ABSTRACT
Astroviruses are small, nonenveloped viruses with a single-stranded positive-sense RNA genome causing acute gastroenteritis in children and immunocompromised patients. Since positive-sense RNA viruses have frequently been found to replicate in association with membranous structures, in this work we characterized the replication of the human astrovirus serotype 8 strain Yuc8 in Caco-2 cells, using density gradient centrifugation and free-flow zonal electrophoresis (FFZE) to fractionate cellular membranes. Structural and nonstructural viral proteins, positive- and negative-sense viral RNA, and infectious virus particles were found to be associated with a distinct population of membranes separated by FFZE. The cellular proteins associated with this membrane population in infected and mock-infected cells were identified by tandem mass spectrometry. The results indicated that membranes derived from multiple cell organelles were present in the population. Gene ontology and protein-protein interaction network analysis showed that groups of proteins with roles in fatty acid synthesis and ATP biosynthesis were highly enriched in the fractions of this population in infected cells. Based on this information, we investigated by RNA interference the role that some of the identified proteins might have in the replication cycle of the virus. Silencing of the expression of genes involved in cholesterol (DHCR7, CYP51A1) and fatty acid (FASN) synthesis, phosphatidylinositol (PI4KIIIβ) and inositol phosphate (ITPR3) metabolism, and RNA helicase activity (DDX23) significantly decreased the amounts of Yuc8 genomic and antigenomic RNA, synthesis of the structural protein VP90, and virus yield. These results strongly suggest that astrovirus RNA replication and particle assembly take place in association with modified membranes potentially derived from multiple cell organelles.
IMPORTANCE Astroviruses are common etiological agents of acute gastroenteritis in children and immunocompromised patients. More recently, they have been associated with neurological diseases in mammals, including humans, and are also responsible for different pathologies in birds. In this work, we provide evidence that astrovirus RNA replication and virus assembly occur in contact with cell membranes potentially derived from multiple cell organelles and show that membrane-associated cellular proteins involved in lipid metabolism are required for efficient viral replication. Our findings provide information to enhance our knowledge of astrovirus biology and provide information that might be useful for the development of therapeutic interventions to prevent virus replication.
INTRODUCTION
Astroviruses are common etiological agents of acute gastroenteritis in children and immunocompromised patients (1, 2). They have also been associated with neurological diseases in humans (3) and other mammals, like minks and cattle (4, 5). In birds, astroviruses cause different pathologies, manifesting as hepatitis in ducks (6) and poultry enteritis mortality syndrome (PEMS) in turkeys (7). There is no vaccine or specific antiviral therapy against astrovirus disease.
Astroviruses are small, nonenveloped viruses with a single-stranded positive-sense RNA [(+)RNA] genome (2). The virus genome has three open reading frames (ORFs), named ORF1a, ORF1b, and ORF2. ORF1a is translated into nsp1a, a nonstructural polyprotein precursor that is subsequently processed by viral and cellular proteases (2). ORF1b encodes nsp1b, the RNA-dependent RNA polymerase (2, 8). ORF2 encodes a precursor structural polyprotein, named VP90 in the human astrovirus (HAstV) serotype 8 (HAstV-8) strain Yuc8, which is proteolytically processed to produce the final capsid viral proteins VP34, VP27, and VP25 through a precursor polypeptide named VP70. ORF2 is expressed from a subgenomic RNA that is coterminal with the genomic RNA (RNAg) at the 3′ end (2, 9).
Viruses interact with a large number of cellular proteins during their replication cycle (10). (+)RNA viruses take advantage of host cells by subverting host protein synthesis and remodeling membranes, co-opting and modulating protein and ribonucleoprotein complexes, and altering cellular metabolic pathways during infection (11). They utilize intracellular membranes to assemble their RNA replication complexes, usually in conjunction with membrane vesicle formation or other membrane rearrangements to generate distinct organelle-like structures, designated viral replication factories (10, 12–14). The association with membranes provides advantages to virus replication: it protects replicating RNA from cellular nucleases; conceals replication-intermediate double-stranded RNA (dsRNA), thus preventing the induction of cellular innate immune responses; provides a scaffold that increases the local concentration of molecules involved in replication; and provides the proper topological orientation of replication complex components (12, 13).
Multiple approaches have been used to study the host factors involved in (+)RNA virus replication, including genome-wide screens (15, 16), riboproteomics (17), yeast two-hybrid screens (18, 19), and copurification of viral and cellular proteins to recognize host factors that are part of viral replication complexes or interact with viral replication proteins (11). Electron microscopy linked to electron tomography has become a powerful tool to determine structural changes in organelles and membrane rearrangements in infected cells (20–22).
Our group previously reported on the association of structural and nonstructural proteins, as well as viral RNA, with membranes derived from astrovirus-infected Caco-2 cells isolated using iodixanol density centrifugation (23), suggesting that astroviral RNA replication occurs in association with membranes. In this work, we further characterized the association of astrovirus with host cell membranes using free-flow zonal electrophoresis (FFZE), a versatile technique that allows the separation of membranes on the basis of the net surface charge during laminar flow through a thin aqueous layer (24, 25). Yuc8 structural and nonstructural proteins, as well as positive- and negative-sense viral RNA and infectious virus, were found to be associated with a distinct population of cell membranes derived from different organelles. A proteomic approach was used to identify the cellular proteins associated with this membrane population, and using RNA interference, the potential role which some of these proteins might have in the replication cycle of the virus in Caco-2 cells was investigated. Knocking down the expression of genes involved in cholesterol and fatty acid synthesis, phosphatidylinositol and inositol phosphate metabolism, and RNA helicase activity decreased the amount of Yuc8 genomic RNA and antigenomic RNA (RNAag) produced and the yield of the structural polyprotein precursor VP90 and of progeny virus, suggesting that astrovirus RNA replication and particle assembly take place in association with potentially different cellular membranes.
MATERIALS AND METHODS
Cells, virus, and reagents.
Cells of the colon adenocarcinoma Caco-2 cell line, obtained from the American Type Culture Collection, were propagated in a 10% CO2 atmosphere at 37°C with advanced Dulbecco modified essential medium (DMEM; Sigma Chemical Co., St. Louis, MO) supplemented with nonessential amino acids (Gibco, Life Technologies, Carlsbad, CA) and 10% fetal bovine serum (FBS; Cansera, Ontario, Canada). HAstV serotype 8 strain Yuc8 was isolated in our laboratory, and it was grown as described previously (26). Small interfering RNAs (siRNAs; smart pools) were purchased from GE Healthcare Dharmacon (Lafayette, CO). Horseradish peroxidase-conjugated goat anti-rabbit polyclonal and anti-mouse immunoglobulin antibodies were purchased from PerkinElmer Life Sciences (Boston, MA). Rabbit polyclonal antiserum to HAstV (Yuc8 strain) was previously described (27). Mouse and rabbit polyclonal antisera to the recombinant astrovirus Yuc8 proteins 1a-3 (amino acid residues 401 to 638 of nsp1a) and 1b-2 (amino acid residues 201 to 362 of nsp1b), which recognize the protease and the RNA polymerase motifs, respectively, were produced in our laboratory (28). Rabbit polyclonal antibodies to the recombinant protein E4 (amino acid residues 666 to 782 of VP90) has been described previously (27).
Infectivity assay.
For the infectivity assay, confluent Caco-2 cell monolayers were infected at a multiplicity of infection (MOI) of 5 with trypsin (Gibco, Life Technologies, Carlsbad, CA)-activated HAstV serotype 8 strain Yuc8 (26) and harvested at 18 h postinfection (hpi), unless otherwise indicated. The infected cells were detected by an immunoperoxidase focus-forming unit assay as described previously (27). Briefly, confluent monolayers of Yuc8-infected Caco-2 cells were fixed with ice-cold 100% methanol for 15 min at room temperature, and intracellular viral antigen was detected with a rabbit hyperimmune serum to Yuc8 virus, followed by incubation with a peroxidase-conjugated goat anti-rabbit immunoglobulin polyclonal antibody. The binding of the secondary antibody was revealed with the substrate 3-amino-9-ethyl-carbazole (Sigma Chemical Co., St. Louis, MO), as previously described (29).
Cell fractionation and FFZE membrane separation.
Membrane protein enrichment using iodixanol gradients was carried out as described previously (23). After sample ultracentrifugation, three visible bands above the 30% iodixanol interface were collected, pooled, and stored at −80°C; this procedure was repeated until a protein concentration of 30 mg/ml was reached. The proteins in the collected samples were analyzed by Western blotting with anti-Yuc8 antibody to confirm the presence of viral proteins. The pooled cellular membranes separated by the density gradient were fractionated by FFZE using a BD free-flow electrophoresis system (BD Proteomics, Germany) as previously described (24). Following separation in the chamber, membrane fractions were continually collected in 96-well deep-well microtiter plates at 4 ml/well (Sunergia Medical, Herndon, VA). The separation of membranes by FFZE was monitored along the run by collecting aliquots in microtiter plates (250 μl/well) at several time points and measuring the protein concentration (by determination of the optical density at 280 nm) using a microplate scanning spectrophotometer (Power WaveX; Bio-Tek Instruments, VT). The viral proteins in fractions separated by FFZE were detected by enzyme-linked immunosorbent assay (ELISA) (see below). Pools of selected consecutive fractions (every two) were concentrated for Western blot analysis by centrifugation at 100,000 × g for 50 min at 4°C. The membrane pellets were resuspended in 150 μl of TN buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) and stored at −80°C until analyzed.
ELISA.
The presence of viral proteins in the 96 fractions separated by FFZE was evaluated by ELISA. Fractions were bound to wells in a 96-well plate overnight at 4°C. The plate was then washed three times with phosphate-buffered saline (PBS)–1% bovine serum albumin (BSA) and blocked with 50 μl of this buffer for 1 h at 4°C. Then, 50 μl of primary antibody (either rabbit polyclonal antibodies to Yuc8 virus, antibodies to the recombinant protein E4, or antibodies to the recombinant protein 1a-3) was added and the mixture was incubated overnight at 4°C. The plates were then washed three times with PBS–1% BSA and incubated for 1 h at 37°C with 50 μl of alkaline phosphatase-labeled anti-rabbit immunoglobulin G conjugate. The plates were subsequently washed three times with PBS–1% BSA, the phosphatase substrate (1 mg/ml of p-nitrophenylphosphate [Sigma] in 1 mM MgCl2, 1% diethanolamine buffer, pH 9.8) was added, and the plates were incubated at room temperature until the absorbance at 408 nm of the positive control, consisting of a lysate of Yuc8-infected Caco-2 cells, reached a value of 1.0.
Immunoblotting.
The detection of proteins by Western blotting was carried out as described previously (23), using the primary antibodies indicated below.
LC-MS/MS.
Combined FFZE fractions 17 and 18, 44 and 45, 46 and 47, 48 and 49, 52 and 53, and 78 and 79 were selected on the basis of the presence or absence of viral RNA. Fractions were precipitated with trichloroacetic acid and acetone. Briefly, 1.2 ml (600 μl of each of the two fractions) was mixed with 240 μl of Tris-EDTA (10 mM Tris HCl, 1 mM EDTA, pH 8.0), 100 μl 0.3% sodium deoxycholate, and 100 μl of 72% trichloroacetic acid. After incubation for 2 h on ice, the samples were centrifuged at 1,500 × g for 20 min at 4°C. The supernatant was then aspirated, and the pellet was resuspended in 90% acetone at room temperature, followed by incubation at −20°C overnight. Following precipitation, the samples were centrifuged as described above, the supernatant was aspirated, and the pellets were dried for 20 min in a Savant integrated SpeedVac system (Thermo Fisher Scientific, Waltham, MA, USA). The samples were sent to the Proteomics Facility at the Institut de Recherches Cliniques (Montreal, Canada). The samples were prepared, digested, and analyzed by nano-liquid chromatography (nano-LC)-tandem mass spectrometry (MS/MS) as previously described (30).
Database search.
Tandem mass spectra were extracted by use of the Mascot Daemon application (version 2.2.2). All MS/MS spectra were analyzed using the Mascot server (Matrix Science, London, United Kingdom) and X! Tandem software (The GPM, version 2007.01.01.1 [thegpm.org]). Mascot was set up to search the ipi_HUMAN_v3_87 database (91,464 entries), assuming that digestion with trypsin was successful. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 15 ppm. The iodoacetamide derivative of cysteine was specified as a fixed modification. The oxidation of methionine was specified as a variable modification (30).
Criteria for protein identification.
Scaffold software (version Scaffold_3.1.2; Proteome Software Inc., Portland, OR) was used to validate the MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they exceeded the specific database search engine thresholds, calculated as −10 log(P), where P is the probability that the observed match between the experimental data and the database sequence is a random event. Mascot identification requires that ion scores be at least greater than the associated identity scores and 20, 15, and 15 for peptides with double, triple, and quadruple charges, respectively. X! Tandem identification requires at least −log(expected) scores of greater than 2.0. Peptide identifications were accepted if they could be established as specified by the Peptide Prophet algorithm at greater than 95% probability (31). Protein identifications were accepted if they could be established at greater than 99% probability and contained at least two unique peptides (32). Proteins that contained similar peptides and could not be differentiated on the basis of MS/MS analysis alone were grouped to satisfy the principles of parsimony; i.e., when assembling peptides into proteins and protein groups, Scaffold software describes the minimum set of protein sequences that adequately accounts for all observed peptides.
Construction of protein-protein interaction networks.
To map the identified proteins into the protein-protein interaction networks, the protein interaction data were gathered from the String database (version 9.1 [http://string.embl.de/]) (33), which includes experimental evidence of protein interactions, protein interaction databases, and text mining co-occurrences. The protein-protein interaction network was visualized using the Cytoscape program (version 3-0-2 [http://cytoscape.org]) (34), and statistical enrichment for specific Gene Ontology (GO) terms was performed using the ClueGO plug-in (version 2.1.3; Laboratory of Integrative Cancer Immunology) (35).
siRNA transfection.
All siRNAs were from GE Healthcare Dharmacon (Lafayette, CO) (siGENOME, SMARTpool siRNA reagents). Caco-2 cells were transfected with siRNAs using an Amaxa electroporation system (Cologne, Germany) following the manufacturer's guidelines. One and a half million cells and 1.5 μg of siRNA were used per electroporation. Seventy-two hours later, the transfection mixture was removed and the cells were washed twice with minimum essential medium (MEM) and infected with astrovirus at an MOI of 5. At 18 hpi the cells were harvested in Laemmli sample buffer for Western blotting or in the TRIzol reagent (Invitrogen, Life Technologies, Carlsbad, CA) for RNA extraction.
LDH cell viability assay.
Cell viability was determined with a commercial kit (Sigma-Aldrich Co., St. Louis, MO). At 72 hpi, the cell culture medium from the different experimental conditions was collected and stored at 4°C. The cell monolayer was then lysed according to the manufacturer's protocol. The supernatant and 1/10 of the cell fraction were independently mixed with the assay mixture and kept at room temperature for 30 min. The lactate dehydrogenase (LDH) activity was determined on the basis of the optical density of the samples, using the total LDH activity (from the medium and cells) as a control, and the data were expressed as a percentage of the total activity.
Viral RNA analysis.
Total extracted cellular RNA was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen, Life Technologies, Carlsbad, CA) using oligonucleotides Mon344(−) or Mon343(+) (see Table S1 in the supplemental material) (23, 36), yielding an amplicon of 316 bp.
qRT-PCR analysis.
Purified total RNA extracted from cell lysates was treated with RNase-free DNase (Roche, Basel, Switzerland) to eliminate possible DNA contamination. Quantitative reverse transcription-PCR (qRT-PCR) was performed with a first-strand cDNA synthesis kit and Maxima SYBR green quantitative PCR master mix (Thermo Fisher Scientific, Waltham, MA, USA) according to the standard protocol described in the instructions accompanying the kit. Quantitative analysis of the data was performed using the Prism 7000 analysis software program (Applied Biosystems, Life Technologies, Carlsbad, CA). The primers used to amplify ARF6, ARL1, DHCR7, CYP51A1, FASN, PI4KIIIα, PI4KIIIβ, CDS2, ITPR3, DDX23, EIF6, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cellular mRNAs and ORF1b astrovirus Yuc8 RNA are listed in Table S1 in the supplemental material. The results were normalized to the level of total GAPDH mRNA detected in each RNA sample. The relative fold changes in gene expression were calculated by the 2ΔCT method, where CT is the threshold cycle (37). The results presented are from three independent experiments, and statistical significance was determined with an unpaired two-tailed Student's t test.
RESULTS
FFZE separates membrane-associated proteins from astrovirus-infected Caco-2 cells in three different populations.
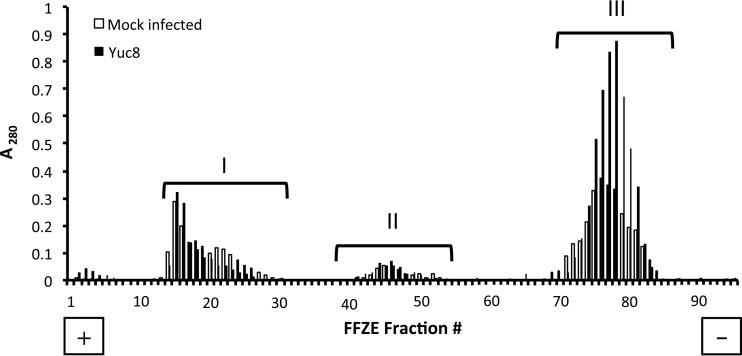
In a previous report, our group described the presence of astrovirus Yuc8 RNA polymerase in low-density fractions of iodixanol gradients of virus-infected Caco-2 cell homogenates, suggesting its association with cellular membranes (23). To further characterize the interaction of infectious astrovirus and astrovirus components with cell membranes, we separated the pooled cellular membranes of Yuc8-infected cells obtained from iodixanol gradients by free-flow zonal electrophoresis (FFZE). Three distinct protein populations (populations I to III) were observed in fractions collected from both infected and mock-infected Caco-2 cells (Fig. 1). Quantification of the proteins present in these populations showed differences in migration and concentration in infected cells from those in noninfected cells, suggesting that a difference in the distribution of cellular membranes between these two experimental conditions may exist. These differences were most obvious in population III (Fig. 1), although this population did not contain viral components (see below).
FIG 1.
Separation of Caco-2 cell membranes by free-flow zonal electrophoresis. The three membrane-containing, low-density bands observed in iodixanol gradients of Yuc8-infected and uninfected lysed Caco-2 cells were pooled and separated by FFZE as described in Materials and Methods. The optical density at 280 nm of each fraction collected from the FFZE fraction collector was measured as an indicator of the protein content. Closed bars, samples from infected cells; open bars, samples from mock-infected cells; I, II, and III, the three different membrane populations. The positions of the cathode (−) and the anode (+) are indicated. The traces shown are representative of those from three biological replicates.
Infectious virus and viral components are present exclusively in FFZE membrane population II.
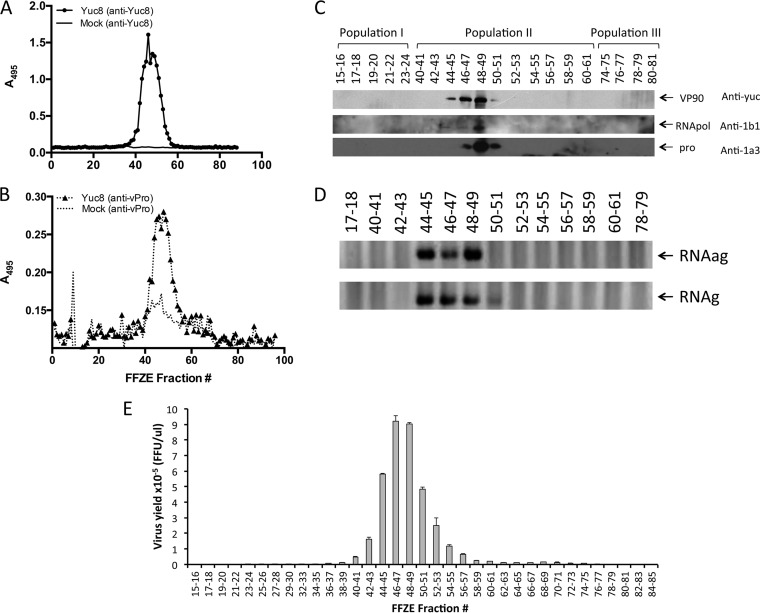
To determine the distribution of viral components among the three membrane populations separated by FFZE, the presence of astrovirus proteins, RNA, and infectious viral particles was determined. The distribution of the structural protein VP90 and the nonstructural protein nsp1a was detected by ELISA with antibodies to purified viral particles (anti-Yuc8) and to the viral protease motif (anti-1a3), respectively. The viral proteins were found only in membrane population II, specifically, between fractions 41 and 54 (Fig. 2A and B). The distribution of viral proteins was also analyzed by immunoblotting using antibodies to VP90 (anti-Yuc8), the viral RNA polymerase (anti-1b1), and the viral protease (anti-1a3). The peak abundance of VP90 was detected in fractions 46 to 49, while the RNA polymerase and protease were most abundantly found in fractions 48 and 49 (Fig. 2C). These results confirm the specific presence of viral proteins detected by ELISA in membrane population II, mainly in fractions 46 to 49.
FIG 2.
Detection of viral RNA, viral proteins, and infectious viral particles in fractions separated by FFZE. Samples obtained by FFZE were analyzed by ELISA for the presence of the structural protein VP90 (anti-Yuc8) (A) and the viral protease (anti-vPro) (B) using anti-Yuc8 and anti-1a3 antibodies, respectively. (C) The FFZE fractions were pooled (2 fractions per pool, as indicated) and analyzed by Western blotting to detect VP90, the RNA polymerase (RNApol), and the viral protease (pro), using anti-Yuc8, anti-1b1, and anti-1a3 antibodies, respectively. (D) RNA was extracted from pooled FFZE fractions, and reverse transcription was carried out with oligonucleotide Mon343(+) (top) to detect the negative-sense RNA or with oligonucleotide Mon344(−) (bottom) to detect the positive-sense RNA; PCR was carried out with the same pair of oligonucleotides. (E) The pooled fractions were treated with trypsin (200 μg/ml) for 1 h at 37°C. The titer of infectious virus was determined by an immunoperoxidase focus-forming assay as described in Materials and Methods. The data shown are representative of those from three biological replicates.
The distribution of negative-sense viral RNA strands (antigenomic RNA [RNAag]) and positive-sense viral RNA strands (genomic RNA [RNAg]) in the combined FFZE fractions of membrane population II, as well as in representative fractions of membrane populations I and III, was also determined by semiquantitative RT-PCR. Reverse transcription was performed with either oligonucleotide Mon343(+) or Mon344(−) (see Materials and Methods) to synthesize cDNA from negative- or positive-strand RNA, respectively. After inactivation of the reverse transcriptase, the synthesized cDNA was amplified by PCR using both the Mon343(+) and Mon344(−) primers in each case (see Materials and Methods). As shown in Fig. 2D, both genomic and antigenomic RNAs were detected in fractions 44 to 49, supporting the idea that astrovirus genome replication occurs in association with membranes (23). Finally, the potential association of infectious particles with FFZE membrane fractions was also evaluated. Infectious virus was mainly detected in fractions 42 to 55, with the highest viral titer being found between fractions 46 and 49 of membrane population II, although negligible amounts of virus (<0.1% of the total infectivity) were also detected in some fractions in population III (Fig. 2E). Altogether, these results show that RNA replication and the assembly of new viral particles occur in association with membrane population II, particularly in fractions 46 to 49, where the relative amount of viral components is enriched. Fractions 44 and 45 are also of interest since they contain high concentrations of RNA, some viral protein, but only a small fraction of the overall viral infectivity.
Proteomic analysis of FFZE membrane population II showed the presence of unique proteins involved in different biological processes.
To identify cellular proteins that might be involved in astrovirus replication, the proteins present in pooled FFZE fractions 44 and 45, 46 and 47, 48 and 49, and 52 and 53 of Yuc8-infected and mock-infected cells were analyzed by shotgun LC-MS/MS. In addition, fractions 17 and 18 (membrane population I) and 78 and 79 (membrane population III) were also analyzed for comparison of their proteins to the proteins associated with the fractions in population II. The resulting fragment ion spectra were searched against those in databases of human proteins. The proteins identified in each of the pooled fractions analyzed are shown in Tables S2 and S3 in the supplemental material for astrovirus-infected and mock-infected cells, respectively. A total of 615 and 660 proteins were identified in membrane population II of infected and mock-infected cells, respectively, with an overlap of 528 proteins. Seventy-nine of these proteins were identified only in infected cells, and 127 were exclusively found in mock-infected cells (see Table S4 in the supplemental material). Our analysis next focused on pooled fractions 48 and 49, where the largest amounts of infectious virus and viral components were found. Out of the 79 proteins identified solely in membrane population II of infected cells, 11 were present uniquely in fractions 48 and 49 (Table 1; see also Table S4 in the supplemental material). On the other hand, when the fragment ion spectra were searched against the spectra in a viral sequence database, the astrovirus capsid protein and the nonstructural polyproteins nsp1a and nsp1b were identified in fractions 44 to 53 of infected cells (confirming the data presented in Fig. 2) but not in fractions 17 and 18 or 78 and 79 of infected cells or in any fraction from uninfected cells analyzed (not shown), confirming that viral proteins are present exclusively in membrane population II.
TABLE 1.
Proteins found exclusively in fractions 48 and 49 of membrane population II from Yuc8-infected Caco-2 cells
| Protein full name | UniProt accession no. |
|---|---|
| Eukaryotic translation initiation factor 6 | P56537 |
| ADP-ribosylation factor 6 | Q5U025 |
| Claudin-6 | P56747 |
| ADP-ribosylation factor-like protein 1 | P40616 |
| Sodium-dependent multivitamin transporter | B4DDE5 |
| Pyrroline-5-carboxylate reductase 1, mitochondrial isoform 2 | P32322 |
| Isoform 1 of ubiquitin-conjugating enzyme E2 variant 1 | Q13404 |
| Isoform 1 of serine/threonine-protein phosphatase 2A, 65-kDa regulatory subunit A beta isoform | P30154 |
| Isoform 1 of phosphatidate cytidylyltransferase 2 | B4E3K6 |
| Inositol 1,4,5-trisphosphate receptor type 3 | Q59ES2 |
| Probable ATP-dependent RNA helicase DDX23 | A8KA56 |
The biological processes and molecular functions of proteins identified in fractions 48 and 49 of infected and uninfected cells were determined and mapped in a protein-protein interaction network using the String database and visualized using the Cytoscape program. Most of the functional characteristics of these proteins, as expected, were shared between infected and uninfected cells, suggesting that astrovirus infection does not perturb, at least significantly, cell metabolism. There were, however, different processes and functions overrepresented in infected cells, including ATP biosynthesis and fatty acid synthesis, processes that have been shown to be relevant for efficient infection of positive-sense viruses (Table 2) (38).
TABLE 2.
Gene Ontology annotation of proteins identified in FFZE fractions 48 and 49 of Yuc8-infected cellsa
| Process or function | Cells for which GO annotation was determined |
|---|---|
| Biological processes | |
| Generation of energy | Mock-infected cells |
| Glutamine metabolic process | Mock-infected cells |
| Hexose metabolic process | Mock-infected cells |
| Regulation of translation | Mock-infected cells |
| Respiratory electron transport chain | Mock-infected cells |
| Cell junction organization | Yuc8-infected cells |
| Cholesterol biosynthetic process | Yuc8-infected cells |
| De novo posttranslational folding | Yuc8-infected cells |
| Mitochondrial ATP synthesis-coupled proton transport | Yuc8-infected cells |
| mRNA metabolic process | Yuc8-infected cells |
| Protein localization in mitochondria | Yuc8-infected cells |
| SRP-dependent cotranslational protein targeting the membrane | Yuc8-infected cells |
| RNA processing | Yuc8-infected cells |
| Vesicle targeting to and from within Golgi apparatus | Yuc8-infected cells |
| Actin filament organization | Both mock-infected and Yuc8-infected cells |
| ATP biosynthetic process | Both mock-infected and Yuc8-infected cells |
| Glucose catabolic process | Both mock-infected and Yuc8-infected cells |
| Protein N-linked glycosylation | Both mock-infected and Yuc8-infected cells |
| Regulation of translation initiation | Both mock-infected and Yuc8-infected cells |
| Molecular functions | |
| Cytochrome c oxidase activity | Mock-infected cells |
| Nucleic acid binding | Mock-infected cells |
| Protein phosphatase binding | Mock-infected cells |
| rRNA binding | Mock-infected cells |
| Actin filament binding | Both mock-infected and Yuc8-infected cells |
| mRNA binding | Both mock-infected and Yuc8-infected cells |
| RNA binding | Both mock-infected and Yuc8-infected cells |
| Translation factor activity | Both mock-infected and Yuc8-infected cells |
| Unfolded protein binding | Both mock-infected and Yuc8-infected cells |
| Acetyl coenzyme A acetyltransferase activity | Yuc8-infected cells |
| ATPase activity to transmembrane movement of ions | Yuc8-infected cells |
| Hydrogen transporter activity | Yuc8-infected cells |
GO annotations were determined using the ClueGO plug-in in the protein-protein interaction network visualized by Cytoscape. SRP, signal recognition particle.
Membranes from different origins cosegregate with astroviral components in infected Caco-2 cells.
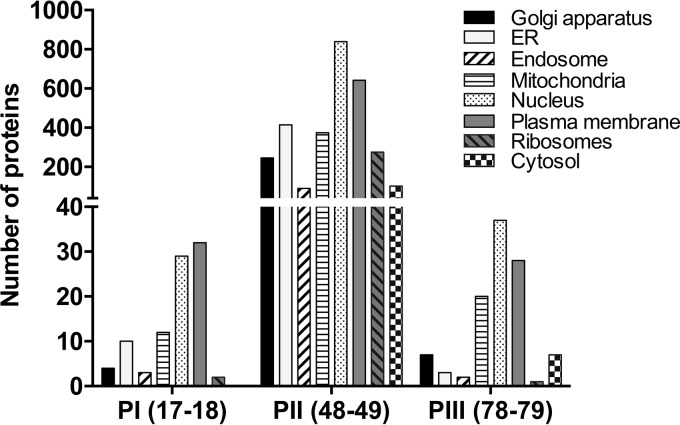
The results of analysis of the cellular compartment GO annotations for the proteins identified in Yuc8-infected membrane populations I (fractions 17 and 18), II (fractions 48 and 49), and III (fractions 78 and 79) are shown in Fig. 3. Analysis of the equivalent fractions from uninfected cells showed a distribution of the origin of cellular membranes very similar to that found in infected cells (not shown). Proteins associated with membranes of different origins were detected in all three populations. Of interest was the diversity of proteins detected in fractions 48 and 49 of population II. We were able to identify proteins from multiple membrane compartments, including the Golgi apparatus, endoplasmic reticulum (ER), mitochondria, and nucleus, as well as from the plasma membrane, which were enriched in fractions 48 and 49; however, the possibility of a specific association of viral components with a specific type(s) of membrane(s) cannot be concluded from these data, and further experiments with a better resolution of cell organelles would be required to make the assignment.
FIG 3.
Gene Ontology annotation for the cell compartment of the proteins from FFZE fractions. The classification of proteins from fractions 17 and 18 (population I [PI]), 48 and 49 (population II [PII]), and 78 and 79 (population III [PIII]) is indicated. The analysis was performed using Scaffold software (version 4) as described in Materials and Methods.
Lipid and phosphoinositol metabolism and a cellular RNA helicase activity are important for astrovirus replication.
Out of the 11 proteins exclusively identified in fractions 48 and 49 of infected cells (Table 3), 6 were selected to further characterize their possible participation in astrovirus replication. Five more proteins present in fractions 48 and 49 of infected cells, although they were not exclusive to these fractions, were also characterized. The selection was based on their involvement in the biosynthetic processes identified to be overrepresented in these FFZE fractions and on the role that similar proteins have been reported to play in the replication of other (+)RNA viruses (39–41). The selected proteins (Table 3) are involved in vesicular traffic (ADP-ribosylation factor 6 [ARF6], ADP-ribosylation factor-like protein 1 [ARL1]), cholesterol synthesis (lanosterol 14-alpha demethylase isoform 1 [CYP51A1], 7-dehydrocholesterol reductase [DHCR7]), fatty acid synthesis (fatty acid synthase [FASN]), phospholipid metabolism (phosphatidylinositol-4-kinase α and β [PI4KIIIα and PI4KIIIβ, respectively], isoform 1 of phosphatidate cytidylyltransferase 2 [CDS2]), phosphoinositol binding (inositol 1,4,5-trisphosphate receptor type 3 [ITPR3]), protein translation (eukaryotic translation initiation factor 6 [EIF6]), and RNA metabolism (ATP-dependent RNA helicase DDX23 [DDX23]).
TABLE 3.
Proteins in Yuc8-infected cells whose expression was knocked down by RNA interference
| Protein full name | Abbreviation | Biological process (GO accession no.) |
|---|---|---|
| ADP-ribosylation factor 6 | ARF6 | Vesicle mediated transport (GO:0016192) |
| ADP-ribosylation factor-like protein 1 | ARL1 | (Golgi vesicle transport (GO:0048193) |
| Lanosterol 14-alpha demethylase isoform 1 | CYP51A1 | Cholesterol biosynthetic process (GO:0006695) |
| Isoform 1 of phosphatidate cytidylyltransferase 2 | CDS2 | Phospholipid metabolic process (GO:0006644) |
| Probable ATP-dependent RNA helicase DDX23 | DDX23 | Helicase activity (GO:0004386) |
| 7-Dehydrocholesterol reductase | DHCR7 | Cholesterol biosynthetic process (GO:0006695) |
| Eukaryotic translation initiation factor 6 | EIF6 | Translation (GO:0006412) |
| Fatty acid synthase | FASN | Fatty acid biosynthetic process (GO:0006633) |
| Inositol 1,4,5-trisphosphate receptor type 3 | ITPR3 | Phosphatidylinositol binding (GO:0035091) |
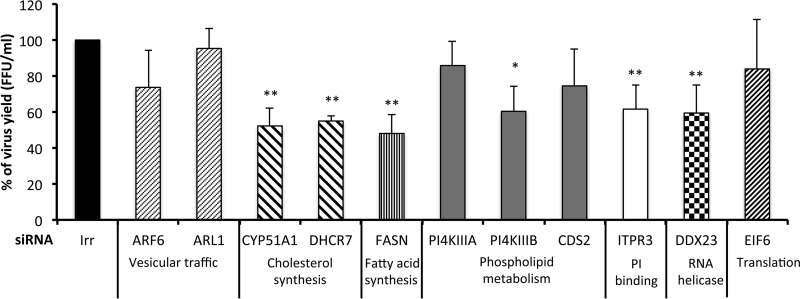
The roles of the above-mentioned proteins in astrovirus replication were evaluated by knocking down the expression of the cognate genes by RNA interference. For this, Caco-2 cells were transfected with the corresponding siRNA or with an irrelevant siRNA as a control. At 72 h posttransfection, the cells were infected with Yuc8 (MOI, 5), and at 18 hpi the cells were subjected to three freeze-thaw cycles and the viral progeny was titrated as described in Materials and Methods. Silencing of the expression of the CYP51A1, DHCR7, FASN, PI4KIIIβ, ITPR3, and DDX23 genes reduced the yield of virus progeny between 40 and 50% (Fig. 4). The viability of the cells, determined by an LDH activity release assay, was not affected, and the reduction in gene expression induced by the corresponding siRNAs ranged from 40 to 80%, as determined by real-time quantitative RT-PCR of the target mRNA (data not shown).
FIG 4.
Yuc8 viral progeny produced in siRNA-transfected cells. Caco-2 cells transfected with the indicated siRNAs were infected at 72 h posttransfection with astrovirus Yuc8 (MOI, 5). Focus-forming units (FFU) were quantified at 18 h after infection by an immunoperoxidase assay (see Materials and Methods). Irr, irrelevant siRNA (nontargeting; Dharmacon). The biological process in which the knocked-down genes are involved is indicated. The arithmetic means ± standard deviations from three independent experiments performed in duplicate are shown. **, P < 0.001; *, P < 0.01.
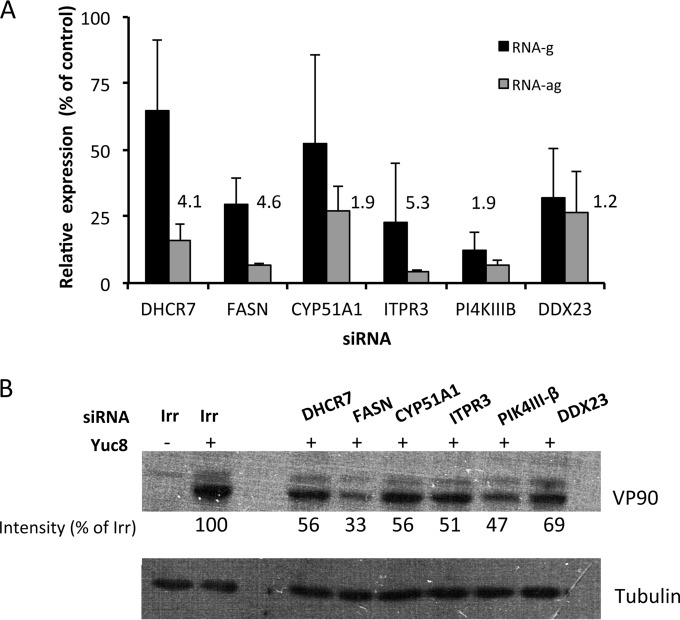
To explore the stage of the astrovirus replication cycle affected by silencing the expression of the various genes, the synthesis of viral genomic RNA (RNAg) and antigenomic RNA (RNAag) was determined by real-time qRT-PCR. For this, Caco-2 cells transfected with siRNAs specific for DHCR7, FASN, CYP51A1, ITPR3, PI4KIIIβ, or DDX23 were infected with astrovirus Yuc8 at an MOI of 5, and the cells were harvested at 12 hpi. The viral RNA was extracted and the abundance of RNAg and RNAag was determined as described in Materials and Methods. Silencing of the expression of these genes decreased the synthesis of RNAag from 75% (CYP51A1 and DDX23) to more than 90% (FASN, ITPR3, and PI4KIIIβ) (Fig. 5A), while the reduction of the level of RNAg was less pronounced, ranging from 35% for DHCR7 and CYP51A1 to about 60% for FASN, ITPR3, and DDX23 and to 80% for PI4KIIIβ. The ratio of RNAg to RNAag ranged from 1.2-fold (DDX23) to 5.3-fold (ITPR3) (Fig. 5A). The ratio of RNAg to RNAag in cells treated with an irrelevant siRNA was about 200. The moderate effect of siRNAs on the reduction of virus yield and genomic and antigenomic viral RNAs observed may be explained by the partial knockdown of the specific gene expression achieved with the various siRNAs. Alternatively, the existence in some cases of redundant enzymatic activities or the presence of enzyme isoforms not affected by the employed siRNA might also contribute to the partial inhibition effect observed.
FIG 5.
(A) Abundance of genomic and antigenomic viral RNA in silenced cells. The indicated siRNAs were transfected into Caco-2 cells, at 72 h posttransfection the cells were infected with astrovirus Yuc8 (MOI, 5), and the RNA was extracted at 12 hpi. The expression of viral RNA was determined by real-time qRT-PCR as described in Materials and Methods. Results represent the percent expression relative to that in cells transfected with an irrelevant siRNA (nontargeting; Dharmacon). The arithmetic means ± standard deviations from three independent experiments performed in duplicate are shown. The numbers represent the ratio of the RNAg concentration/RNAag concentration in the presence of the indicated siRNAs. (B) Effect of siRNAs on the synthesis of the structural polyprotein VP90. The cells were transfected and infected as described above. At 12 hpi, the cell monolayer was solubilized in Laemmli sample buffer and the proteins were analyzed by Western blotting. The structural polyprotein VP90 was detected using anti-Yuc8 serum. Tubulin was used as a loading control. The optical density of the bands was normalized to that of the loading control (tubulin), and the percentage of the VP90 signal of cells transfected with the indicated siRNAs was calculated using the intensity of VP90 from cells transfected with an irrelevant siRNA to be an intensity of 100%. The blots are representative of those from three independent experiments.
The synthesis of the structural polyprotein VP90 in Yuc8-infected siRNA-transfected cells was also evaluated by Western blotting. At 12 hpi, the amount of accumulated VP90 ranged from 33 to 69% compared to the amount that accumulated in cells transfected with an irrelevant siRNA. The greatest effect on VP90 synthesis was observed after the expression of FASN, ITPR3, and PI4KIIIβ was knocked down (Fig. 5B). This observation is in agreement with the finding that the cells transfected with siRNAs corresponding to these three genes showed the lowest level of RNAg. Similarly, there was a correlation between the levels of knockdown of the expression of DHCR7 and CYP51A1, which resulted in a decrease in the amount of RNAg of less than 50% and a reduction in the amount of the accumulated VP90 protein of about 40%.
DISCUSSION
It has been described that (+)RNA viruses replicate in association with membranes, and the membranes that they exploit to generate structures for replication and assembly have been shown to be heterogeneous. Our group previously reported the association of structural and nonstructural astrovirus Yuc8 proteins and both RNAg and RNAag viral RNA species with cell membranes in nucleus-free extracts after density gradient separation in iodixanol (23). Also, a protein corresponding to the carboxy terminus of nsp1a, which has been shown to interact with the viral RNA polymerase, was colocalized with viral RNA and calnexin (42, 43). In this work, we further characterized the association of viral proteins, RNA, and infectious virus with cell membranes using FFZE as an analytical tool.
The membranes of astrovirus-infected and mock-infected Caco-2 cells were separated by FFZE into three clearly distinct populations defined by the presence of cellular proteins in the different fractions. Of interest, viral structural and nonstructural proteins, as well as genomic and antigenomic RNA and infectious viral particles, were found exclusively in FFZE membrane population II, predominantly in fractions 44 to 49, suggesting that astrovirus RNA replication and particle assembly occur in association with the same membranes and probably at the same membrane sites, refining previous observations that these processes occur with membranes in general (23). For some (+)RNA viruses, RNA replication and assembly have been described to take place in different membranous compartments (14, 44–46). Depending on the virus, replication might occur on altered membranes derived from the ER (47), Golgi apparatus (48), mitochondria (49), lipid droplets (50), or even lysosomes (51). For instance, the replication of hepatitis C virus (HCV) has been shown to occur in a replication complex bound either to ER membranes that contain nonstructural viral proteins and RNA (47) or to altered membranes derived from different organelles (51), as has also been observed for poliovirus (52). In the case of astrovirus, membrane population II was found to be enriched in membranes from different organelles, including the Golgi apparatus, ER, mitochondria, plasma membrane, and nucleus, suggesting an arrangement similar to that observed for poliovirus. The finding of a large number of nuclear membrane proteins in population II is of interest, and it may reflect the idea that the nuclear envelope is continuous with the ER membrane. Further experiments are needed, however, to define whether RNA replication and particle assembly occur in the same type of membranous structures and whether the astrovirus RNA replication complex is bound to a single type of membrane or to modified membranes derived from different origins.
Electron microscopy and electron tomography analyses have generated significant information about the three-dimensional architecture of the replication factories of RNA(+) viruses (53). The architecture of membrane rearrangement in astrovirus-infected cells is not known; however, ultrastructural analysis of Yuc8-infected Caco-2 cells by electron microscopy and immunoelectron microscopy showed large groups of viral particles surrounding vesicles (O rings) that were not observed in mock-infected cells and that seemed to contain the viral RNA polymerase (23). These vesicles probably correspond to the double-membrane vesicles observed in Caco-2 cells infected with HAstV-4, beside which viral aggregates were observed (43). Additionally, cisterna-like structures surrounding a dark center have been observed in Caco-2 cells infected with Yuc8 (23), and these structures are similar to those found in poliovirus-infected HeLa cells (52). In both cases, viral particles were observed around these horseshoe-like structures. The presence of structural proteins in the membrane of vesicles surrounded by viral particles has also been described for both HAstV-4 and HAstV-8 (23, 43). Even though it would be necessary to directly study the participation of lipid droplets in the rearrangement of membranes induced by astrovirus, the fact that these organelles are considered to originate from the ER (54) suggests their possible participation in the new membranous structures generated during astrovirus replication.
The evidence that FFZE fractions 48 and 49 from population II of infected cells were highly enriched in Yuc8 proteins and viral RNA suggested that these fractions may be important for viral replication. To investigate this further, we selected from these fractions 11 proteins that were either involved in enriched biosynthetic processes (Table 3) or previously reported to be required for the replication of (+)RNA viruses (55–60) to investigate their possible role in astrovirus replication. RNA interference of the genes encoding these 11 proteins showed that knocking down the expression of 6 of them reduced the yield of infectious virus. These findings reveal that the cholesterol synthesis enzymes DHCR7 and CYP51A1, the fatty acid synthase FASN, the phosphatidylinositol kinase PI4KIIIβ, the inositol 1,4,5-trisphosphate receptor ITPR3, and the RNA helicase DDX23 are all required for the efficient replication of the virus in Caco-2 cells. The moderate effect of siRNAs on the reduction of virus yield and genomic and antigenomic viral RNAs that was observed may be explained by the partial knockdown of the expression of the specific genes achieved with the various siRNAs. Alternatively, the existence in some cases of redundant enzymatic activity or the presence of enzyme isoforms not affected by the siRNA employed might also contribute to the partial inhibition effect observed. Finally, the transfection efficiency of the siRNAs is also a factor to consider, since the untransfected, astrovirus-infected cells produced a normal amount of viral progeny.
The requirement of cholesterol and fatty acid synthesis for astrovirus replication is not unprecedented. In fact, other (+)RNA viruses have also been shown to depend on the synthesis of these compounds and to induce extensive ultrastructural changes in infected cells to provide the platforms required to assemble arrays of replication factories (61). For example, HCV infection induces significant changes in the expression of genes involved in the biosynthesis, degradation, and transport of intracellular lipids (62) and alters lipid metabolism (63). In the case of dengue virus, fatty acid biosynthesis was identified to be a cellular pathway required for virus replication (40, 64–66). The replication of dengue virus was also shown to depend on the production of endogenous cholesterol, since the virus nonstructural proteins associate with lipid rafts and cholesterol depletion reduces the rates of Japanese encephalitis virus and dengue virus infection (67, 68). It has also been reported that West Nile virus modulates host cell cholesterol homeostasis by upregulating cholesterol biosynthesis and redistributing cholesterol to viral replication membranes (69). Furthermore, HCV infection increases the abundance and activity of FASN, with a corresponding increase in the production of fatty acids and neutral lipids (70), and dengue virus induces the relocalization of FASN, recruiting it to the virus replication sites (40) in a process mediated by Rab18, which is inserted into the phospholipid monolayer of lipid droplets and is responsible for the trafficking of lipid droplets inside cells (66, 71).
An additional angle of lipid metabolism in virus-infected cells is related to lipid droplets (72). The association of HCV proteins and viral factories with lipid droplets is necessary for the replication and assembly of virus progeny (45, 73, 74), and the replication of dengue virus also dependd on these organelles (46). In the case of rotavirus, a dsRNA virus, lipid droplets have been shown to be recruited to viral replication complexes in viroplasms (44). Of interest, the neutral lipids stored in these organelles have been shown to be used to generate the energy likely required for the replication of HCV and rotavirus (63, 75). The involvement of pathways of fatty acid biosynthesis found in this work suggests that lipid droplets might also work as an energy source for astrovirus replication.
RNA helicases have important roles in viral life cycles, where they are either encoded by the virus genome or recruited from the host. These enzymes are particularly important for RNA viruses since they are involved in replication of the viral genome and in mRNA translation. Despite the length of the astrovirus genome of 6.8 to 7.9 kb, a virus-encoded RNA helicase has not been identified, and it has been speculated that astrovirus uses a cellular enzyme (8). In this regard, it is of interest that a cellular RNA helicase was identified in our proteomic approach. Its precise role in astrovirus replication needs to determined, but the fact that knockdown of the expression of helicase DDX23 decreases the virus yield, as well as RNA and protein synthesis, indicates that it has a relevant role during the life cycle of the virus. Similar results have been reported for coronaviruses; the nonstructural protein 14 of the coronavirus infectious bronchitis virus interacts with DDX1, a cellular RNA helicase in the DEX(D/H) helicase family. The replication of this virus was decreased by manipulating the expression of DDX1 either by RNA interference or by overexpression of a mutant DDX1 protein (76).
Phosphoinositols (PIs) are phospholipids that mediate signaling cascades in the cell by binding to effector proteins. Reversible phosphorylation of the inositol ring at positions 3, 4, and 5 results in the synthesis of seven different PIs, each of which has a unique subcellular distribution with a predominant localization in subsets of membranes (39). These lipids play a major role in recruiting and regulating the function of proteins at membrane interfaces (77), and the requirement for PI4KIII isoforms α and β (PI4KIIIα and PI4KIIIβ, respectively) for virus replication has been reported. Lim and Hwang (78) showed that PI4KIIIα is indispensable for HCV replication, specifically, for the structural integrity of the membranous structures with which the HCV replication complex interacts. In enterovirus, the recruitment of PI4KIIIβ to membranes increases the synthesis of phosphatidylinositol 4-phosphate (PI4P) lipids, resulting in a PI4P-enriched membrane microenvironment that enhances the recruitment of the viral RNA-dependent RNA polymerase, which in turns initiates RNA synthesis at these membranes (79). PI4P lipid-enriched membranes have also been proposed to generate high-curvature membrane pockets to protect viral proteins and RNA from host defense (80) and/or to provide binding sites to concentrate viral/host proteins at the membranous web for efficient viral RNA synthesis (39). Further studies are required to determine the mechanism through which PI4KIIIβ contributes to the replication of astrovirus.
The precise step of the astrovirus replication cycle that is favored by the various host factors identified in this work is difficult to determine. Upon infection, the genomic RNA is translated into a nonstructural polyprotein, which, after being processed by viral and cellular proteases, renders the RNA-dependent RNA polymerase and other viral proteins needed to form the viral RNA replication complex. This replication complex, in turn, synthesizes the RNAag using RNAg as the template. The RNAag is then copied by the replication complex into additional RNAg molecules and also serves as a template to synthesize the subgenomic RNA that encodes the virus structural proteins. The observation that the synthesis of RNAag was impaired to a greater extent than that of RNAg (RNAg/RNAag ratio, 1.2 to 5.3) in most of the cases evaluated in this work suggests that the proteins identified to be required for astrovirus replication are needed directly or indirectly to favor the synthesis of RNAag. Further work is needed to define in more detail the stage of the replication cycle that requires the identified proteins, although it is not unreasonable to assume that at least the enzymes involved in lipid metabolism could be required to provide the membranous environment required for an efficient RNA replication process and might also be required in a pathway involved in the production of energy from fatty acid breakdown during virus replication, as has been reported for other viruses.
In summary, the results presented in this study strongly suggest that astrovirus replication and virus assembly take place in association with cell membranes possibly derived from different organelles and exploit cellular lipid metabolism for efficient replication in Caco-2 cells. The mechanism through which the genes found in this work contribute to a successful virus infection is unknown, but their identification provides the basis for further studies that will allow us to deepen our knowledge about the replication of this important pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by grants CB07-79574 (to E.M.) and P-178232 (to R.V.-E.) from CONACyT—Mexico and IN219910 (to E.M.) from DGAPA-UNAM. A.M. was the recipient of a scholarship from CONACyT—Mexico.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01225-15.
REFERENCES
- 1.Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Lewis D, Gray JJ. 2005. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J Clin Microbiol 43:3890–3894. doi: 10.1128/JCM.43.8.3890-3894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Méndez EA, Arias CF. 2013. Astroviruses, p 609–628. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol 48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouzalas IG, Wuthrich D, Walland J, Drogemuller C, Zurbriggen A, Vandevelde M, Oevermann A, Bruggmann R, Seuberlich T. 2014. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J Clin Microbiol 52:3318–3324. doi: 10.1128/JCM.01195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, Pan M, Wang X, Xu Y, Xie X, Knowles NJ, Yang H, Zhang D. 2009. Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J Gen Virol 90:1104–1108. doi: 10.1099/vir.0.008599-0. [DOI] [PubMed] [Google Scholar]
- 7.Koci MD, Seal BS, Schultz-Cherry S. 2000. Molecular characterization of an avian astrovirus. J Virol 74:6173–6177. doi: 10.1128/JVI.74.13.6173-6177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci U S A 90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroe SS, Jiang B, Stine SE, Koopmans M, Glass RI. 1993. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J Virol 67:3611–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. 2003. Host factors in positive-strand RNA virus genome replication. J Virol 77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul D, Madan V, Bartenschlager R. 2014. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, Packer J, Schurdak M, Molla A. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 17.Vashist S, Urena L, Chaudhry Y, Goodfellow I. 2012. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J Virol 86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Breton M, Meyniel-Schicklin L, Deloire A, Coutard B, Canard B, de Lamballerie X, Andre P, Rabourdin-Combe C, Lotteau V, Davoust N. 2011. Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol 11:234. doi: 10.1186/1471-2180-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mairiang D, Zhang H, Sodja A, Murali T, Suriyaphol P, Malasit P, Limjindaporn T, Finley RL Jr. 2013. Identification of new protein interactions between dengue fever virus and its hosts, human and mosquito. PLoS One 8:e53535. doi: 10.1371/journal.pone.0053535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kujala P, Ikaheimonen A, Ehsani N, Vihinen H, Auvinen P, Kaariainen L. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendez E, Aguirre-Crespo G, Zavala G, Arias CF. 2007. Association of the astrovirus structural protein VP90 with membranes plays a role in virus morphogenesis. J Virol 81:10649–10658. doi: 10.1128/JVI.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkla BJ, Vera-Estrella R, Pantoja O. 2007. Enhanced separation of membranes during free flow zonal electrophoresis in plants. Anal Chem 79:5181–5187. doi: 10.1021/ac070159v. [DOI] [PubMed] [Google Scholar]
- 25.Hannig K. 1982. New aspects in preparative and analytical continuous free-flow cell electrophoresis. Electrophoresis 3:235–243. doi: 10.1002/elps.1150030502. [DOI] [Google Scholar]
- 26.Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, Arias CF. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol 76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendez E, Salas-Ocampo E, Arias CF. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol 78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez E, Salas-Ocampo MP, Munguia ME, Arias CF. 2003. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J Virol 77:11378–11384. doi: 10.1128/JVI.77.21.11378-11384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arias CF, Romero P, Alvarez V, Lopez S. 1996. Trypsin activation pathway of rotavirus infectivity. J Virol 70:5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkla BJ, Vera-Estrella R, Pantoja O. 2012. Protein profiling of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Proteomics 12:2862–2865. doi: 10.1002/pmic.201200152. [DOI] [PubMed] [Google Scholar]
- 31.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 33.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. 2003. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belliot G, Laveran H, Monroe SS. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch Virol 142:1323–1334. doi: 10.1007/s007050050163. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Heaton NS, Randall G. 2011. Multifaceted roles for lipids in viral infection. Trends Microbiol 19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delang L, Paeshuyse J, Neyts J. 2012. The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem Pharmacol 84:1400–1408. doi: 10.1016/j.bcp.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodgers MA, Villareal VA, Schaefer EA, Peng LF, Corey KE, Chung RT, Yang PL. 2012. Lipid metabolite profiling identifies desmosterol metabolism as a new antiviral target for hepatitis C virus. J Am Chem Soc 134:6896–6899. doi: 10.1021/ja207391q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuentes C, Guix S, Bosch A, Pinto RM. 2011. The C-terminal nsP1a protein of human astrovirus is a phosphoprotein that interacts with the viral polymerase. J Virol 85:4470–4479. doi: 10.1128/JVI.01515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guix S, Caballero S, Bosch A, Pinto RM. 2004. C-terminal nsP1a protein of human astrovirus colocalizes with the endoplasmic reticulum and viral RNA. J Virol 78:13627–13636. doi: 10.1128/JVI.78.24.13627-13636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. 2010. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol 84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 46.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Hage N, Luo G. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol 84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 48.Roulin PS, Lotzerich M, Torta F, Tanner LB, van Kuppeveld FJ, Wenk MR, Greber UF. 2014. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe 16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Miller DJ, Schwartz MD, Ahlquist P. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaunt ER, Cheung W, Richards JE, Lever A, Desselberger U. 2013. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J Gen Virol 94:1310–1317. doi: 10.1099/vir.0.050146-0. [DOI] [PubMed] [Google Scholar]
- 51.Berger KL, Randall G. 2009. Potential roles for cellular cofactors in hepatitis C virus replication complex formation. Commun Integr Biol 2:471–473. doi: 10.4161/cib.2.6.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel A, Giddings TH Jr, Ladinsky MS, Kirkegaard K. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol 70:6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thiam AR, Farese RV Jr, Walther TC. 2013. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A 106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrlich LS, Medina GN, Khan MB, Powell MD, Mikoshiba K, Carter CA. 2010. Activation of the inositol (1,4,5)-triphosphate calcium gate receptor is required for HIV-1 Gag release. J Virol 84:6438–6451. doi: 10.1128/JVI.01588-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu L, Horstmann H, Ng C, Hong W. 2001. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J Cell Sci 114:4543–4555. [DOI] [PubMed] [Google Scholar]
- 58.Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J 31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma M, Sasvari Z, Nagy PD. 2010. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol 84:2270–2281. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma M, Sasvari Z, Nagy PD. 2011. Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology 415:141–152. doi: 10.1016/j.virol.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog 8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. 2010. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol 84:5404–5414. doi: 10.1128/JVI.02529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liefhebber JM, Hague CV, Zhang Q, Wakelam MJ, McLauchlan J. 2014. Modulation of triglyceride and cholesterol ester synthesis impairs assembly of infectious hepatitis C virus. J Biol Chem 289:21276–21288. doi: 10.1074/jbc.M114.582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carvalho FA, Carneiro FA, Martins IC, Assuncao-Miranda I, Faustino AF, Pereira RM, Bozza PT, Castanho MA, Mohana-Borges R, Da Poian AT, Santos NC. 2012. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J Virol 86:2096–2108. doi: 10.1128/JVI.06796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martins IC, Gomes-Neto F, Faustino AF, Carvalho FA, Carneiro FA, Bozza PT, Mohana-Borges R, Castanho MA, Almeida FC, Santos NC, Da Poian AT. 2012. The disordered N-terminal region of dengue virus capsid protein contains a lipid-droplet-binding motif. Biochem J 444:405–415. doi: 10.1042/BJ20112219. [DOI] [PubMed] [Google Scholar]
- 66.Tang WC, Lin RJ, Liao CL, Lin YL. 2014. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol 88:6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee CJ, Lin HR, Liao CL, Lin YL. 2008. Cholesterol effectively blocks entry of flavivirus. J Virol 82:6470–6480. doi: 10.1128/JVI.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rothwell C, Lebreton A, Young Ng C, Lim JY, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA. 2009. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 69.Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Nasheri N, Joyce M, Rouleau Y, Yang P, Yao S, Tyrrell DL, Pezacki JP. 2013. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chem Biol 20:570–582. doi: 10.1016/j.chembiol.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Rasineni K, McVicker BL, Tuma DJ, McNiven MA, Casey CA. 2014. Rab GTPases associate with isolated lipid droplets (LDs) and show altered content after ethanol administration: potential role in alcohol-impaired LD metabolism. Alcohol Clin Exp Res 38:327–335. doi: 10.1111/acer.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herker E, Ott M. 2012. Emerging role of lipid droplets in host/pathogen interactions. J Biol Chem 287:2280–2287. doi: 10.1074/jbc.R111.300202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Filipe A, McLauchlan J. 2015. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med 21:34–42. doi: 10.1016/j.molmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Hope RG, Murphy DJ, McLauchlan J. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem 277:4261–4270. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 75.Crawford SE, Utama B, Hyser JM, Broughman JR, Estes MK. 2013. Rotavirus exploits lipid metabolism and energy production for replication, p 74 Abstr Annu Meet Am Soc Virol, Penn State University, University Park, PA. [Google Scholar]
- 76.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. 2010. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol 84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 78.Lim YS, Hwang SB. 2011. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J Biol Chem 286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stapleford KA, Miller DJ. 2010. Role of cellular lipids in positive-sense RNA virus replication complex assembly and function. Viruses 2:1055–1068. doi: 10.3390/v2051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.