doi: 10.1099/vir.0.81465-0. [DOI] [PubMed] [Google Scholar]
- 20.Teng Y, Hou Z, Gong J, Liu H, Xie X, Zhang L, Chen X, Qin QW. 2008. Whole-genome transcriptional profiles of a novel marine fish iridovirus, Singapore grouper iridovirus (SGIV) in virus-infected grouper spleen cell cultures and in orange-spotted grouper, Epinephulus coioides. Virology 377:39–48. doi: 10.1016/j.virol.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Falkner FG, Moss B. 1990. Transient dominant selection of recombinant vaccinia viruses. J Virol 64:3108–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]