ABSTRACT
Influenza viruses continue to present global threats to human health. Antigenic drift and shift, genetic reassortment, and cross-species transmission generate new strains with differences in epidemiology and clinical severity. We compared the temporal transcriptional responses of human dendritic cells (DC) to infection with two pandemic (A/Brevig Mission/1/1918, A/California/4/2009) and two seasonal (A/New Caledonia/20/1999, A/Texas/36/1991) H1N1 influenza viruses. Strain-specific response differences included stronger activation of NF-κB following infection with A/New Caledonia/20/1999 and a unique cluster of genes expressed following infection with A/Brevig Mission/1/1918. A common antiviral program showing strain-specific timing was identified in the early DC response and found to correspond with reported transcript changes in blood during symptomatic human influenza virus infection. Comparison of the global responses to the seasonal and pandemic strains showed that a dramatic divergence occurred after 4 h, with only the seasonal strains inducing widespread mRNA loss.
IMPORTANCE Continuously evolving influenza viruses present a global threat to human health; however, these host responses display strain-dependent differences that are incompletely understood. Thus, we conducted a detailed comparative study assessing the immune responses of human DC to infection with two pandemic and two seasonal H1N1 influenza strains. We identified in the immune response to viral infection both common and strain-specific features. Among the stain-specific elements were a time shift of the interferon-stimulated gene response, selective induction of NF-κB signaling by one of the seasonal strains, and massive RNA degradation as early as 4 h postinfection by the seasonal, but not the pandemic, viruses. These findings illuminate new aspects of the distinct differences in the immune responses to pandemic and seasonal influenza viruses.
INTRODUCTION
Influenza A virus (IAV) infection causes hundreds of thousands of deaths annually, and individual strains show a large variation in pathogenicities and infection rates. A 20 to 50% attack rate in some countries by the recent 2009 influenza pandemic (1, 2) and the specter of the 1918 pandemic, which killed 2% of the global population (3, 4), makes it important to improve the understanding of differences in the immunological responses to IAV strains and, in particular, differences between seasonal and pandemic IAVs.
The severity of influenza ranges from subclinical illness to severe respiratory involvement and cytokine dysregulation, leading to significant mortality (5–7). Many factors may influence disease severity, including preexisting immunity (e.g., prior vaccination or infection), host genetic determinants, age, health status, and characteristics of the virus strain. Seasonal influenza viruses cause annual epidemics that most commonly result in subclinical to mild illness (8). Indeed, experimental infection of immunocompetent human subjects with a seasonal H1N1 influenza virus strain, A/Texas/36/1991 (TX), resulted in mild clinical symptoms and virus shedding that resolved within 7 days (9, 10). However, seasonal influenza viruses can cause significant morbidity and mortality in high-risk populations (11, 12). Pandemic influenza viruses that show high levels of transmission and infectivity have the potential to be a serious health threat to the global human population. Whereas the pandemic of 1918 caused up to 100 million deaths predominantly in young healthy adults (13, 14), the influenza pandemics in 1957, 1968, and 2009 resulted in much less morbidity and mortality. Despite its high rate of infection, the 2009 pandemic strain showed a global mortality comparable to that of seasonal viruses, although the groups at risk for severe and fatal infections differed between the 2009 and seasonal influenza strains (1, 15).
The immune response to IAV emerges from dynamic interactions of multiple elements, including the interaction of molecular host defense mechanisms in specialized immune cells and viral determinants over time. Dendritic cells (DC) are professional antigen-presenting cells that are important mediators of the innate and adaptive immune responses to IAV infection (16). DC produce type I interferons (IFN) that contribute to the innate immune response. They also present antigens to T cells to initiate adaptive immune responses. Pathogens can impair DC function (17–21), which may influence immunological homeostasis and the clinical outcome of infection.
Individual IAV strains show several differences in sequence features (Table 1), including virulence determinants within the NS1, PB1-F2, and PB2 proteins, which have been implicated in pathogenesis (22). Although NS1 is known to block general cytokine expression (23), the A/California/4/2009 (Cal/09) NS1 protein does not efficiently inhibit host gene expression (24). Regardless, infections with Cal/09 produce lower levels of cytokines and chemokines than infections with seasonal influenza virus strains (25). The outcome of cellular infection, and the clinical outcome of infection, can also be influenced by the activity of the polymerase complex (26–28). Another virulence factor is PB1-F2, an influenza gene product which is initiated by an alternative start codon within the PB1 segment (29, 30). A/Brevig Mission/1/1918 (1918) contains an S66 residue in PB1-F2, which increases its virulence compared to that of other strains with N66 residues in their PB1-F2s, including the seasonal H1N1 influenza virus strains prior to 2009 (31). Cal/09, on the other hand, encodes a truncated PB1-F2 protein of 11 amino acids due to multiple stop codons (32, 33). While differences in viral determinants have been identified, the differences in human host cell responses and their role in infectivity and pathogenesis are incompletely understood.
TABLE 1.
Sequence features of H1N1 influenza A virus strainsa
| Virus | Nature of virus | Yr of isolation | Receptor specificity(ies) | No. of aa in NS1 | CPSF binding | No. of aa in PB1-F2 (mutation)b | PB2 virulence markersc |
|---|---|---|---|---|---|---|---|
| 1918 | Pandemic | 1918 | α-2,6 | 230 | Yes | 90 (S66) | K627 D701 |
| Cal/09 | Pandemic | 2009 | α-2,6 and α-2,3 | 219 | No | 11 | E627 D701 |
| NC | Seasonal | 1999 | α-2,6 | 230 | Yes | 57 | K627 D701 |
| TX | Seasonal | 1991 | α-2,6 | 230 | Yes | 57 | K627 D701 |
aa, amino acids; CPSF, cleavage and polyadenylation specificity factor.
The PB1-F2 virulence marker is N66S.
PB2 virulence markers are E627K and D701N.
The typical DC response to IAV infection is initiated by viral pathogen recognition receptors, such as the cytosolic receptors RIG-I and MDA5, that trigger a transcriptional program, including the production of type I IFNs. These antiviral cytokines signal in both autocrine and paracrine fashion through the JAK-STAT pathway, leading to additional transcription events involving the induction of hundreds of interferon-stimulated genes (ISGs) (5, 7, 34–37). Several studies have examined global transcriptional responses to infection with several IAV strains using animal model and in vitro experimental systems (6, 38–41). However, a comparison of the global transcriptional responses during the initial phase of infection, during which the responses in human DC are studied, and detailed time course data of the response to the highly pathogenic reconstructed 1918 virus have not been previously reported. We investigated whether seasonal H1N1 or pandemic H1N1 influenza virus strains induced specific transcriptional responses in infected human DC.
Here, we present the first comprehensive comparative study of seasonal and pandemic H1N1 viruses at early time points after infection. We compared the transcriptional responses of human-monocyte-derived DC to infection with two pandemic H1N1 viruses (Cal/09 and 1918) and two seasonal H1N1 viruses (A/New Caledonia/20/1999 [NC] and TX). By focusing on H1N1, our analysis identified differences in the primary immune responses between pandemic and seasonal strains, without being confounded by differences among IAV serotypes. Our results revealed that the initial transcriptional response of DC to IAV infection is characterized by a common antiviral program whose timing is strain specific. Notably, we observed strain-specific signatures that were particular to DC infected by seasonal strains. These results provide a basis for understanding the differences in host-pathogen interactions among H1N1 influenza virus strains.
MATERIALS AND METHODS
Differentiation of DC.
All human research protocols for this work were reviewed and approved by the institutional review board (IRB) of the Icahn School of Medicine at Mount Sinai (ISMMS). Both donors used for the microarray experiments were Caucasian males aged 50 to 65 years. All additional donors of peripheral blood mononuclear cells (PBMC) were anonymous, and no demographic information was available. Monocyte-derived DC were obtained from healthy human blood donors by following a standard protocol described elsewhere (42). Briefly, human PBMC were isolated from buffy coat cells by Ficoll density gradient centrifugation and positive immunomagnetic purification for CD14, followed by a 5-day incubation with 500 U/ml human granulocyte-macrophage colony-stimulating factor (hGM-CSF) (PeproTech, Rocky Hill, NJ) and 1,000 U/ml human interleukin 4 (hIL-4) (PeproTech, Rocky Hill, NJ). All experiments were replicated using cells obtained from different donors.
Virus preparation and viral infection.
The human isolates of H1N1 influenza A viruses (see Table SA1 in the supplemental material) A/Brevig Mission/1/1918 (1918), A/California/4/2009 (Cal/09), A/New Caledonia/20/1999 (NC), and A/Texas/36/1991 (TX) were propagated in specific-pathogen-free embryonated hen's eggs (Charles River, North Franklin, CT, USA) (43). All experiments involving 1918 were conducted under high-containment (enhanced biosafety level 3 [BSL3]) laboratory conditions in accordance with guidelines of the ISMMS institutional biosafety office, the National Institutes of Health, and the Centers for Disease Control and Prevention. Infectious titers of influenza viruses were determined by standard plaque assay on Madin-Darby canine kidney (MDCK) epithelial cells. Before experiments described here were carried out, the infectivity of each influenza virus preparation for human DC was measured by NP staining and its titer was adjusted to so that each strain infected approximately 60% of DC in cells obtained from each of 6 anonymous donors. For infection, virus stocks were diluted in serum-free medium and added directly onto pelleted DC at a multiplicity of infection (MOI) of 1. DC from donors were infected in triplicate with the influenza virus strains for 10 min in RPMI medium at 37°C. After infection, cells were centrifuged to remove the viral inoculation medium and resuspended in DC medium. At the time points indicated in the figures, infected DC were pelleted and fixed with RNAprotect cell reagent (Qiagen) for RNA extraction. For imaging flow, samples were fixed with 1% paraformaldehyde (PFA).
RNA extraction.
For the microarray analysis, cells were homogenized by using QIAshredder microcentrifuge spin columns (Qiagen, Dusseldorf, Germany), and RNA was isolated from cells using a Qiagen Micro RNeasy Plus kit by following the manufacturer's protocol (Qiagen, Dusseldorf, Germany). RNA quality was assayed by determination of the RNA integrity number using a model 2100 Bioanalyzer (Agilent). For integrated fluidic circuit (IFC) real-time PCR assays (Fluidigm), RNA was extracted with Agencourt RNAdvance Cell v2 (Beckman Coulter) and RNA quantity was measured with the RiboGreen system (Life Technologies) using a fluorimeter.
Microarray analysis.
RNA samples were processed and hybridized to a HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA). Arrays were processed at Yale's Keck Biotechnology Resource Laboratory, and raw expression data were output by the Illumina GenomeStudio software. Microarray data are available through the Gene Expression Omnibus (GEO) database (accession number GSE55278). The data were log transformed and median normalized. Activities were calculated after collapsing multiple probe identifiers mapping to a single official gene symbol by keeping the probe identifier with the highest average expression. Differential expression was defined for each probe at each infection time point using two criteria: (i) an absolute fold change of at least 2 relative to the value for a time-matched allantoic fluid (AlaF) control and (ii) a significant change in expression by limma (linear models for microarray data) (Bioconductor [44] implementation) after correction for multiple-hypothesis testing (false-discovery rate [FDR] ≤ 0.05). Genes with multiple probes were collapsed to keep probes with the highest average expression within differentially expressed genes. Differentially expressed genes were grouped into clusters using hierarchical clustering with Ward's linkage. The set of genes from each of the clusters was submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to determine the functional annotations. Probe identifications were used as the identifiers with the background set to HumanHT-12_V3_0_R2_11283641_A. All of this analysis was performed using Bioconductor software packages (44) in R.
IFC multiplex real-time PCR.
RNA was reverse transcribed using the VILO master mix (Life Technologies) by a 60-min incubation at 42°C, followed by enzyme inactivation at 85°C for 5 min. cDNA was preamplified using the TaqMan PreAmp mix with a 0.18 μM concentration (each) of all forward and reverse primers for 10 cycles of 95°C for 10 s and 60°C for 4 min. Gene expression was measured on a Biomark instrument using the 96-gene, 96-sample integrated fluidic circuit dynamic array (Fluidigm). Genes for analysis were chosen among the genes with the highest loadings in principal components (PC1 and PC2) in the principal-component analysis (PCA) of the microarray data. PCR primers and universal TaqMan detection probes (Roche) were designed and chosen with commercial Web-based software, Universal ProbeLibrary Assay Design Center (Life Science, Roche). The reaction was carried out according to the Fluidigm protocol with concentrations for primers of 18 μM and for probes of 4 μM.
Functional network generation.
The genes from each of the clusters were used to generate a gene-gene functional network using a publically accessible immunospecific Bayesian network integration of public data (ImmuNet, http://immunet-dev.princeton.edu) (45). Networks were generated using the ImmuNet website and the “Immune Global” context. The minimum relationship confidence was set at 0.1, and the maximum number of genes to infer was set at 10. Unconnected genes were removed from the networks before visualization. Links to interactive versions of the networks are provided at http://tsb.mssm.edu/primeportal/?q=Immunet_interactive_networks.
Time shift analysis.
The timings of ISG expression were estimated for Cal/09, NC, and TX infections, with the 1918 strain as a baseline, using a statistical method adapted from reference 46. The set of ISGs was defined as genes upregulated in DC 2.5 h after stimulation with IFN-β (47) and also significantly upregulated following infection with at least one viral strain. At each potential time shift (±2 h in 15-min intervals), the Pearson correlation was determined using the average expression across replicates. Values at intervals that were not directly measured were generated by linear interpolation. The optimal time shift was determined by maximizing the fractional rank of the position of the gene of interest within a null distribution of correlations. This null distribution was composed of correlations between the ISG of interest and all non-ISGs on the microarray. One minus the fractional rank provides the measure of significance, and time shifts with a P of <0.001 were taken to be significant. For each strain, a probability density function (PDF) was generated from the set of significant shifts by kernel density estimation using a Gaussian kernel (default parameters in the R function statistics for density). The median of the PDFs was taken to be the time shift for the strain. For PCR validation of the time shifts, the same method was used to estimate shifts for Cal/09 and TX infections, with the NC strain as a baseline. The null distribution was composed of correlations between the ISG of interest and all non-ISGs assayed by PCR. Time shifts with a P of <0.1 were taken to be significant.
Transcription factor target identification.
Using the UCSC Genome Bioinformatics site, we downloaded the transcription start site (TSS) data for all human RefSeq genes, defined by the January 2010 refGene table (48). The region ±2 kb around each TSS was identified within a genome-wide multiple alignment of sequences of 45 vertebrate species to the human genome (49), also available through the UCSC Genome Bioinformatics site. In order to identify putative transcription factor binding sites, the human sequences, along with aligned regions from mouse, were masked for repetitive elements using RepeatMasker (http://www.repeatmasker.org) and then analyzed using the TRANSFAC MATCH (50) algorithm, with a cutoff, as defined within the database, chosen to minimize the sum of false positives and false negatives. The analysis was performed for all high-quality vertebrate transcription factor matrices in the 2011.1 release of TRANSFAC (51), and putative binding sites were considered to be evolutionarily conserved if matches were also found at the aligned positions in the mouse sequences and had no gaps present in the multiple alignments between the species being compared. Each TRANSFAC matrix was linked to a set of gene symbols describing potential binding factors using annotations present in the “Binding Factor” field of the database. Only vertebrate TRANSFAC matrices that could be linked to a HUGO Gene Nomenclature Committee (HGNC) gene symbol, either directly or through an alias listed for an NCBI gene, were included.
Assessment of NF-κB translocation after viral infection.
Human DC were infected with Cal/09, NC, or TX for 4, 6, 8, and 10 h. Reactions were stopped with 1% paraformaldehyde, and cells were subsequently permeabilized with methanol and stained with monoclonal antibodies for the transcription factors NF-κB and IRF3 (both from Abcam) and a nuclear stain, Hoechst 33342 (Sigma). Cells were imaged with the ISX imaging flow cytometer (Amnis). The Pearson correlation coefficient was computed over the masked portion of image pixel intensities of the nuclear stain and the transcription factor. Nuclear translocation was assessed using the log2-transformed and control-normalized value of the computed Pearson correlation coefficient.
Viral RNA analysis.
Primers were developed and tested for efficient amplification of NS1 mRNA, NS2 mRNA, NS viral RNA (vRNA), NP mRNA, and NP vRNA from all four viruses. For analysis, each of the available triplicate RNA samples for each time point after infection by each of the four viruses was diluted in reverse transcriptase buffer and split into seven aliquots, one with 50% of the RNA, one with 25% of the RNA, and five with 10% each. Reverse transcription (RT) of the large aliquot was carried out using random priming. Sham RT was carried out on the 25% sample. One NS1 mRNA-, NS2 mRNA-, NS vRNA-, NP mRNA-, or NP vRNA-specific primer was added to the remaining aliquots. The same RT primer was used for NS1 and NS2 mRNAs. The large aliquot was used to measure rRNA and control housekeeping genes. The cDNA samples and the sham samples were diluted 10-fold and amplified by PCR using a supplemented reverse transcription primer and the specific reverse primer for the target RNA. In every experiment, at least 97% of the amplification was dependent on reverse transcription with the specific primer. For NS1, the RT and PCR primer for both NS1 and NS2 was CAAGTCCTCCGATGAGGACCCCAA. The specific NS1 mRNA reverse primer was TTCACCATTGCCTTCTCTTCCAGG, and the specific NS2 mRNA reverse primer was CCGTGTCAAGCTTTCAGGACATACT. The RT and PCR primer for NS vRNA was CCCGAGCAAATAACATTTATGCAAGC, and the reverse primer was GCCCATCTCTTGTTCTACTTCAA. For NP, the RT and PCR primer for NP mRNA was GGTTCCGACGGATGCTCTGATTTC, and the reverse primer was ATGGCGTCTCAAGGCACCAAACG. The RT and PCR primer for NP vRNA was CCCCTTCCTTTGACATGAGTAATGAA, and the reverse primer was AATACTCCTCTGCATTGTCTCCGAA.
Pre-mRNA analysis.
RT-PCR for pre-mRNAs for the uninduced TGFBI and CD74 genes was carried out across the polyadenylation sites of each gene with the RT primer 3′ of the site. Quantitative RT-PCR (qRT-PCR) for mRNAs used primers 5′ of the polyadenylation sites. All data were normalized against rRNA. Copy numbers for pre-mRNAs and mRNA of each of the four human genes were determined in triplicate for all four viruses for each time point. For TGFBI, the RT and PCR primer for mRNA was CAGGGGAGCCAGGAGGGTCAA, and the reverse primer was ACAGCCATTGACCTTTTCAGACAA. The RT and PCR primer for pre-mRNA was CACACTGTACCATGCACACAGGA, and the reverse primer was GACAGAGCCATGGTGTGTTTGTAA. For CD74, the RT and PCR primer for mRNA was GGGCAGTTGCTCATTGTTGGAGATAA, and the reverse primer was CAGGAGCTGTCGGGAAGATCAGAA. The RT and PCR primer for pre-mRNA was CTGGATCTCAACAGAAGAGCCAGGAG, and the reverse primer was CCCTAGTTCCCTCTGCTCAGCCAA.
RESULTS
DC transcriptional responses to infection with pandemic or seasonal influenza viruses share a core.
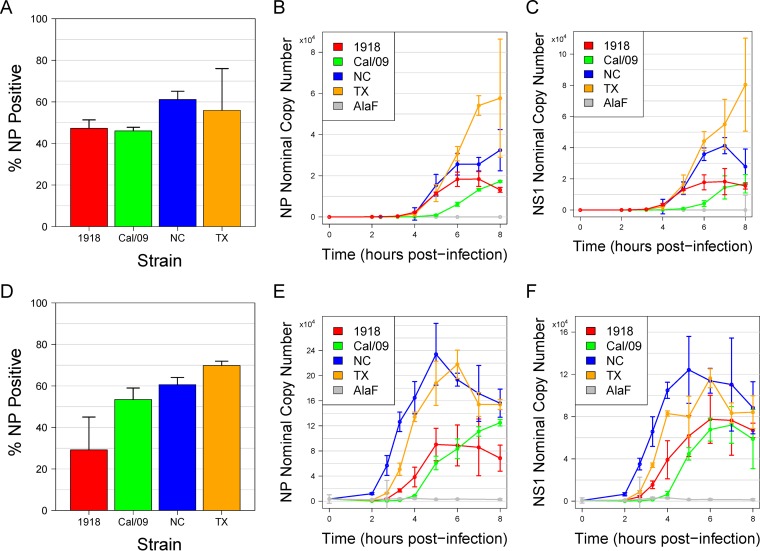
To identify differences in the host transcriptional responses to infection with pandemic or seasonal H1N1 influenza A viruses, we conducted independently replicated experiments in which monocyte-derived DC from two human donors were infected in vitro at an MOI of 1 with a pandemic strain (1918 or Cal/09) or a seasonal strain (TX or NC) (Table 1). Viral transcripts for NS1 and NP increased over time for all strains and were somewhat higher for the two seasonal strains (Fig. 1). Overall, the four strains displayed similar infectivity levels, facilitating comparative analysis of their responses. Genome-wide transcriptional profiling on samples collected at multiple time points up to 8 h postinfection was performed. Differentially expressed genes were identified by comparing expression values at each time point with those for a time-matched control (treatment with allantoic fluid [AlaF]).
FIG 1.
Infectivity of seasonal and pandemic strains. Human DC were infected with the 1918, Cal/09, NC, or TX influenza strain. (A) Infectivity was measured by using flow cytometry to assess viral NP production 8 h postinfection. Data are the averages of results from three replicates, and error bars indicate the standard deviations. (B to C) PCR was used to measure viral NS1 mRNA (B) or viral NP mRNA (C) expression during an 8-h-long time course. Data are the medians of results from three replicates, and error bars indicate the standard deviations. (D) Infectivity of donor 2. (E to F) PCR results for donor 2.
The expression of 400 genes was significantly increased at one or more time points by more than 2-fold following infection with one or more IAV strains (FDR < 0.05) (Fig. 2A). More than half of the upregulated genes were uniquely induced by infection with only one of the virus strains. The 1918 infection induced the largest number of unique transcripts (126 unique genes). A core response program comprising 58 genes was significantly induced by all four strains at one or more time points (Fig. 2B). Functional analysis indicated that these core immune response genes were enriched for antiviral pathways, including interferon-dependent responses.
FIG 2.
The transcriptional response to infection with seasonal and pandemic influenza strains share a core. Differentially expressed genes were identified by comparing their levels of expression at each time point with those of the time-matched control genes (absolute fold change, ≥2; FDR < 0.05). The numbers of up- or downregulated genes in DC infected with the 1918 (red), Cal/09 (green), NC (blue), and TX (orange) strains at each time point are indicated. (B) A common core of 58 genes was identified by analyzing the overlap among genes that were upregulated at any time point for each strain (a complete list of genes is provided in Table SA2 in the supplemental material).
Infections with seasonal and pandemic viruses induce differential global responses.
The various numbers of differentially expressed genes at each time point postinfection suggested that each virus strain induced a distinct kinetic profile. The fastest and most robust response was induced by the 1918 strain. In this case, 143 genes were upregulated by 4 h postinfection compared with 23, 52, and 29 genes upregulated by Cal/09, NC, and TX infection, respectively (Fig. 2A). Although the response to Cal/09 lagged behind all of the other strains, the total number of upregulated genes at 8 h was similar to that of 1918. Notably, there is also a large number of transcripts downregulated by the seasonal viruses (Fig. 2A).
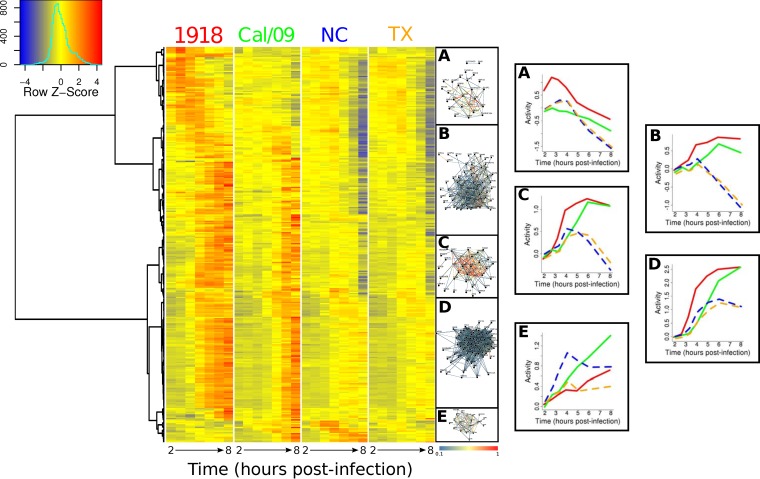
Hierarchical clustering analysis of expression profiles of the 400 genes significantly induced by at least one strain revealed five distinct temporal patterns that were associated with different functions (Fig. 3). Two clusters (Fig. 3, clusters B and C) displayed qualitatively similar patterns, with genes induced by all of the virus strains during the first few hours postinfection. Whereas infection with the seasonal viruses induced a downregulation of these genes at 4 h, infection with the pandemic viruses induced a consistent upregulation of these genes after 4 h. In cluster B, 13 genes could be associated with the intracellular signaling cascade (Expression Analysis Systematic Explorer [EASE] score, 16.7%; P < 0.01); 11 could be associated with immune response (15.4%; P < 0.001), 9 could be associated with regulation of programmed cell death (10.3%, P < 0.001), and 7 could be associated with the regulation of the I-κB kinase/NF-κB cascade (9%; P < 0.00001) (Table 2). In cluster C, we found 5 genes associated with responses to organic substances (10.9%. P = 0.06), 3 associated with protein stimulus (6.5%; P = 0.02), and 3 associated with response to virus (6.5%; P = 0.09), as well as 3 associated with transcription from the RNA polymerase II promoter (Table 2). Genes in cluster D exhibited sustained induction following infection with all 4 viruses. However, with the seasonal viruses, peak fold changes were reached in ∼6 h, while with pandemic viruses, gene expression continued to increase following infection, resulting in a 2-fold difference in expression levels from those of the seasonal viruses at later time points. In this cluster, 29 genes were associated with immune response (33.3%; P = 2e–18), 17 were associated with response to virus (19.5%; P = 1.6e–19), 9 were associated with the macromolecule metabolic process (10.3%; P < 0.05), and 9 were associated with regulation of programmed cell death (10.3%; P = 0.06) (Table 2). In all of these clusters (B to D), the 1918 strain induced the strongest response. Cluster E contained early-response genes that were dominated by NC until 4 h. In this cluster, seasonal viruses showed a short downregulation at 4 h and stayed stable in expression after 6 h. In contrast, infection with the pandemic viruses led to upregulation of this cluster throughout the first 8 h. In cluster E, we identified 9 genes associated with defense responses (34.6%; P = 1.4e–06), 8 associated with regulation of transcription (30.8%; P < 01), 7 associated with response to virus (26.9%; P = 8.4e–09), and 7 associated with immune responses (26.9%; P = 3.6e–06) (Table 2). We used immunospecific Bayesian data integration to infer the functional relationships among members of each cluster in the context of a massive compendium of publically accessible data (45). This analysis demonstrated a high level of functional connectivity of the genes comprising each cluster (Fig. 3) and identified significant enrichment for ISGs in clusters B to E. Two additional clusters that contained genes that appeared to be upregulated at the latest time points were detected. Analysis of the trajectories and annotations for these genes suggested that these clusters were artifacts resulting from the widespread mRNA decreases caused by the seasonal viruses and were not considered further (data not shown).
FIG 3.
Temporal clustering identifies five distinct gene expression patterns. Hierarchical clustering of the expression profiles for upregulated genes (fold change, ≥ 2; FDR < 0.05 with respect to the time-matched controls) identified five groups (A to E). Temporal profiles for individual genes are depicted in the heatmap by row-normalized fold changes from profiles after allantoic fluid stimulation, while the rightmost set of plots shows the average activity of each cluster, estimated by QuSAGE, following infection by pandemic strains (solid lines indicate 1918 [red] and Cal/09 [green]) and seasonal strains (dashed lines indicate NC [blue] and TX [orange]). Immunospecific functional interactions between the genes in each cluster were predicted using ImmuNet (networks adjacent to the heatmap). See http://tsb.mssm.edu/primeportal/?q=Immunet_interactive_networks.
TABLE 2.
Clusters of genes expressed after infection with IAV

Notably, cluster A contained genes that were almost exclusively significantly upregulated by the 1918 strain (57 of 62 genes were exclusively differentially expressed following 1918 infection). This cluster was enriched for 15 genes linked to the regulation of transcription (3% of all genes; P = 0.04), 12 linked to regulation of programed cell death (2.4%, P = 0.01), and 9 linked to immune responses (1.8%; P = 0.002), as well as 9 linked to the intracellular signaling cascade (1.8%; P = 0.05) (Table 2). An upstream regulator analysis (Ingenuity Pathway Analysis) of the genes in cluster A identified the transcription factors SP1, SP3, and EGR-1 as potential regulators (see Discussion). The patterns of all the clusters were similar in donor 2, although the time of activation was ∼1.1 h earlier than with donor 1 (data not shown). Consistently with this earlier activation, cluster A was not detected in donor 2, as the first sampling at 2 h was later than the anticipated upregulation for these genes (due to faster kinetics of the transcriptional response). Overall, these results demonstrate commonalities with and differences from infection of DC by the different IAV strains.
Principal-component analysis (PCA) of the upregulated genes identified strong commonalities between the initial transcriptional responses to infection by all virus strains (Fig. 4A). In order to compare the evolutions of these transcription changes, lines were drawn through the average response at each time point. Strikingly, the transcriptional responses diverged 4 h after infection when the seasonal and the pandemic strains were compared. Principal-component 1 (PC1) captured the common response, with all four strains moving along this axis until ∼4 h. The responses of the pandemic strains (1918, Cal/09) continued to move along this axis until 8 h, while the seasonal-strain responses (NC, TX) abruptly changed to track together along PC2. The same qualitative pattern was observed in the transcriptional responses of DC from donor 2, with the divergence occurring earlier, as is consonant with the earlier gene responses to all viruses observed with infection of DC from that donor (Fig. 4B). Further validation of the microarray time courses of the responses to Cal/09, NC, and TX in DC derived from four additional donors was performed by IFC real-time PCR assay of 63 key genes. Notably, despite these assays being performed on different platforms, these experiments show similar levels of divergence between the responses to the pandemic Cal/09 virus and those to the seasonal strains in all six donors (Fig. 4D; Table A1).
FIG 4.
Divergent trajectories of seasonal and pandemic infection. (A) PCA was carried out using the set of 400 upregulated genes (fold change, ≥2; FDR < 0.05 with respect to the time-matched controls). Each point is an individual microarray sample, and solid lines (AlaF control [gray], 1918 [red], Cal/09 [green], NC [blue], and TX [orange]) connect replicate averages at each time point, with 4 and 8 h postinfection indicated. (B) Expression data from donor 2 were plotted on the PCA axes from donor 1. (C) The average log fold change was used to quantify the activities of 181 genes annotated to the gene ontology (GO) term “Defense Response to Virus” (GO:0051607), as well as a set of 50 housekeeping genes. Lines connect the estimated activities at each time point in each infection (AlaF control [gray], 1918 [red], Cal/09 [green], NC [blue], TX [orange]). Results for donor 2 were similar (not shown). (D) Sixty-three key genes with the highest loadings in PC1 and PC2 from the experiment whose results are shown in panel A were measured by IFC real-time PCR in DC from four additional individuals (donors 3 to 6) following infection with Cal/09, NC, and TX. The expression patterns of these 63 genes were projected onto the first two principal components computed from the PCA of donor 1. The microarray results from the original two donors for these 63 genes, as well as the results obtained with these genes for four other donors, are plotted. Note that the overall patterns observed and the differences between the pandemic and seasonal viruses are similar in cells from all donors.
To gain insight into the different temporal patterns exhibited by the pandemic and seasonal strains in the PCA, functional analysis was carried out on the contributions (loadings) of each gene to PC1 and PC2 using the limma (linear models for microarray data) gene set test method (Bioconductor [44] implementation) with gene sets from KEGG, Reactome, and GO (Molecular Signatures Database [MSigDB], version 4.0). Genes in PC1 were associated with antiviral functions, such as interferon response, as well as other innate immune response pathways (RIG-I-like pathway, Toll-like receptor [TLR] pathways, cytosolic DNA sensing pathway; P < 0.05). No functional annotations were significantly associated with PC2. Because the genes in PC1 were antivirally related and the genes in PC2 appeared global, we tested whether these PCA trajectories could be recapitulated using an axis specific for antiviral defense and an axis specific for housekeeping genes. The plot of the average log fold change for the antiviral and for the housekeeping genes showed a pattern for each virus that was similar to that obtained by PCA (Fig. 4C). These results indicate that all viruses share an early antiviral gene induction program (PC1) that develops fastest in 1918 and slowest in Cal/09 and that the two seasonal viruses share a nonspecific gene program (PC2) initiating at about 4 h.
IAV strains differentially induce the ISG response.
Type I interferons (including multiple IFN-α genes and IFN-β) that are induced and released in response to viral infection stimulate Jak-Stat pathway signaling, leading to the upregulation of a complex interferon-stimulated gene (ISG) program (36, 52). We selected 633 ISGs previously shown to be upregulated following DC exposure to IFN-β (53) (Table A2). Among these ISGs, only 163 genes (26%) were stimulated by at least one of the four influenza viruses studied, suggesting modification of the responses by these IAVs. The earliest and most robust upregulation of the canonical ISG, mx1, occurred with infection by the 1918 strain (Fig. 5A). Surprisingly, the upregulation of ifnb1 by 1918 did not precede that of the other strains, and its magnitude was the smallest (Fig. 5B). The relative timing of the interferon response by infection with the four virus strains was further investigated using time lag correlation analysis (see Materials and Methods). This analysis confirmed that the earliest ISG response occurred following 1918 infection. The ISG response following NC infection occurred ∼30 min after the 1918 response, while the TX response was delayed by an additional ∼15 min (Fig. 5C). The slowest ISG response occurred following infection with Cal/09, which occurred ∼75 min after the 1918 response. The amplitude of the overall ISG response was largest with the two pandemic strains (Fig. 5D). The differences in the relative timings of the ISG responses following infection with NC, TX, and Cal/09 were confirmed in 4 additional donors by analysis of the IFC multiplex real-time PCR data set (Fig. 5E to H; Table A2). Thus, the relative times of ISG induction by the different strains were not correlated with the relative timing or magnitude of ifnb1 induction. These results show that the timings and magnitudes of the ISG response varied across strains and that the ISG response appeared to be disconnected from the induction of ifnb1, especially for the 1918 strain (see Discussion).
FIG 5.
Temporal shift in the antiviral IFN-β response. Temporal mRNA expression profiles of the ISG mx1 (A) and ifnb1 (B) (1918 [red], Cal/09 [green], NC [blue], TX [orange]). (C) The timing of ISG expression was calculated for the Cal/09, NC, and TX strains relative to that of the 1918 strain. Each curve indicates the estimated probability density function for the time shift, with the median shift indicated by the dashed vertical lines. (D) Average log fold changes were used to quantify the activities of ISGs at each time point in each infection. (C and D) The set of ISGs was defined as genes upregulated in DC 2.5 h after stimulation with IFN-β. (E to H) The timing of ISG expression was calculated for the Cal/09 and TX strains relative to that of the NC strain using 63 key genes from donors 3 to 6, assayed by integrated fluidic circuit real-time PCR.
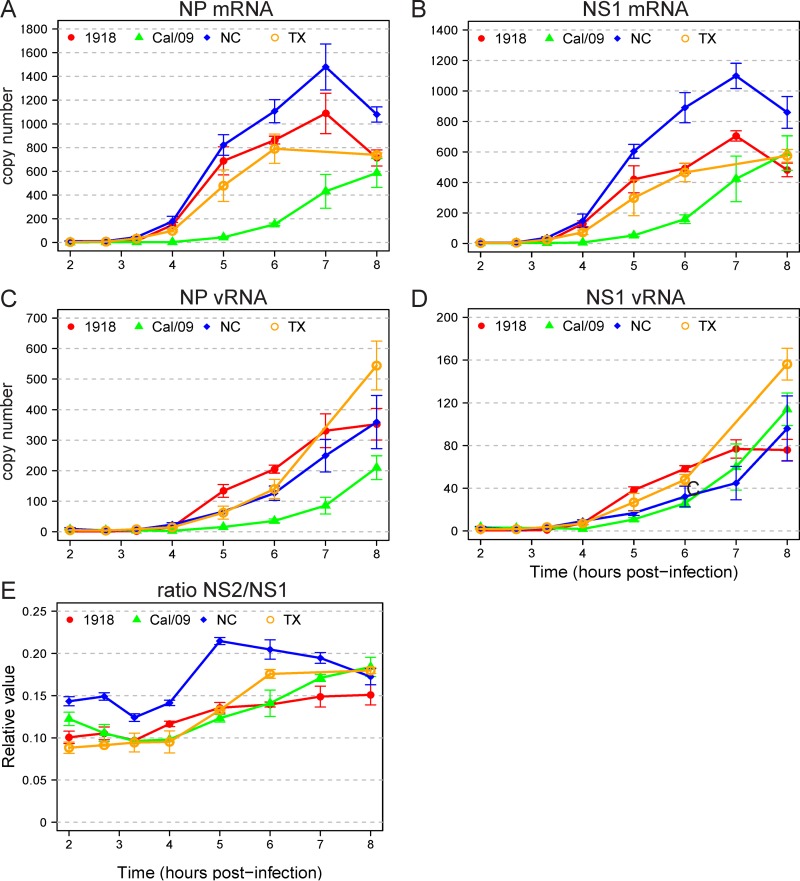
To further characterize the virus-host interactions for the different strains, we measured the expression of viral mRNAs and viral genomic RNAs. Viral mRNAs, which provide a template for the synthesis of virus proteins, are positive-sense RNAs and viral genomic RNAs (vRNAs) are negative-sense RNAs. Therefore, we used strand-specific primers to measure the timing of synthesis of positive-strand NS1 and NP mRNAs and negative-strand NS1 and NP vRNAs for DC infected with each of the four viruses. The 1918 vRNAs were first transcribed at the same time, but to a higher level, than the seasonal virus vRNAs, and the Cal/09 vRNAs were delayed (Fig. 6A to D). The nuclear export protein NEP (NS2) is generated by splicing the same virus transcript that encodes the virus immune antagonist NS1. Differences in the rates of splicing among the viruses might alter the timing of responses after infection. We therefore also assayed the relative levels of spliced NS2 mRNA (encoding NEP) and unspliced NS1 mRNA. The ratio of NS2 mRNA to NS1 mRNA remained between 0.1 and 0.15 for all four viruses at all time points up to 4 h (Fig. 6E). Overall, we found that the very early ISG response following infection with the 1918 strain did not correspond with the relative timing of infb1 induction or viral mRNA or viral genomic RNA production.
FIG 6.
Expression of virus-encoded RNAs. Copy numbers of virus-encoded RNAs were determined by qPCR with the same RNA sample used in the microarray time course experiment. Relative copy numbers for NP mRNA (A), NS1 mRNA (B), NP vRNA (C), and NS1 vRNA (D) for each of the four viruses are depicted. (E) Ratio of spliced NS2 mRNA to unspliced NS1 mRNA.
NC infection uniquely stimulated NF-κB signatures.
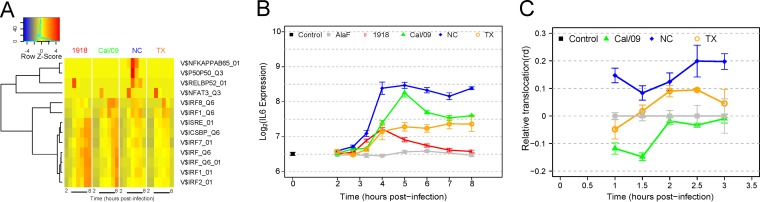
The activities of transcription factors are reflected by alterations in the levels of the genes that they regulate. In order to detect changes in transcription factor activity after infection with different influenza virus strains, we tested for significant overrepresentation of transcription factor targets among upregulated genes (see Materials and Methods). As expected, significant activities of IRF family transcription factors, including IRF7 and IRF9, were detected following infection with all strains (FDR < 0.05). In contrast, NF-κB activity was strain specific, with significant changes in target genes observed only following NC infection (Fig. 7A). This pattern was exemplified by the expression pattern of il6, an NF-κB target gene (54), which was most significantly induced in DC infected with the NC strain (P < 0.01) (Fig. 7B). We tested these computationally predicted differences in NF-κB activity by measuring the levels of NF-κB nuclear translocation in response to infection by different strains using imaging flow cytometry. Supporting the computational results, DC infected with NC showed a much higher early nuclear translocation of NF-κB after infection than did DC infected with Cal/09 or TX (Fig. 7C).
FIG 7.
Infection with the NC strain induces a stronger NF-κB response than those induced by the other viruses. (A) Active transcription factors were detected by overrepresentation of targets among upregulated genes at each time point. Putative targets were identified computationally by scanning the promoter regions for the presence of TRANSFAC binding sites, and the significance of overrepresentation was determined by a hypergeometric test. Colors are row-normalized log10 P values, with red indicating the highest significance. (B) Average mRNA expression profiles of IL-6 following infection with each of the influenza strains. Error bars indicate standard deviations across the microarray replicates. (C) DC were infected with the Cal/09, NC, and TX strains and fixed after 1, 1.5, 2, 2.5, and 3 h. Cells were permeabilized and then stained for NF-κB, and the degree of nuclear translocation was assayed with imaging flow cytometry.
Seasonal strains induce marked mRNA loss.
PCA of the time course data revealed a striking divergence of the responses to infection with the pandemic and with the seasonal strains that occurred after 4 h (Fig. 4). Examination of the temporal trajectories of individual genes showed that the expression of a large number of genes began to decrease at 4 h following infection with the seasonal strains (Fig. 3). The time course of the overall mRNA expression levels showed that the two seasonal viruses caused a dramatic decrease in global mRNA expression, compared with that caused by the pandemic strains, beginning at 4 h after infection (Fig. 8A). The viral mRNAs did not show this pattern, indicating that the decrease in global mRNA was restricted to host genes (Fig. 1).
FIG 8.
Seasonal H1N1 influenza virus infection induces RNA degradation. (A) Distribution of median-normalized mRNA expression intensities across each microarray sample. Boxes indicate the upper 95th and lower 90th percentiles of expression. (B) Levels of pre-mRNA and mRNA for TGFB1. (C) Levels of pre-mRNA and mRNA for CD74. Expression values were normalized to 1.0 at 2 h after infection.
The increase in the global loss of mRNA beginning at 4 h after infection by the seasonal viruses is consistent with an increased rate of mRNA degradation. mRNA levels depend on both synthesis and degradation. Nuclear processing of mRNA is known to be affected by the influenza virus NS1 protein (55). However, an effect on transcription cannot be the predominant driver of mRNA loss with the seasonal virus infections because the median half-life of human mRNAs is on the order of 10 h, which, if unchanged, would not lead to such a rapid loss of transcripts (56). For example, analysis of two representative genes known to have long half-lives, TGFBI and CD74, show a decrease of about 50% of expression between 4 and 6 h after seasonal virus infection, a change that cannot be explained by inhibition of synthesis or RNA processing (Fig. 8B and C). When we measure the levels of pre-mRNA for these genes, we detect an early spike in levels that is consistent with virus-mediated inhibition of pre-mRNA processing and export (Fig. 8B and C). However, the rapidity of mRNA loss supports the presence of widespread mRNA degradation as the mechanism for decreased expression. We also measured the stability of β-actin mRNA (actb) in uninfected DC after exposure to actinomycin D and found that no decrease in mRNA could be detected over a 4-h experiment (data not shown). In contrast, the levels of actb decrease by about 50% between 4 and 8 h after infection by each of the seasonal viruses assayed by both microarray analysis and PCR (data not shown), which therefore must represent induction of mRNA degradation by the seasonal viruses.
The in vitro response to influenza infection mimics the in vivo response.
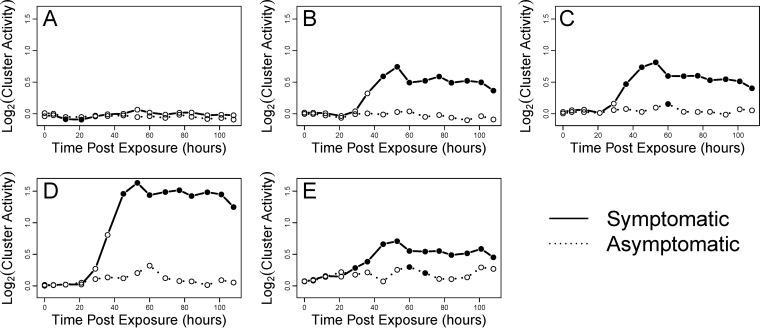
We were interested in determining the relationship of the gene program that we identified in human DC in vitro to the effects of influenza virus infection in humans in vivo. We compared the clusters identified in our data (Fig. 3) to the time course data obtained from human volunteers infected with the H3N2/Wisconsin seasonal influenza virus strain (57). Notably, clusters B, C, D, and E, which show robust upregulation following in vitro infection of DC (Fig. 3), were also found to be activated in blood during in vivo infection (Fig. 9). Interestingly, the significant increase in gene activity of those clusters was almost entirely restricted to the infected subjects who became symptomatic. Furthermore, we found that the one cluster that was specific to the 1918 virus infection in vitro was not activated during the in vivo infection by a seasonal virus of much lower pathogenicity. This result further supports this cluster as a unique signature for the response to the 1918 virus. Overall, these results support the relevance of the comparative study of these viruses in vitro to the systemic responses to influenza virus infection in vivo.
FIG 9.
The DC response to influenza infection is specifically associated with symptomatic influenza virus infection in humans. (A to E) The temporal activities of influenza virus response gene expression clusters were measured in blood samples from a cohort of 17 subjects infected with the seasonal influenza virus strain H3N2/Wisconsin. Subjects were split into symptomatic and asymptomatic groups. Activity was defined by the average fold change of genes in the cluster across the subject group. Significant activity (indicated by filled circles) was determined using QuSAGE, with a threshold P of <0.05.
DISCUSSION
In this work, we compared four H1N1 influenza viruses by studying a detailed time course of the global gene expression changes elicited in human DC. Here, we compared the immune responses to infection with two seasonal and two pandemic viruses. Furthermore, we compared the response elicited by infection with highly pathogenic 1918 to those elicited by infection with the more benign Cal/09 viruses. We found both common and distinguishing features in the responses elicited by the different strains. Among the distinguishing differences were a time shift in the onset of the ISG response and global degradation of mRNA which was caused by infection with the two seasonal influenza strains.
Our experiments were performed with human monocyte-derived DC in vitro, and the limitations of this experimental system must be considered in attempting to make inferences about human infection. In order to gain a better sense of the relevance of our results to human in vivo infection, we compared the data from infected DC to measurements in blood obtained from healthy volunteers infected with a seasonal H3N2 influenza virus. Four of the five gene clusters identified in our experiments (Fig. 3) show a sustained increase in expression following in vivo infection (Fig. 9). Among infected healthy volunteers, only some individuals became symptomatic (57). Notably, these four influenza virus-regulated clusters in DC were all significantly activated in blood cells from symptomatic, but not from asymptomatic, individuals (Fig. 9). Furthermore, cluster A, which is restricted to highly pathogenic 1918 infection in DC, is not regulated in vivo by the influenza strain utilized, which is not highly pathogenic. Thus, there is a striking correspondence between the gene changes observed with DC infection in vitro and with influenza virus infection in vivo.
Interestingly, we do not find strong evidence for overall gene sets that were unique to the seasonal or to the pandemic viruses. Using expression cutoff criteria, we found induced genes that are restricted to both seasonal viruses or both pandemic viruses (Fig. 2). However, further examination of the regulatory changes of these genes showed that most genes appeared to be induced by all viruses but reached the significance threshold in only one of the groups.
However, the responses to individual viruses in DC showed unique features. The NC infection caused the highest activation of NF-κB-regulated genes, a finding that was supported by high levels of NF-κB nuclear translocation induced by this virus (Fig. 7). A rapidly induced gene cluster was unique to the 1918 virus (Fig. 3). Genes comprising this response included the CCL3 gene, which is involved in recruitment and activation of polymorphonuclear leukocytes (58), and the IRAK2 gene, which is implicated in inflammation (59). The gene for transcription factor SP1 was identified by upstream regulatory analysis of the 1918 specific cluster. The induction of SP1 in epithelial cells has been linked to exacerbated responses to IAV infection (60). This association and the presence of inflammatory genes in the 1918-specific cluster are interesting in view of the early and high levels of inflammation associated with 1918 infection in in vivo models (61).
PCA showed commonalities of the seasonal and of the pandemic viruses and overall differences between the two groups (Fig. 3). The pandemic virus Cal/09 induced behavior similar to that of the 1918 virus, although the kinetics of ISG expression was slower than in 1918. Therefore, we characterized the relative timings of the responses to the different influenza strains. The overall ISG response is most rapid following 1918 infection, intermediate for the seasonal viruses, and slowest following Cal/09 infection (Fig. 5). These relative timings do not correspond with the microarray-detected relative timing of ifnb1 induction (Fig. 5B). infb1 is upregulated after infection by all strains by 2 h. Single-cell studies of infb1 induction and its response in conjunction with mathematical modeling suggest that very low levels of ifnb1 generated by a few activated cells, which may not be detected in biochemical assays, may nevertheless be adequate to initiate an ISG response (62, 63). A similar mechanism may underlie the apparent lack of correspondence of ifnb1 levels and timing with the levels and timing of ISG in 1918 infections, unlike in infections with the other viruses. Another possibility is that the seasonal viruses are better at delaying the response to interferon signaling than the 1918 virus is. This possibility is consonant with the high levels of inflammation induced by the 1918 virus, which has been proposed to have contributed to its high mortality.
Overall, these findings identify a core gene response induced by all influenza viruses studied in DC in vitro that also corresponds to the blood transcription gene changes reported with human influenza infection. The presence of this core response in vitro and in vivo suggests the potential relevance of these studies for understanding aspects of human influenza virus infection. The most striking and surprising finding was that the overall responses to the pandemic and seasonal viruses were globally different due to the widespread mRNA loss induced during infection by the seasonal viruses. This global difference in human cellular response to seasonal and pandemic viruses has not been previously identified. Further studies to elucidate the underlying virus and host cell mechanisms for these differences and to investigate their role in influenza pathogenesis are warranted.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIAID contract HHSN272201000054C and grant U19 AI117873.
We thank Yongchao Ge for statistical advice and Irina Nudelman for assistance with the time shift analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01523-15.
REFERENCES
- 1.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Mølbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 2.Opatowski L, Fraser C, Griffin J, de Silva E, Van Kerkhove MD, Lyons EJ, Cauchemez S, Ferguson NM. 2011. Transmission characteristics of the 2009 H1N1 influenza pandemic: comparison of 8 Southern Hemisphere countries. PLoS Pathog 7:e1002225. doi: 10.1371/journal.ppat.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed R, Oldstone MB, Palese P. 2007. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol 8:1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worobey M, Han GZ, Rambaut A. 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A 111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SM, Chan RW, Gardy JL, Lo CK, Sihoe AD, Kang SS, Cheung TK, Guan YI, Chan MC, Hancock RE, Peiris MJ. 2010. Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir Res 11:147. doi: 10.1186/1465-9921-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee N, Wong CK, Chan PK, Chan MC, Wong RY, Lun SW, Ngai KL, Lui GC, Wong BC, Lee SK, Choi KW, Hui DS. 2011. Cytokine response patterns in severe pandemic 2009 H1N1 and seasonal influenza among hospitalized adults. PLoS One 6:e26050. doi: 10.1371/journal.pone.0026050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau LL, Cowling BJ, Fang VJ, Chan KH, Lau EH, Lipsitch M, Cheng CK, Houck PM, Uyeki TM, Peiris JS, Leung GM. 2010. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis 201:1509–1516. doi: 10.1086/652241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. 1998. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Investig 101:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy AW, Platts-Mills TA, Lobo M, Hayden F. 1998. Respiratory nitric oxide levels in experimental human influenza. Chest 114:452–456. doi: 10.1378/chest.114.2.452. [DOI] [PubMed] [Google Scholar]
- 11.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention (CDC). 2010. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb Mortal Wkly Rep 59:1057–1062. [PubMed] [Google Scholar]
- 13.Johnson NP, Mueller J. 2002. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med 76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 14.Taubenberger JK, Morens DM. 2006. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1209.05-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, Zaki SR, Hayden FG, Hui DS, Kettner JD, Kumar A, Lim M, Shindo N, Penn C, Nicholson KG, Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza. 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med 362:1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 16.Reis e Sousa C. 2004. Activation of dendritic cells: translating innate into adaptive immunity. Curr Opin Immunol 16:21–25. doi: 10.1016/j.coi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Servet-Delprat C, Vidalain PO, Bausinger H, Manie S, Le Deist F, Azocar O, Hanau D, Fischer A, Rabourdin-Combe C. 2000. Measles virus induces abnormal differentiation of CD40 ligand-activated human dendritic cells. J Immunol 164:1753–1760. doi: 10.4049/jimmunol.164.4.1753. [DOI] [PubMed] [Google Scholar]
- 18.Chougnet C, Gessani S. 2006. Role of gp120 in dendritic cell dysfunction in HIV infection. J Leukoc Biol 80:994–1000. doi: 10.1189/jlb.0306135. [DOI] [PubMed] [Google Scholar]
- 19.Moutaftsi M, Mehl AM, Borysiewicz LK, Tabi Z. 2002. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 99:2913–2921. doi: 10.1182/blood.V99.8.2913. [DOI] [PubMed] [Google Scholar]
- 20.Mariotti S, Teloni R, Iona E, Fattorini L, Giannoni F, Romagnoli G, Orefici G, Nisini R. 2002. Mycobacterium tuberculosis subverts the differentiation of human monocytes into dendritic cells. Eur J Immunol 32:3050–3058. doi:. [DOI] [PubMed] [Google Scholar]
- 21.Hanekom WA, Mendillo M, Manca C, Haslett PA, Siddiqui MR, Barry C III, Kaplan G. 2003. Mycobacterium tuberculosis inhibits maturation of human monocyte-derived dendritic cells in vitro. J Infect Dis 188:257–266. doi: 10.1086/376451. [DOI] [PubMed] [Google Scholar]
- 22.Medina RA, García-Sastre A. 2011. Influenza A viruses: new research developments. Nat Rev Microbiol 9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kochs G, García-Sastre A, Martínez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, García-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osterlund P, Pirhonen J, Ikonen N, Rönkkö E, Strengell M, Mäkelä SM, Broman M, Hamming OJ, Hartmann R, Ziegler T, Julkunen I. 2010. Pandemic H1N1 2009 influenza A virus induces weak cytokine responses in human macrophages and dendritic cells and is highly sensitive to the antiviral actions of interferons. J Virol 84:1414–1422. doi: 10.1128/JVI.01619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Imai M, Watanabe S, Shinya K, Hatta M, Li C, Neumann G, Ozawa M, Hanson A, Zhong G, Fukuyama S, Kawakami E, Simmons HA, Schenkman D, Brunner K, Capuano SV III, Weinfurter JT, Kilander A, Dudman SG, Suresh M, Hungnes O, Friedrich TC, Kawaoka Y. 2012. Characterization in vitro and in vivo of pandemic (H1N1) 2009 influenza viruses isolated from patients. J Virol 86:9361–9368. doi: 10.1128/JVI.01214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, Webster RG, Hoffmann E. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med 203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc Natl Acad Sci U S A 104:6806–6811. doi: 10.1073/pnas.0701849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 77:7214–7224. doi: 10.1128/JVI.77.13.7214-7224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 31.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trifonov V, Racaniello V, Rabadan R. 2009. The contribution of the PB1-F2 protein to the fitness of influenza A viruses and its recent evolution in the 2009 influenza A (H1N1) pandemic virus. PLoS Curr 1:RRN1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, García-Sastre A, Tumpey TM, Palese P. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J Virol 84:4442–4450. doi: 10.1128/JVI.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng XW, Lu J, Wu CL, Yi LN, Xie X, Shi XD, Fang SS, Zan H, Kung HF, He ML. 2011. Three fatal cases of pandemic 2009 influenza A virus infection in Shenzhen are associated with cytokine storm. Respir Physiol Neurobiol 175:185–187. doi: 10.1016/j.resp.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Oldstone MB, Rosen H. 2014. Cytokine storm plays a direct role in the morbidity and mortality from influenza virus infection and is chemically treatable with a single sphingosine-1-phosphate agonist molecule. Curr Top Microbiol Immunol 378:129–147. doi: 10.1007/978-3-319-05879-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolen CR, Ding S, Robek MD, Kleinstein SH. 2014. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XX, Du N, Zhou JF, Li Z, Wang M, Guo JF, Wang DY, Shu YL. 2010. Gene expression profiles comparison between 2009 pandemic and seasonal H1N1 influenza viruses in A549 cells. Biomed Environ Sci 23:259–266. doi: 10.1016/S0895-3988(10)60061-X. [DOI] [PubMed] [Google Scholar]
- 39.Ramírez-Martínez G, Cruz-Lagunas A, Jiménez-Alvarez L, Espinosa E, Ortíz-Quintero B, Santos-Mendoza T, Herrera MT, Canché-Pool E, Mendoza C, Bañales JL, García-Moreno SA, Morán J, Cabello C, Orozco L, Aguilar-Delfín I, Hidalgo-Miranda A, Romero S, Suratt BT, Selman M, Zúñiga J. 2013. Seasonal and pandemic influenza H1N1 viruses induce differential expression of SOCS-1 and RIG-I genes and cytokine/chemokine production in macrophages. Cytokine 62:151–159. doi: 10.1016/j.cyto.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SM, Gardy JL, Cheung CY, Cheung TK, Hui KP, Ip NY, Guan Y, Hancock RE, Peiris JS. 2009. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS One 4:e8072. doi: 10.1371/journal.pone.0008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. 2014. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. mBio 5(1):e01102–01113. doi: 10.1128/mBio.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Sesma A, Marukian S, Ebersole BJ, Kaminski D, Park MS, Yuen T, Sealfon SC, Garcia-Sastre A, Moran TM. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol 80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, García-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorenshteyn D, Zaslavsky E, Fribourg M, Park CY, Wong AK, Tadych A, Hartmann BM, Albrecht RA, Garcia-Sastre A, Kleinstein SH, Troyanskaya OG, Sealfon SC. Interactive comprehensive human data resource to elucidate immune pathways and diseases. Immunity, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, Navarro G, Roach JC, Rosenberger CM, Rust AG, Yudkovsky N, Aderem A, Shmulevich I. 2008. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol 4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakar J, Hartmann B, Marjanovic N, Sealfon S, Kleinstein S. 2015. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol 16:46. doi: 10.1186/s12865-015-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conde L, Vaquerizas JM, Ferrer-Costa C, de la Cruz X, Orozco M, Dopazo J. 2005. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res 33:W501–W505. doi: 10.1093/nar/gki476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, Haussler D, Miller W. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 69:912–920. [PubMed] [Google Scholar]
- 53.Hartmann BM, Marjanovic N, Nudelman G, Moran TM, Sealfon SC. 2014. Combinatorial cytokine code generates anti-viral state in dendritic cells. Front Immunol 5:73. doi: 10.3389/fimmu.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Libermann TA, Baltimore D. 1990. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol 10:2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hale BG, Randall RE, Ortín J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 56.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13:1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, Øien NC, McClain MT, Varkey JB, Nicholson B, Carin L, Kingsmore S, Woods CW, Ginsburg GS, Hero AO. 2011. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza A infection. PLoS Genet 7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cook DN. 1996. The role of MIP-1 alpha in inflammation and hematopoiesis. J Leukoc Biol 59:61–66. [DOI] [PubMed] [Google Scholar]
- 59.Wan Y, Kim TW, Yu M, Zhou H, Yamashita M, Kang Z, Yin W, Wang JA, Thomas J, Sen GC, Stark GR, Li X. 2011. The dual functions of IL-1 receptor-associated kinase 2 in TLR9-mediated IFN and proinflammatory cytokine production. J Immunol 186:3006–3014. doi: 10.4049/jimmunol.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbier D, Garcia-Verdugo I, Pothlichet J, Khazen R, Descamps D, Rousseau K, Thornton D, Si-Tahar M, Touqui L, Chignard M, Sallenave JM. 2012. Influenza A induces the major secreted airway mucin MUC5AC in a protease-EGFR-extracellular regulated kinase-Sp1-dependent pathway. Am J Respir Cell Mol Biol 47:149–157. doi: 10.1165/rcmb.2011-0405OC. [DOI] [PubMed] [Google Scholar]
- 61.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, García-Sastre A, Swayne DE, Katze MG. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J, Nudelman G, Shimoni Y, Kumar M, Ding Y, Lopez C, Hayot F, Wetmur JG, Sealfon SC. 2011. Role of cell-to-cell variability in activating a positive feedback antiviral response in human dendritic cells. PLoS One 6:e16614. doi: 10.1371/journal.pone.0016614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu J, Sealfon SC, Hayot F, Jayaprakash C, Kumar M, Pendleton AC, Ganee A, Fernandez-Sesma A, Moran TM, Wetmur JG. 2007. Chromosome-specific and noisy IFNB1 transcription in individual virus-infected human primary dendritic cells. Nucleic Acids Res 35:5232–5241. doi: 10.1093/nar/gkm557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.