ABSTRACT
The vaccinia virus (VACV) E3 protein has been shown to be important for blocking activation of the cellular innate immune system and allowing viral replication to occur unhindered. Mutation or deletion of E3L severely affects viral host range and pathogenesis. While the monkeypox virus (MPXV) genome encodes a homologue of the VACV E3 protein, encoded by the F3L gene, the MPXV gene is predicted to encode a protein with a truncation of 37 N-terminal amino acids. VACV with a genome encoding a similarly truncated E3L protein (VACV-E3LΔ37N) has been shown to be attenuated in mouse models, and infection with VACV-E3LΔ37N has been shown to lead to activation of the host antiviral protein kinase R pathway. In this report, we present data demonstrating that, despite containing a truncated E3 homologue, MPXV phenotypically resembles a wild-type (wt) VACV rather than VACV-E3LΔ37N. Thus, MPXV appears to contain a gene or genes that can suppress the phenotypes associated with an N-terminal truncation in E3. The suppression maps to sequences outside F3L, suggesting that the suppression is extragenic in nature. Thus, MPXV appears to have evolved mechanisms to minimize the effects of partial inactivation of its E3 homologue.
IMPORTANCE Poxviruses have evolved to have many mechanisms to evade host antiviral innate immunity; these mechanisms may allow these viruses to cause disease. Within the family of poxviruses, variola virus (which causes smallpox) is the most pathogenic, while monkeypox virus is intermediate in pathogenicity between vaccinia virus and variola virus. Understanding the mechanisms of monkeypox virus innate immune evasion will help us to understand the evolution of poxvirus innate immune evasion capabilities, providing a better understanding of how poxviruses cause disease.
INTRODUCTION
Monkeypox virus (MXPV) is a member of the family Poxviridae and was first reported to cause disease in humans in the Congo region of Africa in 1972 (1). MPXV is endemic in the western and central areas of Africa, and the mortality rate from MXPV infections is approximately 10%, whereas that from variola virus (VARV) infections is 30% (2, 3). While MPXV is pathogenic in humans, it spreads poorly from human to human (4). The first outbreak of MPXV in the Western Hemisphere occurred in 2003, with 37 confirmed cases occurring in the midwestern United States (5); however, no fatalities were associated with this outbreak. DNA sequencing of the U.S. MPXV isolates showed they were more closely related to the less pathogenic West African strains of MPXV than the more virulent Central African strains. This may offer one explanation as to why no human fatalities were associated with this outbreak of MPXV in 2003 (5).
With the eradication of smallpox and the cessation of the human smallpox vaccination program, there is a growing concern that VARV and MPXV could be employed as biological weapons. This is especially disconcerting, given that today's human population has relatively low or nonexistent immunity to smallpox (1, 6). The use of a vaccinia virus (VACV; smallpox vaccine) to protect against an MPXV infection has been shown to be 85% effective in preventing disease (7), but it is highly unlikely that it will be used as a preexposure vaccine, due to the side effects associated with vaccination. For these reasons, there is an increasing interest in gaining a better understanding of how MPXV is able to evade innate immunity and cause disease in humans.
The majority of what is known about orthopoxvirus innate immune evasion has come through the study of VACV and how it inhibits the host's antiviral immune response. The VACV E3L gene has been shown to play a critical role in inhibiting the cellular interferon (IFN) antiviral immune response (8). The E3 protein is able to bind double-stranded RNA (dsRNA) and sequester it away from known pattern recognition receptors (PRRs; e.g., protein kinase R [PKR], RIG-I, MDA-5, and OAS), thereby preventing their activation (8–10).
The VACV E3 protein contains two conserved domains: an N-terminal Z-nucleic acid (Z-NA) binding domain (BD) and a C-terminal dsRNA-binding domain. Both of these domains have been shown to be necessary for wild-type virus pathogenesis in a mouse model, as well as full IFN resistance (IFNr) in vitro (11, 12). Both the N-terminal and C-terminal domains of the VACV E3 protein have been shown to be necessary for the complete inhibition of the PKR pathway (13). In vitro studies show that two VACV recombinants, VACV-E3LΔ83N (in which the first 83 amino acids of the N terminus of E3L are deleted) and VACV-E3LΔ37N (which encodes only the smaller of the two proteins [p20] translated from E3L mRNA) have a host range and IFNr phenotype similar to those of wild-type (wt) VACV in most cells in culture (14, 15). However, in a mouse model, both VACV-E3LΔ83N and VACV-E3LΔ37N were 100- to 1,000-fold less pathogenic than wt VACV (16). Phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α) was detected in culture for cells infected with either virus at late times postinfection (∼9 h postinfection [hpi]), while phosphorylation of eIF2α was not present in wt VACV-infected HeLa cells (13). Phosphorylation of eIF2α has also been detected in mice after infection with VACV-E3LΔ83N (13). This suggests that the N terminus of the E3 protein may be required to inhibit the activation of the host's antiviral response.
Genomic comparisons between MPXV and VACV show sequence identity between these two viruses. The MPXV F3 protein is a homologue of the VACV E3 protein, with the two proteins having nucleotide and protein sequence identities of 92% and 88%, respectively. The genomic sequence of MPXV suggests that the F3 protein contains a complete, functional, C-terminal dsRNA-binding domain, while the first 37 amino acids of the N terminus of the Z-NA binding domain are deleted. Since the entire VACV E3 protein has been shown to be necessary for the full inhibition of PKR in both cells in culture and whole animals and for pathogenesis in mice, we were interested in characterizing the inhibition of innate immunity and replication in MPXV-infected cells. We show that even though the MXPV genome encodes an altered E3 homologue, MPXV has an IFNr and host range phenotype similar to that of wt VACV and it is able to inhibit the cellular antiviral immune response to a greater extent than a VACV mutant containing an equivalent N-terminal truncation in E3. These data suggest that MPXV can suppress phenotypes associated with the lack of an N-terminal domain on its E3 homologue. Replacement of E3L in VACV with F3L did not lead to full inhibition of PKR and did not rescue replication in cells in culture, indicating that the suppression of the lack of an N-terminal domain on F3 is extragenic in nature.
MATERIALS AND METHODS
Sequence analysis.
The amino acid sequences of the MPXV F3 (GenBank accession number AY603973, VACV E3 (GenBank accession number M35027), and VARV E3 (GenBank accession number L22579) were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) and aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/).
Cells and viruses.
All cell lines were obtained from ATCC, with the exception of HeLa F2 (used for Western blot analyses of expression and phosphorylation), which was a kind gift of George Pavlakis, NCI. Vero-E6 cells, baby hamster kidney (BHK-21) cells, rabbit kidney (RK) RK13 cells, and RK-E3L cells (RK13 cells stably transfected with a plasmid expressing the E3L gene, using the Tet-Off system from Clontech) were maintained in Eagle's minimal essential medium (MEM; Cellgro) supplemented with 5% fetal bovine serum (FBS; HyClone). HeLa F2 cells and BSC-40 cells were maintained in Dulbecco's modified minimal essential medium (DMEM; Cellgro) supplemented with 5% FBS and 2 mM l-glutamine. JC (murine adenocarcinoma) cells were maintained in RPMI (ATCC) supplemented with 10% heat-inactivated FBS. All cells were incubated at 37°C in the presence of 5% CO2. For IFN treatment, cells were pretreated with the appropriate amount of human alpha A/D interferon (IFN-α A/D; PBL Assay Science) at 37°C for 18 h. Vaccinia virus Copenhagen (VC-2) and Western Reserve (WR) strains, designated VACV, were used as the parental viruses for all the recombinant viruses used throughout this study. VACV with an E3L deletion (VACVΔE3L) and VACV containing a 37-amino-acid N-terminal truncation of E3L (VACV-E3LΔ37N) were generated as previously described (15, 17). All experiments with MPXV (strain WR 7-61) were performed in accordance with the protocols approved by Arizona State University and the Centers for Disease Control and Prevention (CDC). For all HeLa and Vero-E6 cell infections, Copenhagen strains of vaccinia virus were used, whereas for all JC cell infections, Western Reserve strains were used. Encephalomyocarditis virus (EMCV) was obtained from ATCC.
Overlapping PCR.
VACV E3L-flanking DNA regions of about 200 bp in length were amplified from the VACV Copenhagen genome. The upstream flanking region was amplified with forward primer 5′-ATGTTACAACGCAATCGATACATGAA-3′ and reverse primer 5′-TTTTAGAGAGAACTAACACAACCAGCAAT-3′. The downstream flanking region was amplified with forward primer 5′-CCTCGTGCAAATATCCAAACGC-3′ and reverse primer 5′-CTGATTCTAGTTATCAATAACAGTTAGTAGTTT-3′. The F3L gene of MPXV, including the 5′ untranslated region homologous to VACV E3L, was amplified from MPXV DNA through PCR with primers containing sequences overlapping the amplified flanking regions of VACV E3L. The forward overlapping primer for F3L was 5′-GGTTGTGTTAGTTCTCTCTAAAAAAATTTCTAAGATCTATATTGA-3′, and the reverse primer was 5′-CTAACTGTTATTGATAACTAGAATCAGAATCTAATGATGACATAACTAAG-3′. All PCR products were purified using a Wizard SV Gel and PCR cleanup system (Promega) according to the manufacturer's instructions.
Transfection and in vivo recombination.
In vivo recombination was performed in BHK-21 cells as described previously (16). Transfections were performed using the Lipofectamine reagent (Invitrogen) in conjunction with the Plus reagent (Invitrogen) to enhance the transfection efficiency. BHK-21 cells were transfected with the F3L PCR product according to the manufacturer's protocol and infected with VACVΔE3L at a multiplicity of infection (MOI) of 0.01. At 48 hpi, the cells were harvested and plaques were allowed to form in BSC-40 cells. All viruses were amplified in BHK-21 cells. The recombinant VACV expressing the F3 protein of MPXV is referred to as VACVΔE3L::F3L (VACV-F3L). VACV-F3L was generated for both the Copenhagen and WR strains of VACV. Figure 1 indicates the placement of the F3L gene in the VACV genome relative to that in the VACV-E3LΔ37N and the MPXV genomes.
FIG 1.
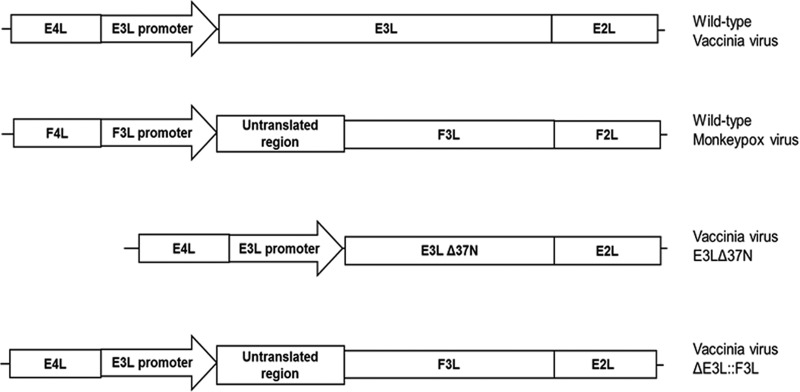
Schematic of viruses used in this study.
Interferon sensitivity assay.
BSC-40 and RK13 cells were seeded in 6-well plates and pretreated with 0, 1, 10, 100, or 1,000 IU/ml of IFN-α A/D (cross-reactive with human, monkey, mouse, bovine, rat, cat, pig, rabbit, guinea pig, or hamster cells; catalog number 11200-2; PBL). Cells were infected with an estimated amount of 100 PFU of wt VACV, VACVΔ37N, VACV-F3L, MPXV, or EMCV and incubated at 37°C in the presence of 5% CO2 for 72 h for MPXV and 48 h for the other viruses. Plaques were visualized by staining with 0.1% crystal violet in 20% ethanol solution.
Multicycle growth kinetics.
JC cells were seeded in 6-well plates so that they were 80% confluent at the time of infection. The cells were infected with VACV, VACV-E3LΔ37N, MPXV, or VACV-F3L at an MOI of 0.01. Infected cells were harvested at 3, 12, 24, 48, and 72 hpi. The titers of VACV, the VACV recombinants, and MPXV were determined as described above.
Real-time PCR.
JC cells were infected at an MOI of 5 with VACV, VACV-E3LΔ37N, and MPXV. Total RNA was extracted at 1, 2, 4, 6, 8, 10, and 12 hpi using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. cDNA was generated with 500 ng of total RNA diluted in 17.5 μl of H2O (RNase and DNase free), followed by addition of 1 μl oligo(dT) (500 μg/ml; Promega) and incubation at 70°C for 5 min. The samples were then chilled on ice and a reverse transcription mix (10 μl of 5× Moloney murine leukemia virus [M-MLV] real-time PCR buffer [Promega], 20 μl of 1 mM deoxynucleoside triphosphates, 1 μl of M-MLV reverse transcriptase [200 U/ml; Promega], 0.5 μl of RNasin Plus RNase inhibitor [40 U/μl; Promega]) was added. The samples were incubated at 37°C for 1 h and 95°C for 10 min and then placed on ice. Real-time PCR was performed with an MJ Mini thermocycler (Bio-Rad) under the following conditions: 95°C for 10 min and 35 cycles at 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Two hundred nanograms of cDNA was added to the real-time PCR mix (12.5 μl of 2× iQ SYBR green Supermix [Bio-Rad], 2.5 μl of 10 mM forward and reverse real-time primers, and 5.5 μl of double-distilled H2O for a total volume of 25 μl). VACV and MPXV M1L gene transcripts were detected with primer pairs M1L-F (5′-AAC GGA CCA CAT CCT TCT TC-3′) and M1L-R (5′-ATC CAA ACG CGT GTGATA AA-3′). VACV and MPXV G8R gene transcripts were detected with the primer pair G8R-F (5′-GCG GAT CTG TAA ACA TTT GG-3′) and G8R-R (5′-CCT TGGACA CAG GAA GAT TAA A-3′) and the MPXV-specific primer MPXV-G8R-R (5′-CCT TGG ACA CTG GAA GGT TAA A-3′), respectively. VACV A5L gene transcripts were detected with the primer pair A5L-F (5′-TTT CCA TCC GAT TGT TGT GT-3′ and A5L-R (5′-AGT TCA CTC CTT CCA GCG TT-3′), and MPXV A5L gene transcripts were detected with the MPXV-specific primer pair MPXV-A5L-F (5′-CTT CCA TCC GAT TGT TGTGT-3′) and MPXV-A5L-R (5′-AGT ACA CTC CTT CCA GCG TT-3′). For VACV and MPXV genome real-time PCR, 500 ng of DNA was added to the real-time PCR mix, and the same thermocycling conditions and G8R primer sets described above were used. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the internal loading control, and standardization among samples was done using the primer pair GAPDH-F (5′-CCT GTT CGA CAG TCA GCC G-3′) and (5′-GA CCA AAT CCG TTG ACT CC-3′).
PKR and eIF2α phosphorylation.
Subconfluent HeLa cell monolayers were infected with VACV, VACVΔE3L, VACV-E3LΔ37N, MPXV, or VACV-F3L at an MOI of 5 in the presence or absence of 1,000 IU/ml of IFN-α A/D interferon (PBL). At the indicated times postinfection, cell lysates were prepared in radioimmunoprecipitation assay (RIPA) buffer. Cells were scraped into 1 ml of 1× phosphate-buffered saline (PBS) and pelleted by centrifugation at 1,000 × g and 4°C for 10 min. The cell pellet was resuspended in 75 μl of RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 200 mM NaF, 2 mM Na3VO4) for 10 min, followed by centrifugation at 10,000 × g for 10 min. The supernatants were removed, and an equal volume of 2× SDS-PAGE loading buffer (125 mM Tris-HCl, 20% glycerol, 4% SDS, 0.001% bromphenol blue, 0.2% 2-mercaptoethanol) was added. The samples were boiled for 5 min and analyzed on 12% SDS-polyacrylamide gels. Proteins were transferred to a nitrocellulose membrane at 100 V for 60 min in 10 mM N-cyclohexyl-3-aminopropanesulfonic acid, pH 11, with 20% methanol. The membranes were blocked with TTBS (20 mM Tris-HCl, pH 7.8, 180 mM NaCl, 0.05% Tween 20) with 3% milk (Carnation nonfat dry milk) for 1 h. The membranes were probed with rabbit anti-phospho-PKR (1:1,000; Abcam), mouse anti-PKR (1:1,000; Santa Cruz), rabbit anti-phospho-eIF2α (1:1,000; Cell Signaling), and rabbit anti-eIF2α (1:1,000; Santa Cruz). Secondary goat anti-rabbit IgG and goat anti-mouse IgG conjugated to horseradish peroxidase (1:10,000; Santa Cruz) were added, followed by chemiluminescence. All lysates were also probed by Western blotting with goat anti-rabbit immunoglobulin GAPDH (Sigma) to confirm equal loading of the samples.
RESULTS
MPXV produces a truncated version of the VACV E3 protein.
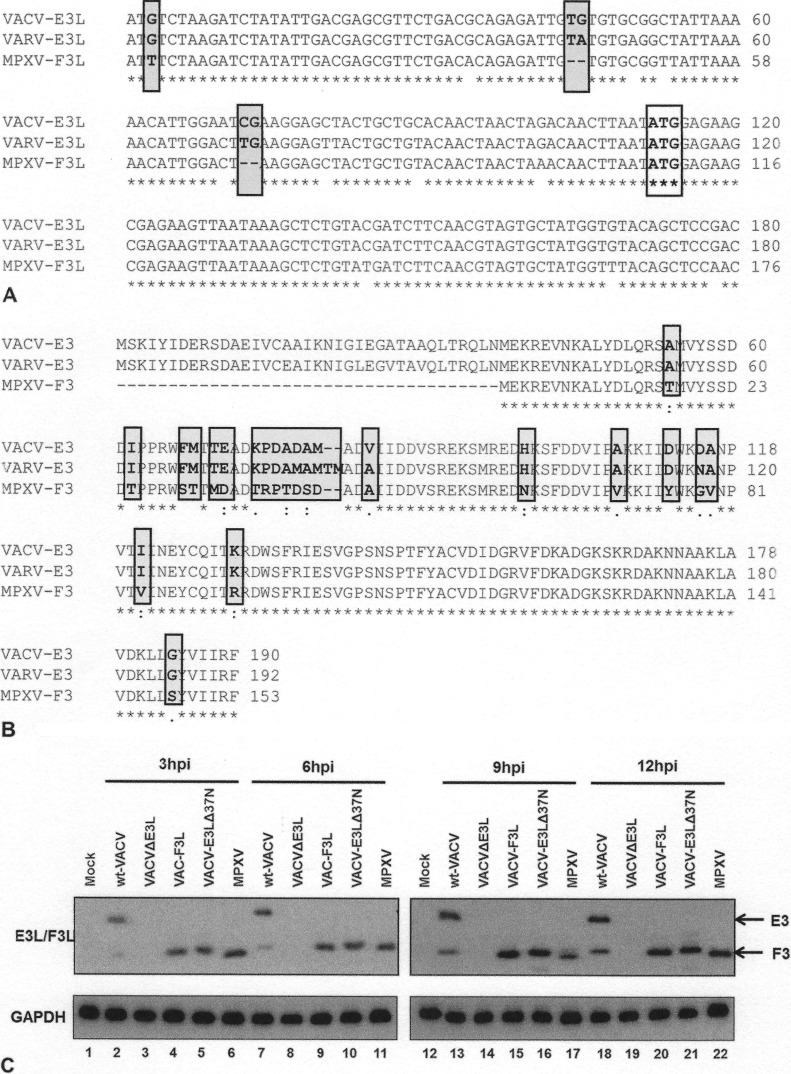
Both VACV and VARV contain an open reading frame (ORF) approximately 570 nucleotides long encoding E3-like proteins (Fig. 2A). The major product of this open reading frame in VACV is a protein that has been called p25 (8). However, the open reading frames in both viruses contain initiator AUG codons at position 38, which in the case of VACV has been shown to lead to the synthesis of small amounts of an N-terminally truncated protein, p20 (9). The ORF of the MPXV E3 homologue, encoded by the MPXV F3L gene, is shorter than the corresponding ORFs in VACV and VARV. The AUG codon that is presumed to lead to initiation of translation in the VACV and VARV E3L genes is instead AUU in MPXV, and there are two 2-bp deletions in the 5′ region of F3L that would lead to a frameshift, even if translation did initiate at the AUU sequence in MPXV (Fig. 2A). The second AUG codon in MPXV F3L is intact (Fig. 2A) and is predicted to encode a protein equivalent to the VACV p20 protein (Fig. 2B). To determine if the MPXV F3 protein is indeed truncated compared to the VACV E3 protein, HeLa cells were infected with wt VACV, VACV-E3LΔ37N, or MPXV (Fig. 1) at an MOI of 5 PFU/cell and protein lysates were harvested at 3, 6, 9, and 12 hpi. Western blot analysis with antiserum that recognizes the VACV E3 proteins showed that VACV produces both the p25 and p20 products of E3L, while VACV-E3LΔ37N generated only the p20 product of E3L (Fig. 2C). As predicted from the DNA sequence, MPXV produced a protein that is similar in size to the protein produced by VACV-E3LΔ37N, a virus engineered to produce a protein with a deletion of the first 37 amino acids. The MPXV E3-related protein ran with a slightly higher mobility than the VACV p20 protein (Fig. 2C, lanes 6, 11, 17, and 22). All viruses expressed E3-like proteins with similar kinetics, with protein being detected as early as 3 hpi (Fig. 2C). While the major E3L-encoded protein in VACV-infected cells is p25, with only smaller amounts of p20 accumulating, large amounts of p20 were made in either MPXV- or VAVC-E3LΔ37N-infected cells. This is consistent with the somewhat weak Kozak sequence of the first initiator AUG (18), while the AUG at codon 38 of the VACV E3L gene is predicted to have a strong Kozak sequence (18).
FIG 2.
(A) VACV-E3L homologue alignment. The first 160 nucleotides of the VACV E3L gene homologues of VACV, VARV, and MPXV were aligned by use of the ClustalW program. The sequences begin at nucleotide 1 of each gene, which is the start codon in the case of VACV and VARV. The C-terminal dsRNA BD is highly conserved among all three viruses. Nonhomologous nucleotide differences in MPXV that lead to the inability to generate p25 are shown in gray boxes. The conserved AUG codon that leads to the generation of p20 (the only product expressed by MPXV) is boxed only. Asterisks indicate the nucleotide positions of the consensus sequence among all three sequences. (B) Protein sequence alignment. The VACV E3 protein homologues of VACV, VARV, and MPXV were aligned by use of the ClustalW program. Asterisks, amino acid positions of the consensus sequence among all three sequences; amino acids highlighted in gray, changes in sequences among MPXV, VARV, and VACV; dots, degree of similarity, with one dot being less conservative than two. The N-terminal Z-NA BD for VACV and VARV is highly conserved, whereas the E3 homologue of MPXV (F3) is predicted to contain a 37-amino-acid N-terminal truncation. (C) MPXV produces a p20 form of VACV-E3. HeLa cells were infected with wt VACV, VACV-E3LΔ37N, VACV-F3L, and MPXV at an MOI of 5. At 3 (lanes 2 to 6), 6 (lanes 7 to 11), 9 (lanes 13 to 17), and 12 (lanes 18 to 22) hpi, protein lysates were isolated and analyzed by Western blotting with antibodies specific for the C terminus of E3.
MPXV possesses an IFN-resistant phenotype.
To determine if MPXV is able to inhibit the IFN antiviral immune response and replicate in the presence of IFN, cells were treated with increasing amounts of IFN-α A/D (0 to 1,000 U/ml) for 18 h. After IFN treatment, the cells were infected with 100 PFU of MPXV, and 48 h later the plaques were stained with crystal violet. Similar to VACV plaque formation, MPXV plaque formation was unaffected by IFN treatment (Fig. 3A), while EMCV plaque formation was decreased by IFN treatment (Fig. 3B). Similar results were seen in Vero-E6 cells and HeLa cells (data not shown).
FIG 3.
MPXV replication in the presence of IFN. (A) RK13 cells were treated with increasing amounts of IFN-α A/D for 18 h and then infected with 100 PFU of MPXV. Cells were stained at 48 hpi with crystal violet. (B) Subconfluent BSC-40 cells were treated with the indicated amount of recombinant IFN for 18 h prior to infection. Treated cells were infected with approximately 100 PFU of VACV, EMCV, or MPXV. Cells were stained with crystal violet at 48 hpi.
MPXV prevents phosphorylation of PKR and eIF2α.
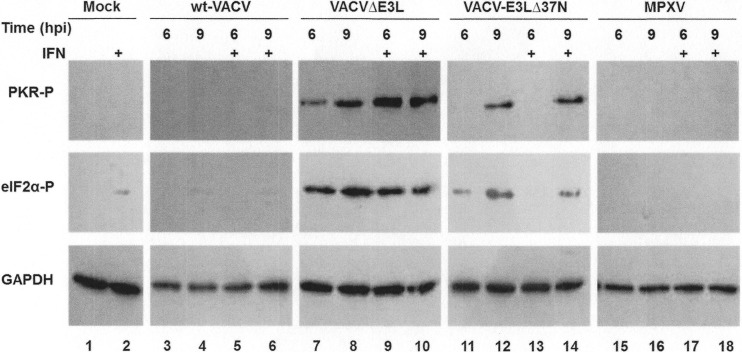
The VACV E3 protein inhibits the IFN antiviral immune response by preventing the phosphorylation of PKR and eIF2α (8). The entire N-terminal domain of the VACV E3 protein is required for full inhibition of PKR activation both in HeLa cells and in the mouse model (13). To determine if the lack of a full N-terminal domain encoded by the MPXV F3L gene also yields a protein that fails to fully inhibit the activation of PKR, HeLa cells were either mock infected or infected with wt VACV, VACV-E3LΔ37N, VACVΔE3L, or MPXV in the presence or absence of IFN treatment. Protein lysates were harvested at 6 and 9 hpi and analyzed by Western blotting with antiserum that recognizes the phosphorylated forms of PKR and eIF2α. Mock-infected and wt VACV-infected cells showed undetectable levels of phosphorylated PKR and eIF2α (Fig. 4, lanes 1 to 6). In contrast, VACVΔE3L-infected cells showed high levels of phosphorylated PKR and eIF2α at both 6 and 9 hpi (Fig. 4, lanes 7 to 10), irrespective of IFN treatment. VACV-E3LΔ37N induced the phosphorylation of PKR and eIF2α at 9 hpi (Fig. 4, lanes 12 and 14) but not 6 hpi (Fig. 4, lanes 11 and 12), corresponding to the late activation of the PKR pathway, as previously described (13). Most notably, MPXV did not cause the phosphorylation of PKR and eIF2α at either time point regardless of IFN treatment (Fig. 4, lanes 15 to 18). These data suggest that despite encoding an N-terminally truncated E3 homologue, MPXV can fully inhibit the activation of PKR more similarly to wt VACV than to VACV-E3LΔ37N.
FIG 4.
Inhibition of PKR pathway by MPXV. HeLa cells were either mock infected (lanes 1 and 2) or infected with wt VACV (lanes 3 to 6), VACVΔE3L (lanes 7 to 10), VACV-E3LΔ37N (lanes 11 to 14), or MPXV (lanes 15 to 18) in the presence (+) (lanes 2, 5, 6, 9, 10, 13, 14, 17, and 18) or absence (lanes 1, 3, 4, 7, 8, 11, 12, 15, and 16) of 1,000 U/ml of IFN at an MOI of 5. Protein lysates were isolated at 6 and 9 hpi and analyzed by Western blotting with antibodies specific to the phosphorylated forms of PKR (PKR-P) and eIF2α (eIF2α-P). Detection of GAPDH was used to ensure equal loading of proteins.
MPXV replicates in JC cells.
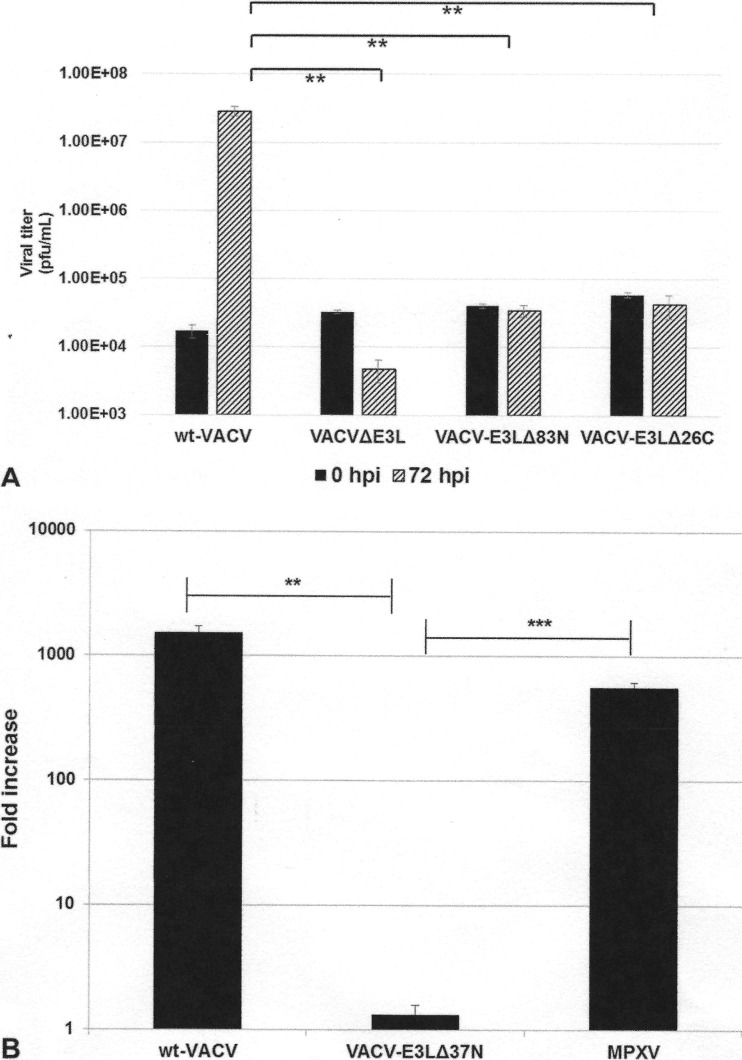
While the N terminus of VACV E3 is required for full inhibition of PKR both in cells in culture and in the mouse model (13), until recently the N terminus has not been shown to be necessary for replication in any cells in culture (our recent data [11] have shown that the N terminus is necessary for IFNr in mouse embryonic fibroblast [MEFs]). We have identified murine JC cells to be a model where a full N-terminal domain is required for the replication of VACV (Fig. 5). VACVs containing either N-terminal or C-terminal truncations in E3 (i.e., VACV-E3LΔ37N [Fig. 5B] or VACV-E3LΔ83N and VACV-E3LΔ26C [Fig. 5A]) replicate poorly in JC cells. Considering that MPXV contains a natural N-terminal truncation in F3L, we hypothesized that this virus would not be able to replicate in these cells. Therefore, we performed multistep growth kinetics assays in JC cells, comparing the replication of wt VACV, VACV-E3LΔ37N, and MPXV. At 72 hpi, wt VACV replicated to high titers, whereas VACV-E3LΔ37N replicated poorly in JC cells (Fig. 5B). Remarkably, despite expressing an N-terminally truncated F3 protein, MPXV was able to replicate to titers similar to those of wt VACV in these cells (Fig. 5B). This suggests either that the amino acid changes present in the MPXV F3 protein (see Fig. 2B) may compensate for the lack of a full-length N-terminal Z-NA binding domain (BD) or that the virus has evolved an alternative mechanism or mechanisms to counteract the host antiviral innate immune response.
FIG 5.
Replication in JC cells. (A) JC cells were infected with the indicated viruses at an MOI of 0.01 PFU/cell. Infected cells were harvested at 0 and 72 hpi, and the titer was determined by plaque assay in RK-E3L cells. Data are presented as means with standard errors from three experiments. **, P ≤ 0.01. (B) JC cells were either mock infected or infected with wt VACV, VACV E3LΔ37N, or MPXV at an MOI of 0.01 PFU/cell. Viruses were harvested at 72 hpi, and the titer was determined by plaque assay in BSC-40 cells. Data represent the fold increase in viral growth at 72 hpi and are presented as means with standard errors from multiple experiments. Statistical analyses were done by using an unpaired t test comparing wt VACV and MPXV to VACV-E3LΔ37N. **, P ≤ 0.01; ***, P ≤ 0.001.
Accumulation of viral early, intermediate, and late transcripts in VACV- and MPXV-infected cells.
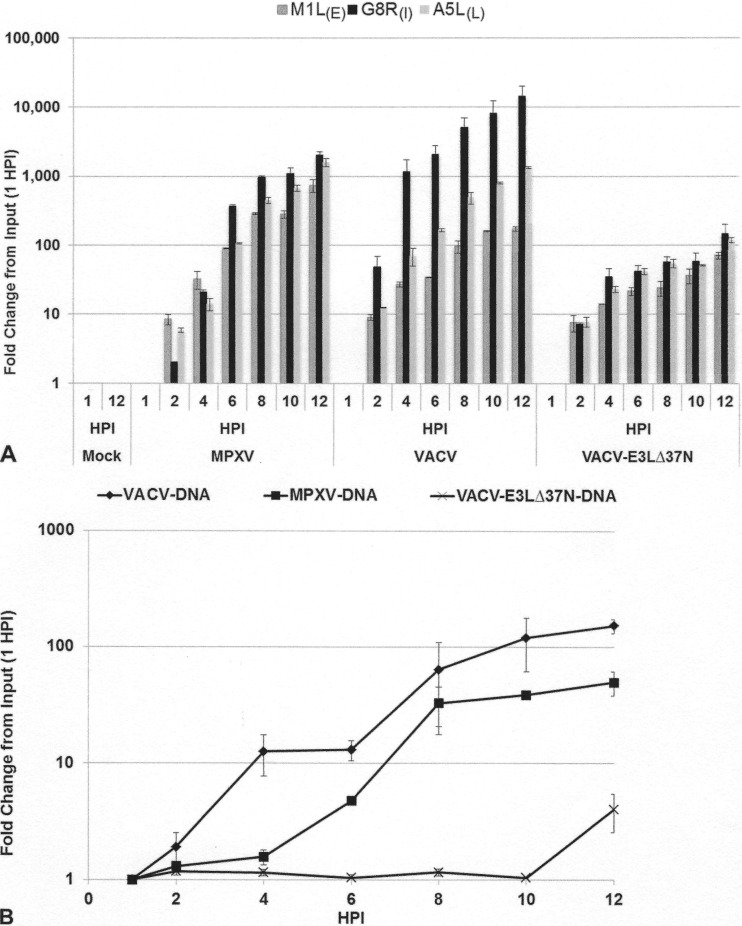
To determine the stage at which VACV with N-terminal mutations in E3 is blocked, we examined the production of early, intermediate, and late viral transcripts using real-time quantitative PCR with primers specific for early (M1L), intermediate (G8R), and late (A5L, WR strain) viral transcripts (Fig. 6). In VACV-infected JC cells at 2 hpi, viral transcripts were induced by 10-fold and continued to increase to more than 1,000-fold for the intermediate and late genes by 12 hpi (Fig. 5A and B). In the cells infected with VACV-E3LΔ37N, all three genes were induced at 2 hpi, but intermediate and late transcripts failed to be induced to levels comparable to those in VACV-infected cells at later times postinfection (Fig. 6A), correlating with the inability of VACV-E3LΔ37N to replicate to wild-type titers within these cells. MPXV early and late viral transcripts were induced to levels similar to those of VACV early and late viral transcripts at all time points; however, the amounts of intermediate viral transcripts were 10-fold higher in VACV-infected cells at late times postinfection (Fig. 6A). In addition, we performed quantitative PCR to measure viral genomic replication initiation using the G8R-specific primer set (Fig. 6B). Similar to the findings for viral G8R transcription initiation, VACV genomic replication initiated at about 2 hpi and continued to increase until 12 hpi (Fig. 6B, closed diamonds). MPXV DNA replication (Fig. 6B, closed squares) was delayed approximately 2 h but reached the same level as VACV DNA replication by 8 hpi. Very little DNA replication was detected in JC cells infected with VACVΔ37N, consistent with a lack of intermediate and late gene expression. These data suggest that VACV-E3LΔ37N is restricted in JC cells at the transition from early to intermediate gene expression.
FIG 6.
(A) Gene expression in JC cells. JC cells were either mock infected or infected with MPXV, VACV-E3LΔ37N, or VACV at an MOI of 5. RNA was extracted at 1, 2, 4, 6, 8, 10, and 12 hpi. Real-time quantitative PCR was performed with primers specific for M1L (early [E]), G8R (intermediate [I), and A5L (late [L]) viral gene transcripts. The transcript level of each gene was graphed as the fold change from the amount input (at 1 hpi). Data represent means with standard errors from multiple experiments. (B) Genomic replication in JC cells. Cells were infected with VACV, VACV-E3LΔ37N, or MPXV at an MOI of 5. Cells were harvested at 1, 2, 4, 6, 8, 10, and 12 hpi, and total DNA was extracted with phenol chloroform. Total DNA (500 ng) was used for real-time PCR with VACV G8R gene-specific primers. The graph presents the fold change in genomic DNA as the fold change from the amount input (at 1 hpi), along with transcript levels from Fig. 5A. Data represent means with standard errors from multiple experiments.
The MPXV F3 protein recues IFN resistance in RK13 and BSC-40 cells but does not rescue the VACV-E3LΔ37N phenotype in JC cells.
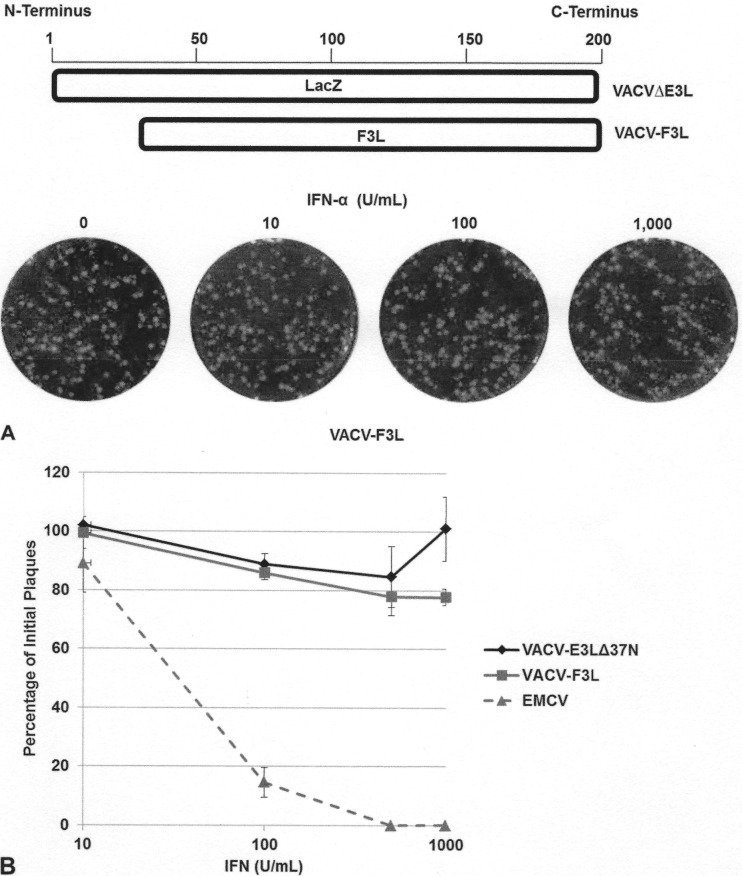
To determine if the amino acid sequence changes present in the MPXV F3 protein are compensating for the lack of a full-length N-terminal Z-NA BD (Fig. 2B), we generated a VACV that expresses the F3 protein of MPXV by inserting the F3L gene in the place of E3L (VACV-F3L; Fig. 1). This virus expressed a 20-kDa E3-like protein with kinetics of expression similar to those of the F3 protein from MPXV (Fig. 2C). We then asked if F3 could restore the IFNr phenotype of VACVΔE3L. A plaque reduction assay in the presence of increasing amounts of IFN in RK13 and BSC-40 cells showed that the F3 protein was able to restore the IFNr phenotype of a VACV in which the E3 protein was deleted (Fig. 7A and B). These data suggest that the MPXV F3 protein is sufficient for conferring the IFNr phenotype of MPXV.
FIG 7.
The MPXV F3 protein restores the IFNr phenotype. (A) Through in vitro recombination, the F3L gene of MPXV was inserted into the E3L locus of VACVΔE3L, generating a recombinant VACV that expresses the MPXV F3 protein (VACV-F3L). RK13 cells were treated with increasing amounts of IFN-α A/D for 18 h, prior to infection with 100 PFU of VACV-F3L. At 48 hpi, the cells were stained with crystal violet. (B) Subconfluent BSC-40 cells were treated with the indicated amount of recombinant IFN at 18 hpi. Treated cells were infected with approximately 100 PFU of VACV E3LΔ37N, VACV-F3L, or EMCV. Cells were stained with crystal violet at 48 hpi.
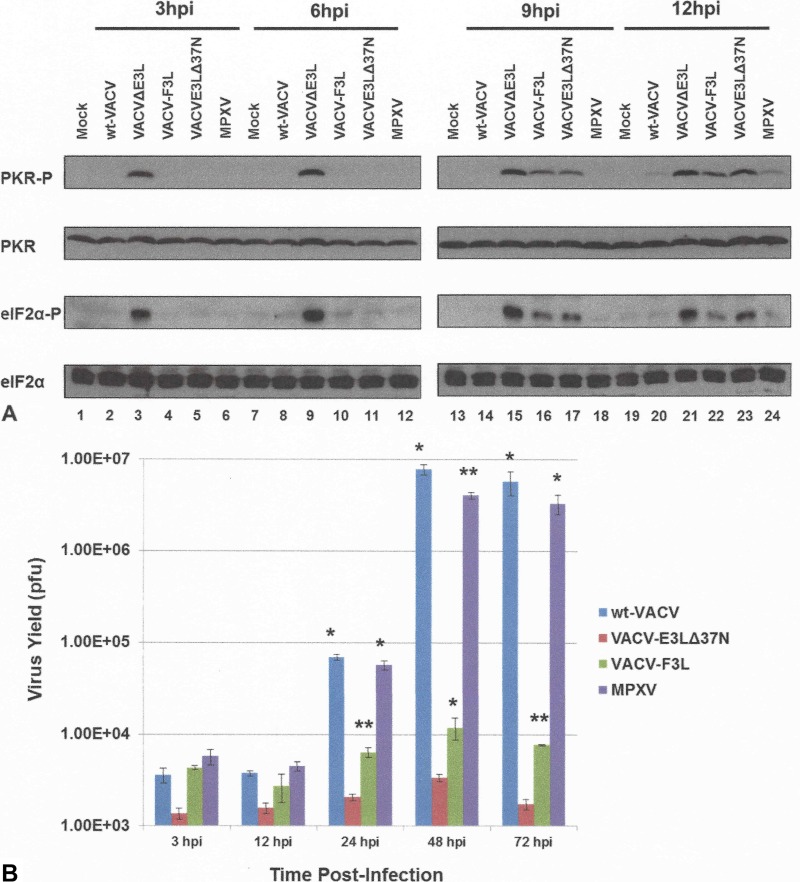
To determine if the F3 protein could restore a fully wild-type phenotype to VACVΔE3L, we examined PKR and eIF2α phosphorylation at late times postinfection in HeLa cells. HeLa cells were infected with wt VACV, VACV-E3LΔ37N, VACVΔE3L, or VACV-F3L at an MOI of 5 PFU/cell. At 3, 6, 9, and 12 hpi, protein lysates were obtained and analyzed by Western blotting for the phosphorylated form of PKR and eIF2α. As demonstrated above, no detectable levels of phosphorylated PKR or eIF2α were detected in wt VACV-infected cells, whereas cells infected with VACVΔE3L showed high levels of PKR and eIF2α phosphorylation from 3 to 12 hpi (Fig. 8A, lanes 3, 9, 15, and 21). Cells infected with VACV-E3LΔ37N showed PKR and eIF2α phosphorylation at 9 and 12 hpi (Fig. 8A, lanes 17 and 23), whereas cells infected with MPXV showed no PKR and eIF2α phosphorylation at 9 hpi and low levels of phosphorylation at 12 hpi (Fig. 8A, lanes 18 and 24). The cells infected with VACV-F3L also showed detectable levels of PKR and eIF2α phosphorylation at only 9 hpi and 12 hpi (Fig. 8A, lanes 16 and 22), suggesting that the amino acid changes present in the F3 protein were not able to fully inhibit the activation of PKR in a VACV background.
FIG 8.
(A) Late activation of the PKR pathway by VACV-F3L. HeLa cells were either mock infected (lanes 1, 7, 13, and 19) or infected with wt VACV (lanes 2, 8, 14, and 20), VACVΔE3L (lanes 3, 9, 15, and 21), VACV-F3L (lanes 4, 10, 16, and 22), VACV-E3LΔ37N (lanes 5, 11, 17, and 23), or MPXV (lanes 6, 12, 18, and 24) at an MOI of 5. Protein lysates were isolated at 3, 6, 9, and 12 hpi and analyzed by Western blotting with antibodies specific to the phosphorylated forms of PKR and eIF2α. (B) VACV-F3L replication is inhibited in JC cells. JC cells were infected with wt VACV, VACV E3LΔ37N, VACV-F3L, or MPXV at an MOI of 0.01 PFU/cell. Viruses were harvested at 3, 12, 24, 48, and 72 hpi, and the titer in BSC-40 cells was determined by plaque assay. Data represent means with standard errors from multiple experiments. Statistical significance was determined by comparison of the results for each group against those for VACV-E3LΔ37N using a multiple t test. *, P ≤ 0.05; **, P ≤ 0.01.
We then asked if the F3 protein of MPXV could restore VACVΔE3L replication in the JC cells. Multicycle growth kinetic assays were performed with VACV-F3L in JC cells. The titers of VACV-F3L were similar to those of VACV-E3LΔ37N, suggesting that VACV-F3L replicated poorly in these cells (Fig. 8B). Therefore, the amino acid changes present in the F3 protein could not compensate for the lack of a full-length N-terminal domain on F3. These data suggest that an alternative viral mechanism(s) is compensating for the lack of a full-length N-terminal Z-NA BD, allowing full inhibition of PKR activation in HeLa cells and replication in JC cells.
DISCUSSION
MPXV is capable of causing a severe smallpox-like disease in humans and, following the eradication of smallpox in 1977, has become the most problematic virus within the genus Orthopoxvirus in regard to human health (19). This is surprising given that MPXV is predicted to express a homologue of the VACV innate immune evasion protein, E3, which is missing 37 N-terminal amino acids (Fig. 2B). Similar truncations of E3 in VACV have been shown to prevent full inhibition of the host antiviral PKR pathway (13) and to attenuate virus-induced pathogenesis in mice (12, 16). Therefore, to begin to understand the disease associated with an MPXV infection, we have begun an investigation of how MPXV subverts the host's antiviral immune response. In this study, we have demonstrated that despite expressing a truncated homologue of E3, MPXV can fully inhibit activation of the PKR pathway and can replicate in cells in which a full-length E3 is required for replication of VACV. This suggests that MPXV contains a gene or genes that suppress the phenotypes normally associated with a lack of a full-length E3 in VACV. Suppression appears to be extragenic in nature, since VACV containing MPXV F3L in place of E3L was phenotypically indistinguishable from VACV-E3LΔ37N.
Suppression of the N-terminal truncation of the MPXV E3 homologue may be a factor in the pathogenesis of MPXV in humans and nonhuman primates. Similar truncations in VACV E3 lead to the activation of PKR in cells in culture (13), to the phosphorylation of eIF2α in infected mice (13), to IFN sensitivity in MEFs (11), and to a 100-fold reduction in virulence in mice (16). IFN sensitivity in MEFs is fully reversed by ablation of PKR, suggesting that in MEFs the sole function of the N terminus is to inhibit PKR activation (11). Thus, suppression of PKR activation during MPXV infection may be a factor in promoting pathogenesis in humans and nonhuman primates. On the other hand, MPXV is not pathogenic in most strains of inbred mice, unless IFN signaling is abrogated (20). Similarly, VACV in which the region coding for the N-terminal domain of E3 is deleted is as pathogenic as wt VACV in IFNAR−/− mice (11). Thus, the lack of a full-length E3 homologue may be one factor that limits the pathogenesis of MPXV in mice. Earl et al. (21) showed that in MPXV-infected (intranasal route) inbred strains of mice, such as BALB/c and C57/Blk, there is a large IFN-λ and CCL5 response in the lungs which is absent in MPXV-infected CAST/EiJ mice, a strain susceptible to MPXV. Further, they showed that inbred strains with mutated IFN-λ or IFN-λ receptor genes displayed an intermediate phenotype in MPXV susceptibility between wild-type strains and CAST/EiJ mice, suggesting that there were additional host factors impacting pathogenesis (21). Since the natural host of MPXV is thought to be a rodent (22), it is possible that MPXV has evolved several mechanisms to limit pathogenesis in the native host, including by increasing sensitivity to type I IFNs through partial truncation of the MPXV E3 homologue.
The N-terminal Z-NA binding domain of VACV E3 is not necessary for replication in most cells in culture (13). However, we have recently shown that the N terminus is necessary for IFN resistance in primary MEFs (11), and in this study, we have shown that the N terminus is necessary for the replication of VACV in murine JC cells. VACV-E3LΔ37N-infected JC cells accumulated early mRNA and early protein but demonstrated reduced levels of intermediate and late mRNAs and did not initiate DNA replication. The replication of E3L mutants of VACV in JC cells correlated with the predicted ability to bind to Z-NA (23). MPXV replicates efficiently in JC cells, despite lacking a full N-terminal Z-NA binding domain. Again, this appears to be due to suppressors at a second site, since VACV with the truncated F3L in place of E3L replicates as poorly in JC cells as VACV-E3LΔ37N does.
The nature of the suppressors in MPXV with a truncation of F3L is unclear. We have shown that MPXV makes less dsRNA than VACV (unpublished data). The synthesis of decreased amounts of dsRNA has been associated with resistance to the antipoxvirus drug isatin-beta-thiosemicarbazone (IBT) (24), and MPXV is in fact more resistant to IBT than VACV is (unpublished). Since the bulk of our data suggest that the N terminus of E3 functions to antagonize double-helical RNA (11), it is tempting to speculate that there is a cause-and-effect relationship between suppression of the lack of a full-length E3 homologue, decreased synthesis of dsRNA, and IBT resistance (IBTr). However, as of yet no IBTr mutations in a VACV-E3LΔ37N background have restored replication in JC cells (our unpublished observations). Mapping of the genes that allow replication in JC cells may shed light on the mechanism of suppression of the lack of a full-length E3L homologue in MPXV.
Of the chordopoxviruses that contain an E3L homologue, only MPXV and the leporipoxviruses myxoma virus (MYXV) and shope fibroma virus lack a full-length homologue (25–28). Like MPXV, MYXV pathogenesis is restricted to only certain species and has little or no capability for pathogenesis in all others (29, 30). For both MYXV and MPXV, restriction in mice requires signaling through the type I and type II IFN pathways (20, 30). This is similar to the restriction in pathogenesis that we have seen with VACV lacking an intact E3 N-terminal domain (11).
In conclusion, we have shown that MPXV contains a suppressor of the lack of a full-length N-terminal domain on its E3 homologue, which allows full inhibition of PKR and allows replication in JC indicator cells. Thus, MPXV appears to have evolved other, yet undiscovered mechanisms to limit activation of the host's antiviral immune response to compensate for the loss of the N terminus of F3.
ACKNOWLEDGMENTS
This work was supported in part by grant R01AI095394 from the National Institute of Allergy and Infectious Diseases.
We thank Connie Chamberlain and Nobuko Fukushima for excellent technical assistance.
REFERENCES
- 1.Marennikova SS, Seluhina EM, Mal'ceva NN, Cimiskjan KL, Macevic GR. 1972. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull World Health Organ 46:599–611. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, Li G, Liszewski MK, Atkinson JP, Jahrling PB, Feng Z, Schriewer J, Buck C, Wang C, Lefkowitz EJ, Esposito JJ, Harms T, Damon IK, Roper RL, Upton C, Buller RM. 2005. Virulence differences between monkeypox virus isolates from West Africa and the Congo Basin. Virology 340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huhn GD, Bauer AM, Yorita K, Graham MB, Sejvar J, Likos A, Damon IK, Reynolds MG, Kuehnert MJ. 2005. Clinical characteristics of human monkeypox, and risk factors for severe disease. Clin Infect Dis 41:1742–1751. doi: 10.1086/498115. [DOI] [PubMed] [Google Scholar]
- 4.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. 1988. Smallpox and its eradication, vol 6 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, Kazmierczak JJ, Stratman EJ, Li Y, Fairley JA, Swain GR, Olson VA, Sargent EK, Kehl SC, Frace MA, Kline R, Foldy SL, Davis JP, Damon IK. 2004. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med 350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- 6.Breman JG, Arita I. 1980. The confirmation and maintenance of smallpox eradication. N Engl J Med 303:1263–1273. doi: 10.1056/NEJM198011273032204. [DOI] [PubMed] [Google Scholar]
- 7.Fine PE, Jezek Z, Grab B, Dixon H. 1988. The transmission potential of monkeypox virus in human populations. Int J Epidemiol 17:643–650. doi: 10.1093/ije/17.3.643. [DOI] [PubMed] [Google Scholar]
- 8.Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A 89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. 1993. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology 195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 10.Watson JC, Chang HW, Jacobs BL. 1991. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- 11.White SD, Jacobs BL. 2012. The amino terminus of the vaccinia virus E3 protein is necessary to inhibit the interferon response. J Virol 86:5895–5904. doi: 10.1128/JVI.06889-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt TA, Jacobs BL. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J Virol 75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langland JO, Jacobs BL. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324:419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Shors ST, Beattie E, Paoletti E, Tartaglia J, Jacobs BL. 1998. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J Interferon Cytokine Res 18:721–729. doi: 10.1089/jir.1998.18.721. [DOI] [PubMed] [Google Scholar]
- 15.Chang HW, Uribe LH, Jacobs BL. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J Virol 69:6605–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandt T, Heck MC, Vijaysri S, Jentarra GM, Cameron JM, Jacobs BL. 2005. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology 333:263–270. doi: 10.1016/j.virol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Shors T, Kibler KV, Perkins KB, Seidler-Wulff R, Banaszak MP, Jacobs BL. 1997. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology 239:269–276. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. 1981. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res 9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breman JG. 2000. Monkeypox: an emerging infection for humans?, p 45–67. In Scheld WM, Craig WA, Hughes JM (ed), Emerging infections 4. ASM Press, Washington, DC. [Google Scholar]
- 20.Stabenow J, Buller RM, Schriewer J, West C, Sagartz JE, Parker S. 2010. A mouse model of lethal infection for evaluating prophylactics and therapeutics against monkeypox virus. J Virol 84:3909–3920. doi: 10.1128/JVI.02012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl PL, Americo JL, Moss B. 2012. Lethal monkeypox virus infection of CAST/EiJ mice is associated with a deficient gamma interferon response. J Virol 86:9105–9112. doi: 10.1128/JVI.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodakevich L, Jezek Z, Messinger D. 1988. Monkeypox virus: ecology and public health significance. Bull World Health Organ 66:747–752. [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A 100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennington TH. 1977. Isatin-beta-thiosemicarbazone causes premature cessation of vaccinia virus-induced late post-replicative polypeptide synthesis. J Gen Virol 35:567–571. doi: 10.1099/0022-1317-35-3-567. [DOI] [PubMed] [Google Scholar]
- 25.Bratke KA, McLysaght A, Rothenburg S. 2013. A survey of host range genes in poxvirus genomes. Infect Genet Evol 14:406–425. doi: 10.1016/j.meegid.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao JX, Macaulay C, Willer D, Evans D, McFadden G. 1999. The complete DNA sequence of myxoma virus. Virology 264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 27.Shchelkunov SN, Totmenin AV, Babkin IV, Safronov PF, Ryazankina OI, Petrov NA, Gutorov VV, Uvarova EA, Mikheev MV, Sisler JR, Esposito JJ, Jahrling PB, Moss B, Sandakhchiev LS. 2001. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett 509:66–70. doi: 10.1016/S0014-5793(01)03144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer DO, McFadden G, Evans DH. 1999. The complete genome sequence of shope (rabbit) fibroma virus. Virology 264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 29.Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. 2013. Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLoS Pathog 9:e1003465. doi: 10.1371/journal.ppat.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Barrett JW, Shao Q, Gao X, Dekaban GA, McFadden G. 2009. Myxoma virus selectively disrupts type I interferon signaling in primary human fibroblasts by blocking the activation of the Janus kinase Tyk2. Virology 387:136–146. doi: 10.1016/j.virol.2009.02.013. [DOI] [PubMed] [Google Scholar]