ABSTRACT
Human papillomavirus 18 (HPV18) is the second most carcinogenic HPV type, after HPV16, and it accounts for approximately 12% of squamous cell carcinoma (SCC) as well as 37% of adenocarcinoma (ADC) of the cervix worldwide. We aimed to evaluate the worldwide diversity and carcinogenicity of HPV18 genetic variants by sequencing the entire long control region (LCR) and the E6 open reading frame of 711 HPV18-positive cervical samples from 39 countries, taking advantage of the International Agency for Research on Cancer biobank. A total of 209 unique HPV18 sequence variants were identified that formed three phylogenetic lineages (A, B, and C). A and B lineages each divided into four sublineages, including a newly identified candidate B4 sublineage. The distribution of lineages varied by geographical region, with B and C lineages found principally in Africa. HPV18 (sub)lineages were compared between 453 cancer cases and 236 controls, as well as between 81 ADC and 160 matched SCC cases. In region-stratified analyses, there were no significant differences in the distribution of HPV18 variant lineages between cervical cancer cases and controls or between ADC and SCC. In conclusion, our findings do not support the role of HPV18 (sub)lineages for discriminating cancer risk or explaining why HPV18 is more strongly linked with ADC than SCC.
IMPORTANCE This is the largest and most geographically/ethnically diverse study of the genetic variation of HPV18 to date, providing a comprehensive reference for phylogenetic classification of HPV18 sublineages for epidemiological and biological studies.
INTRODUCTION
High-risk human papillomavirus (HPV) types are the etiological agents of cervical cancer. HPV18 was first described in 1984 (1) and is the prototype member of the alpha-7 HPV species. Based upon its enrichment in cervical cancer compared to the level in cytologically normal women (2) and its presence in 16% of cervical cancers worldwide (3), HPV18 is widely accepted as the second most carcinogenic HPV type after HPV16.
HPV18 is known to be present in a higher proportion of cervical adenocarcinomas (ADC) (∼37%) than cervical squamous cell carcinomas (SCC) (∼12%), an attribute that is shared by other members of the alpha-7 species (3). This suggests a phylogenetic trait denoting a tendency to cause ADC, and previous studies have suggested that the association with ADC is driven by particular HPV18 variant lineages (4–6).
Most HPV18 infections are asymptomatic and are cleared by the immune system. Factors that favor a small proportion of HPV18 infections to progress to cervical cancer are poorly understood, but studies have implicated a role for HPV18 genetic variation (7–11).
Based upon common phylogenetic patterns of single-nucleotide polymorphisms (SNPs) in the L1 viral genomic region, HPV18 variants originally were classified as European (E), Asian Amerindian (AA), or African (AFR) (12). This classification has been superseded by a whole viral genome sequencing approach that has defined three major lineages (A, B, and C) and additional sublineages (A1 to A5 and B1 to B3) (13) that can be largely translated to the historical nomenclature (A1 and A2 = AA, A3 to A5 = E, and B/C = AFR) (14). In the whole-genome approach, differences of ∼1.0% define variant lineages, and differences of 0.5 to 0.9% define sublineages.
Using a multicenter case-control study design based upon HPV18-positive samples from the biobank at the International Agency for Research on Cancer (IARC), the aim of the current study was to evaluate the genetic diversity of HPV18 worldwide and to explore the association between HPV18 genetic variants and the risk for cervical cancer in geographically and ethnically diverse female populations.
MATERIALS AND METHODS
Origin of clinical specimens.
The IARC has coordinated cervical cancer case series, cervical cancer case-control studies, and population-based HPV prevalence surveys in a large number of countries around the world (15–35 and an as-yet-unpublished study from Rwanda). The collection of samples has spanned a period of over 25 years from 1989 until 2014 and predates the introduction of HPV vaccines. Informed consent was obtained from all participants, and the studies were approved by the IARC Ethical Review Committee. Cervical samples (exfoliated cells or tissue biopsy specimens) derived from these studies have been comprehensively genotyped for HPV type by using a standardized and well-validated protocol (general primer GP5+/6+ PCR-enzyme immunoassay followed by reverse line blot assay) (36) in one centralized laboratory (Molecular Pathology Unit, Department of Pathology, VU University Medical Center, Amsterdam, The Netherlands). All HPV18-positive cervical samples in the IARC biobank were selected for the current analysis, without exclusion, and were categorized into the following regions: northern Africa, sub-Saharan Africa, eastern Asia and Pacific, South/Central Asia, Europe, North America, and South America/Central America.
PCR and DNA sequencing.
DNA extraction from stored samples was performed using a High-Pure PCR template preparation kit (Roche, Mannheim, Germany) or a QIAamp DNA minikit (Qiagen, Hilden, Germany). PCR was performed using a series of M13-tagged HPV18-specific primer pairs that were designed to amplify overlapping regions of the HPV18 long control region (LCR) and E6 open reading frame in order to cover the entire region (see Table S1 in the supplemental material). If a set of three overlapping primers was not able to amplify a sample, then a set of five overlapping primers creating smaller amplicons was used. Each sample underwent two rounds of PCR for each primer pair: 2 μl of DNA in a 10-μl PCR followed by 2.5 μl of PCR product in a 25-μl PCR using the Qiagen multiplex PCR master mix (Qiagen, Hilden, Germany). PCR success was evaluated on a 1.5% agarose gel, and successful reactions were Sanger sequenced in both directions at GATC Biotech (Constance, Germany) using universal M13 sequencing primers, the sequences of which had been incorporated into the PCR products.
The forward and reverse sequence traces of three or five amplicons for each sample were compiled to provide one sequence encompassing the entire HPV18 LCR and E6 open reading frame (nucleotides 7137 to 581) using Geneious v 6.1.8 (Biomatters, Auckland, New Zealand) and compared to the corresponding region of the HPV18 reference sequence (NCBI GenBank accession number X05105 (37), revised as previously described (38) and downloaded from PaVE (pave.niaid.nih.gov) (39). Single nucleotide polymorphisms (SNPs) were confirmed with examination of the sequence chromatograms. All unique sequences were compared pairwise by clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2) using default settings.
Case-control analysis.
Samples were classified as either controls (including normal cells [n = 171], atypical squamous cells of undetermined significance [ASCUS, n = 4], low-grade intraepithelial lesion [LSIL, n = 42], or cervical intraepithelial neoplasia grade 1 [CIN1, n = 6]) or cases (squamous cell carcinoma [n = 339], adenocarcinoma [n = 56], adenosquamous cell carcinoma [n = 25], or unspecified invasive cervical cancer [n = 33]). Samples from population-based HPV prevalence studies for which histology and cytology were unavailable also were classified as controls (n = 13). Samples reported as cervical intraepithelial neoplasia (CIN) grade 2 or 3 or high-grade squamous intraepithelial lesion (HSIL) were excluded from the case-control analysis (n = 19), as were three samples for which sublineage was not able to be determined (two cases and one control). Region-specific associations between variant (sub)lineage and case-control status were assessed by 2-sided P values arising from Fisher's exact test without combining sublineages. Region-specific odds ratios (ORs) and 95% exact confidence intervals (CIs) for the A1 sublineage were calculated against the combination of all other A sublineages.
Case-case analysis.
ADC cases included samples diagnosed as adenocarcinoma (n = 56) or adenosquamous cell carcinoma (n = 25). Two SCC cases were matched by country to each ADC case and, as close as possible, by age. For two of the ADC cases, there was only one SCC case available for matching. The average age was 45.8 and 46.7 years for ADC and SCC cases, respectively. Statistical analysis was performed as described above for the case-control analysis.
Nucleotide sequence accession numbers.
Sequences determined in the course of this work are available from GenBank under accession numbers KP749485 to KP749680.
RESULTS
Sequencing.
The entire LCR and E6 open reading frame were sequenced for a total of 711 HPV18-positive cervical samples from 39 countries, including two countries in northern Africa, nine in sub-Saharan Africa, nine in eastern Asia and the Pacific, five in South/Central Asia, three in Europe, two in North America, and nine in South/Central America (Table 1).
TABLE 1.
Geographic distribution of 711 HPV18-positive cervical samples
| Region and country | No. of samples |
|||
|---|---|---|---|---|
| Case | Control | HSIL/CIN2/3 | Total | |
| Northern Africa | 48 | 5 | 0 | 53 |
| Algeria | 26 | 4 | 0 | 30 |
| Morocco | 22 | 1 | 0 | 23 |
| Sub-Saharan Africa | 75 | 41 | 7 | 123 |
| Benin | 1 | 0 | 0 | 1 |
| Guinea | 2 | 9 | 1 | 12 |
| Kenya | 18 | 0 | 2 | 20 |
| Mali | 16 | 1 | 0 | 17 |
| Nigeria | 1 | 12 | 2 | 15 |
| Rwanda | 0 | 18 | 2 | 20 |
| South Africa | 18 | 1 | 0 | 19 |
| Tanzania | 12 | 0 | 0 | 12 |
| Uganda | 7 | 0 | 0 | 7 |
| Eastern Asia and Pacific | 184 | 90 | 7 | 281 |
| China | 0 | 13 | 5 | 18 |
| Fiji | 0 | 11 | 0 | 11 |
| Indonesia | 24 | 0 | 0 | 24 |
| South Korea | 8 | 4 | 0 | 12 |
| Mongolia | 6 | 24 | 0 | 30 |
| Philippines | 76 | 2 | 0 | 78 |
| Thailand | 69 | 15 | 0 | 84 |
| Vanuatu | 1 | 12 | 2 | 15 |
| Vietnam | 0 | 9 | 0 | 9 |
| South/Central Asia | 31 | 74 | 5 | 110 |
| Bhutan | 5 | 53 | 3 | 61 |
| India | 24 | 16 | 2 | 42 |
| Iran | 1 | 1 | 0 | 2 |
| Nepal | 0 | 4 | 0 | 4 |
| Pakistan | 1 | 0 | 0 | 1 |
| Europe | 24 | 14 | 0 | 38 |
| Georgia | 10 | 9 | 0 | 19 |
| Poland | 9 | 5 | 0 | 14 |
| Spain | 5 | 0 | 0 | 5 |
| North America | 9 | 0 | 0 | 9 |
| Canada | 8 | 0 | 0 | 8 |
| United States | 1 | 0 | 0 | 1 |
| South/Central America | 84 | 13 | 0 | 97 |
| Argentina | 8 | 7 | 0 | 15 |
| Bolivia | 2 | 0 | 0 | 2 |
| Brazil | 15 | 3 | 0 | 18 |
| Chile | 7 | 2 | 0 | 9 |
| Colombia | 3 | 0 | 0 | 3 |
| Cuba | 3 | 0 | 0 | 3 |
| Panama | 11 | 0 | 0 | 11 |
| Paraguay | 26 | 0 | 0 | 26 |
| Peru | 9 | 1 | 0 | 10 |
| Total | 455 | 237 | 19 | 711 |
A total of 189 variations were identified among the 1,302 bases in the LCR and E6 region of the HPV18 genome (14.5% variable nucleotide positions), resulting in 209 unique sequences, or variants, of which 196 were not previously described (see Table S2 in supplemental material). In the LCR there were 126 SNPs, 12 deletions (only three of which were present in more than one sample), and one duplication of 82 bp (present in one sample only). In E6 there were 50 SNPs, 23 of which could result in amino acid changes. The maximum pairwise difference of the LCR and E6 open reading frame sequence between any two variants was 2.72%.
Phylogenetic analysis.
The variants clustered into three main groups (Table 2) corresponding to the previously described lineages A, B, and C (13). In the A lineage, 35 variants, representing 248 samples, were of the same A1 sublineage as the HPV18 reference sequence. Four variants, representing six samples, corresponded to the previously reported A2 sublineage. A3 and A4 sublineages accounted for 86 variants representing 303 samples. A3 appeared to be uniquely defined by the SNP C7486T and A4 to be uniquely defined by the SNP A41G (see Table S2 in supplemental material), but this observation is not consistent with the previously described A3 whole-genome sequence GQ180786 (13), which has 7468C and 41G. Hence, A3 and A4 sublineage variants were combined into one sublineage for the course of this analysis. Twenty-three variants, representing 38 samples, corresponded to the previously reported A5 sublineage.
TABLE 2.
Description of HPV18 variant (sub)lineages based on distinguishing positions in the LCR and E6a
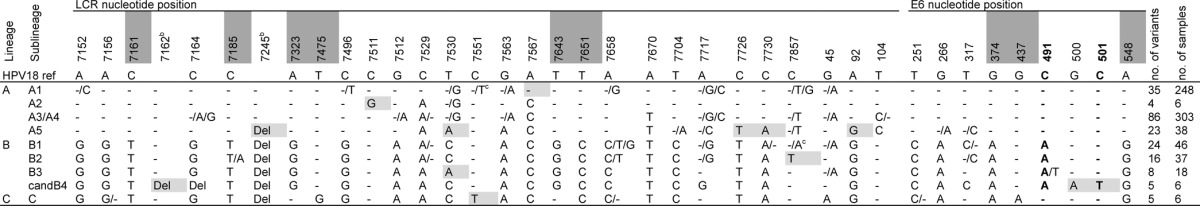
Only SNPs that are consistent for one or more sublineages are included. Nucleotide positions that are diagnostic for a lineage are shaded dark gray. SNPs that are shaded light gray discriminate the sublineage from other sublineages within the same lineage. Boldface indicates an SNP resulting in a change of amino acid: N129K at nucleotide position 491 for the C-to-A SNP and H133Y at nucleotide position 501. Three samples of undetermined sublineage are not shown.
The deletions (Del) are positions 7162 to 7168 and 7245 to 7251.
We did not observe a T at position 7551 for A1 isolates or an A at position 7857 for B1 isolates, but both are reported in the literature (13).
In the B lineage we observed 24 variants, representing 46 samples, that corresponded to the previously reported B1 sublineage; 16 variants, representing 37 samples, corresponded to the previously described B2 sublineage; and eight variants, representing 18 samples, corresponded to the previously described B3 sublineage. In addition, we observed five variants representing six samples that appeared to form a new candidate, sublineage B4 (Table 2). This sublineage was seen in samples from Kenya, Rwanda, Cuba, and Brazil and is approximately 1% different from the other B sublineages in the LCR and E6 region.
We observed five variants representing six samples in the C lineage, with no clear indication of sublineages. Only three variants, each represented by one sample, did not meet the classification described above. Two variants appeared B-like in LCR but A-like in E6 (possibly the amplification of two different variants present in the same sample), and a third variant shared sequence similarity with both the A5 and the A3/A4 sublineages (see Table S2 in supplemental material).
In total, there were 38 nucleotide positions in the LCR and E6 region that could discriminate at least two sublineages from each other (Table 2). Only two of these resulted in an amino acid change (N129K at nucleotide position 491 and H133Y at nucleotide position 501). At the lineage level, nine SNPs were diagnostic, that is, consistently present and unique (dark gray shaded nucleotide positions in Table 2). Each lineage had at least one diagnostic SNP (e.g., C at 7161 for A lineage, G at 7323 for B lineage, and A at 437 for C lineage). Most sublineages had one or more positions that discriminated a sublineage from the other sublineages within the same lineage (light gray positions in Table 2), but only A1 (A at position 7567), A2 (G at position 7511), and candidate B4 (deletion from 7162 to 7168, 500A and 501T) sublineages had diagnostic SNPs that were unique for only that sublineage.
Many other positions were polymorphic within one or more (sub)lineages but did not appear to define phylogenetic subgroups (see Table S2 in supplemental materials). Some potential phylogenetic stratification was observed within A1 (four variants representing 21 samples, all with A89C and A218T), A3/A4 (13 variants representing 72 samples, all with C7164A and C7184T; 21 variants representing 48 samples, all with T7657C), and A5 (seven variants representing nine samples, all with A536C and T553G).
Geographical distribution.
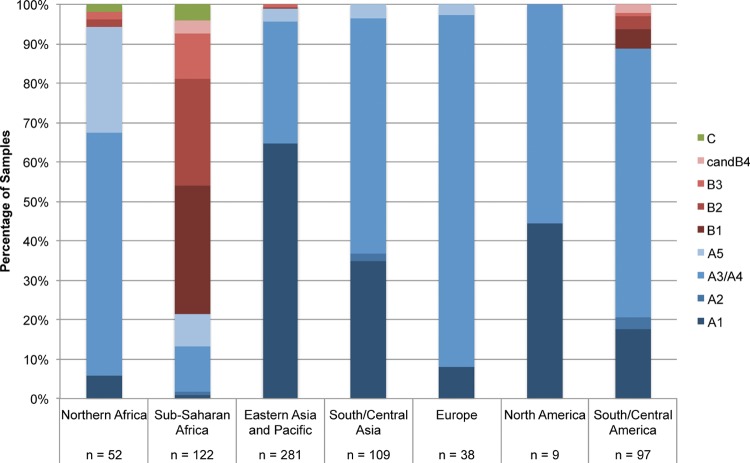
The distribution of HPV18 (sub)lineages varied by geographical region (Fig. 1), with a predominance of the A lineage in most regions except sub-Saharan Africa, where the B lineage predominated. Sub-Saharan Africa showed the broadest range of (sub)lineages. The B lineage also accounted for a small fraction of samples from northern Africa and South/Central America. The C lineage was specific to Africa.
FIG 1.
Distribution of HPV18 (sub)lineages by geographical region, irrespective of cervical diagnosis.
In the A lineage, the A1 sublineage predominated in eastern Asia and the Pacific, whereas the A3/A4 sublineage strongly predominated in Europe but also in northern Africa, South/Central Asia, and South/Central America. The majority of A5 isolates were detected in Africa.
Case-control analysis.
The distribution of HPV18 variant (sub)lineages was compared between invasive cervical cancer cases (n = 453, all histologies) and controls (n = 236) after exclusion of two cases and one control of undetermined sublineage. Within each region, the distribution of the variant lineages (A, B, and C) did not differ significantly between the cases and controls (Table 3).
TABLE 3.
Distribution and statistical comparison of HPV18 lineages between invasive cervical cancer cases and controls
| Region | No. of samples by lineage |
P valuea | ORb (95% CI), A1 vs A2-A5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case |
Control |
|||||||||||
| A1 | A2-A5 | B | C | Total | A1 | A2-A5 | B | C | Total | |||
| Northern Africa | 1 | 43 | 2 | 1 | 47 | 2 | 3 | 0 | 0 | 5 | 1.000 | 0.0 (0.0–1.0) |
| Sub-Saharan Africa | 1 | 15 | 56 | 2 | 74 | 0 | 9 | 29 | 3 | 41 | 0.501 | ∞ (0.0-∞) |
| Eastern Asia and Pacific | 133 | 48 | 3 | 0 | 184 | 45 | 45 | 0 | 0 | 90 | 1.000 | 2.8 (1.6–4.9) |
| South/Central Asia | 4 | 27 | 0 | 0 | 31 | 32 | 41 | 0 | 0 | 73 | 0.2 (0.0–0.6) | |
| Europe | 2 | 22 | 0 | 0 | 24 | 1 | 13 | 0 | 0 | 14 | 1.2 (0.1–75.0) | |
| North America | 4 | 5 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | ||
| South/Central America | 15 | 58 | 11 | 0 | 84 | 2 | 11 | 0 | 0 | 13 | 0.351 | 1.4 (0.3–14.5) |
| Total | 160 | 218 | 72 | 3 | 453 | 82 | 122 | 29 | 3 | 236 | NAc | NAc |
Comparing the distribution in the three lineages (A, B, and C) in cases and controls. Fisher's exact test was used.
Exact confidence intervals.
NA, not appropriate due to strong heterogeneity in lineage distribution by region.
At the sublineage level, A1 variants were overrepresented in cases compared to controls in eastern Asia and the Pacific (OR, 2.8; 95% CI, 1.6 to 4.9) (Table 3). However, the opposite was true in South/Central Asia (OR, 0.2; 95% CI, 0.0 to 0.6), driven by a significant overrepresentation of A3/A4 variants in cases versus controls (see Table S3 in supplemental material). No marked differences in the distribution of sublineages were observed by case-control status in other regions (see Table S3).
Results of the case-control analysis were not materially affected upon the reinclusion of the 19 excluded HSIL/CIN2/3 as cases or in sensitivity analyses restricting to cases with single HPV18 infection or SCC histology only.
Case-case analysis.
Upon comparison of 81 ADC and 160 matched SCC cases, the distribution of variant lineages did not differ between ADC and SCC (Table 4), either overall or within any geographical region. Furthermore, no significant differences were observed when variants were classified at the sublineage level or when the 56 adenocarcinomas and 25 adenosquamous cell carcinomas were analyzed separately (data not shown).
TABLE 4.
Distribution and statistical comparison of HPV18 lineages between ADC and SCC: a matched case-case analysis
| Region | No. of samples by lineage |
Fisher's exact test P value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Adeno |
Squamous |
||||||||
| A | B | C | Subtotal | A | B | C | Subtotal | ||
| Northern Africa | 4 | 0 | 0 | 4 | 7 | 0 | 1 | 8 | 1.000 |
| Sub-Saharan Africa | 2 | 10 | 0 | 12 | 5 | 18 | 0 | 23 | 1.000 |
| Eastern Asia and Pacific | 40 | 2 | 0 | 42 | 84 | 0 | 0 | 84 | 0.109 |
| South/Central Asia | 6 | 0 | 0 | 6 | 11 | 0 | 0 | 11 | |
| Europe | 4 | 0 | 0 | 4 | 8 | 0 | 0 | 8 | |
| North America | 2 | 0 | 0 | 2 | 4 | 0 | 0 | 4 | |
| South/Central America | 9 | 2 | 0 | 11 | 19 | 3 | 0 | 22 | 1.000 |
| Total | 67 | 14 | 0 | 81 | 138 | 21 | 1 | 160 | 0.629 |
DISCUSSION
Among 711 HPV18-positive cervical samples, with a high proportion of isolates from Africa and Asia, we observed 209 unique variants in the LCR and E6 region, which is greater than that of any previous report (13). In addition to previously described lineages and sublineages (A1 to A5, B1 to B3, and C), we saw evidence of a candidate B4 sublineage. We also saw potential sublineages within the A lineage, the most common of these being 13 A3-type variants (representing 72 samples), all with C7164A and C7184T, a combination of SNPs that also has been recorded in A3-type variants elsewhere (10, 40, 41).
The LCR was confirmed to contain much more phylogenetic information than E6 and distinguished nine (sub)lineages (A1, A2, A3/A4, A5, B1, B2, B3, candidate B4, and C) without the requirement for E6. However, even LCR and E6 combined did not allow us to clearly distinguish A3 from A4 variants. A3 appeared to be defined by C7486T and A4 by A41G, but this classification did not hold true for the previously described A3 whole-genome sequence GQ180786 (13). For epidemiological studies based on the detection of SNPs in E6 and/or the LCR, we propose a practical classification of variant lineages using 38 nucleotide positions (29 and nine positions in LCR and E6, respectively) that can each distinguish at least two of the nine (sub)lineages described above from each other (Table 2). Nevertheless, there is much redundancy in this classification, so that not all of these positions are required for classification. Indeed, there are a smaller number of diagnostic SNPs that are specific for a given (sub)lineage.
Our LCR/E6 classification fits with other HPV18 sequences reported in the literature (10, 12, 40–50) and is consistent with lineage definitions based on the gold standard approach of HPV18 whole-genome sequencing (13). Although based on LCR and E6 only, our analysis included many novel variants, particularly among previously underrepresented A5 and B (sub)lineages, which would warrant sequencing the whole genomes in order to strengthen the full picture of HPV18 genetic evolution.
As we have shown previously for HPV16 (51), HPV33 (52), and HPV45 (53), the distribution of HPV18 variant (sub)lineages around the world was confirmed to be geographically/ethnically specific (Fig. 1). Consistent with previous multiregional reports (5, 12, 13), we saw a predominance of B lineages (previously named African variants [14]) in sub-Saharan Africa, whereas there was a strong predominance of A3/A4 lineages (previously named European) in Europe. Consistent with the previous nomenclature of A1/A2 lineages as Asian-Amerindian variants (14), the A1 sublineage was found predominantly in eastern Asia and the Pacific and also was frequent in South/Central Asia and the Americas. We additionally showed that C lineage variants were highly specific for Africa, although they were rarely detected in our series and appear to have been present in only a few cases in previous studies (12, 13, 46, 47). Of note, the A5 sublineage, which has only recently been reported in a single isolate from Thailand (13, 54), was found in a higher proportion of samples from northern Africa than elsewhere. The fact that the LCR of A5 sublineage variants share many features with B and C lineage variants suggests that northern African HPV18 isolates represent a previously understudied branch of HPV18 evolution, as observed for HPV16 (51).
Due to the geographic heterogeneity of variant (sub)lineages, we performed case-control comparisons stratified by region. Using this approach, we were not able to observe any difference in cervical cancer risk between HPV18 variants at the lineage level (Table 3).
Other than one small study that detected a nonsignificant difference of lineage for controls versus CIN3+ cases in Spain (45), previous studies have compared HPV18 variants and clinical outcome using the older E/AA/AFR nomenclature. No significant difference in CIN3+ risk was observed between E/AA and AFR variants during 7 years of follow-up of 221 HPV18-positive women from Costa Rica (55), and no differences were observed in the distribution of HPV18 E/AA versus AFR lineages between 47 controls and 51 cancers in the United States (40) or in two smaller comparisons of cancers and controls (4, 46).
A U.S. study reported that HPV18 variants are more likely to be detected (56), persist (56), and progress to CIN3+ (11) in a host whose race indicates an ancestral geographic distribution that was once shared with that of the variant. However, such a race-risk interaction was not apparent in Brazil (57), and we saw no clear evidence that B/C lineages were more likely to cause cancer in Africa or that A lineages were more likely to cause cancer in Europe. However, there was a significant overrepresentation of A1 variants in cancer in eastern Asia and the Pacific. As A1 variants account for the large fraction of the formerly named AA lineage (together with the rarely detected A2), this is consistent with the U.S. study that reported the risk of developing CIN3+ to be significantly higher for AA than AFR variants (11). However, we did not see this pattern in other regions, and A1 variants actually were significantly underrepresented in cases from South/Central Asia. Such regional differences could be considered consistent with the type of race-associated risk differences described above. Alternatively, these differences may be chance findings driven by small numbers at the sublineage level and the fact that there is residual heterogeneity between the countries grouped together by region (see Table S3 in supplemental materials). Whatever the underlying cause, such apparent regional differences reveal an inherent complexity in studies of HPV variants and cervical cancer risk and give a warning about the extent to which data can be pooled across countries/regions.
Previous studies suggested that the distribution of HPV18 variants differ between ADC and SCC (4–6). However, these studies either included small numbers of HPV18-positive cancers, for which interpretations were based largely on extrapolations of findings on HPV16 (4–6), or were based upon pooling of cases from multiple continents (5). In our much larger comparison, including 81 ADCs, each carefully matched by country and age to two SCCs, we observed no differences in HPV18 variant distribution, either overall or in any individual region. This finding is consistent with a U.S. study in which E/AA/AFR variants were equally distributed among 15 ADC and 10 SCC (40) and suggests that the overrepresentation of HPV18 in ADC (3) is not due to the specific glandular tropism of any HPV18 genetic variant lineage.
We did not observe any changes in amino acids in E6 that are predicted to be critical for zinc binding (cysteine residues 32 and 35, 65 and 68, 105 and 108, and 138 and 141) or the suspected LXXLL binding motif involving Y56 and I130 (see Table S1 in supplemental materials). There were no variants with SNPs at the E6* splice sites involving nucleotides 229 to 233 and 416 to 420 (58), but SNPs at the neighboring nucleotide positions 226 and 227 were observed in 5 samples and 1 sample, respectively. Interestingly, there were 31 samples (12 variants) in the A3/A4 sublineages for which the PDZ binding motif RETQV at amino acid residues 154 to 158 was changed to RATQV (SNP A568C). However, the change from glutamic acid (E) to alanine (A) in the second position of the motif is not predicted to be critical (reviewed in reference 59). Lastly, a C104T SNP in the E6 promoter that has been linked with significantly reduced YY1 transcription factor binding and tumor recurrence (9) was commonly present in A3/A4 and always in A5 variants (Table 2).
Our study was limited by the number of HPV18-positive controls rather than invasive cervical cases, given that it mainly included geographical regions with few or no cervical screening programs. Nevertheless, the reliance on 20 years of IARC studies in a uniquely diverse set of populations from around the world meant that the number of HPV18-positive invasive cervical cancers and controls in the current study was by far the largest studied to date.
In conclusion, the present study provides a practical approach for phylogenetic classification for use in epidemiological studies of the natural history and carcinogenicity of HPV18 genetic variants worldwide. Although some possible differences in carcinogenicity between A sublineages were identified in different regions of Asia, our results suggest that HPV18 variants are not useful for discriminating cancer risk (at least not at the lineage level) and cannot explain why HPV18 is more strongly linked with ADC than SCC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from The Association for International Cancer Research, United Kingdom (project grant 08-0213), the Institut National du Cancer, France (collaboration agreement 07/3D1514/PL-89-05/NG-LC), and the Fondation Innovations en Infectiologie (FINOVI) (project AO1-project 2). The work of A.A.C. was undertaken during the tenure of a postdoctoral fellowship from the International Agency for Research on Cancer, partially supported by the European Commission FP7 Marie Curie Actions–People–Co-funding of Regional, National, and International Programmes (COFUND).
We have no conflicts of interest to declare.
We thank Vanessa Tenet and Jerome Vignat for technical assistance with sample and/or data management, as well as Ikbal Fathallah for assistance with performing sequencing reactions.
The members of the HPV Variant Study Group include the previous IARC staff (N. Muňoz, R. Herrero, and X. Bosch) and local study coordinators in the following countries: Algeria (D. Hammouda), Argentina (D. Loria and E. Matos), Bhutan (U. Tshomo), Bolivia (J. L. Rios-Dalenz), Brazil (J. Eluf-Neto), Canada (P. Ghadirian), Chile (C. Ferreccio and J. M. Ojeda), China (M. Dai, L.K. Li, and R. F. Wu), Cuba (M. Torroella), Fiji (N. Pearce), Georgia (T. Alibegashvili and D. Kordzaia), Guinea (N. Keita and M. Koulibaly), India (T. Rajkumar and R. Rajkumar), Indonesia (Sarjadi), Iran (N. Khodakarami), Kenya (P. Gichangi and H. De Vuyst), South Korea (D.-H. Lee and H. R. Shin), Mali (S. Bayo), Mongolia (B. Dondog), Morocco (N. Chaouki), Nepal (A. T. L. Sherpa), Nigeria (J. O. Thomas), Panama (E. de los Rios), Paraguay (P. A. Rolon), Peru (E. Caceres and C. Santos), Philippines (C. Ngelangel), Poland (A. Bardin and W. Zatonski), Rwanda (F. Ngabo and M. Gatera), South Africa (D. Moodley), Spain (S. de Sanjose and X. Castellsague), Tanzania (J. N. Kitinya), Thailand (S. Chichareon and S. Tunsakul), Uganda (H. R. Wabinga), and Vanuatu (B. Aruhuri and I. H. Frazer).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01747-15.
REFERENCES
- 1.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. 1984. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J 3:1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan P, Clifford GM, Franceschi S. 2013. Human papillomavirus types in glandular lesions of the cervix: a meta-analysis of published studies. Int J Cancer 132:248–250. doi: 10.1002/ijc.27663. [DOI] [PubMed] [Google Scholar]
- 3.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. 2011. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer 128:927–935. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 4.Burk RD, Terai M, Gravitt PE, Brinton LA, Kurman RJ, Barnes WA, Greenberg MD, Hadjimichael OC, Fu L, McGowan L, Mortel R, Schwartz PE, Hildesheim A. 2003. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res 63:7215–7220. [PubMed] [Google Scholar]
- 5.De Boer MA, Peters LA, Aziz MF, Siregar B, Cornain S, Vrede MA, Jordanova ES, Fleuren GJ. 2005. Human papillomavirus type 18 variants: histopathology and E6/E7 polymorphisms in three countries. Int J Cancer 114:422–425. doi: 10.1002/ijc.20727. [DOI] [PubMed] [Google Scholar]
- 6.Lizano M, Berumen J, Guido MC, Casas L, Garcia-Carranca A. 1997. Association between human papillomavirus type 18 variants and histopathology of cervical cancer. J Natl Cancer Inst 89:1227–1231. doi: 10.1093/jnci/89.16.1227. [DOI] [PubMed] [Google Scholar]
- 7.De la Cruz-Hernandez E, Garcia-Carranca A, Mohar-Betancourt A, Duenas-Gonzalez A, Contreras-Paredes A, Perez-Cardenas E, Herrera-Goepfert R, Lizano-Soberon M. 2005. Differential splicing of E6 within human papillomavirus type 18 variants and functional consequences. J Gen Virol 86:2459–2468. doi: 10.1099/vir.0.80945-0. [DOI] [PubMed] [Google Scholar]
- 8.Hecht JL, Kadish AS, Jiang G, Burk RD. 1995. Genetic characterization of the human papillomavirus (HPV) 18 E2 gene in clinical specimens suggests the presence of a subtype with decreased oncogenic potential. Int J Cancer 60:369–376. doi: 10.1002/ijc.2910600317. [DOI] [PubMed] [Google Scholar]
- 9.Rose BR, Thompson CH, Zhang J, Stoeter M, Stephen A, Pfister H, Tattersall MH, Cossart YE. 1997. Sequence variation in the upstream regulatory region of HPV 18 isolates from cervical cancers. Gynecol Oncol 66:282–289. doi: 10.1006/gyno.1997.4740. [DOI] [PubMed] [Google Scholar]
- 10.Sichero L, Franco EL, Villa LL. 2005. Different P105 promoter activities among natural variants of human papillomavirus type 18. J Infect Dis 191:739–742. doi: 10.1086/427825. [DOI] [PubMed] [Google Scholar]
- 11.Xi LF, Koutsky LA, Hildesheim A, Galloway DA, Wheeler CM, Winer RL, Ho J, Kiviat NB. 2007. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol Biomarkers Prev 16:4–10. doi: 10.1158/1055-9965.EPI-06-0670. [DOI] [PubMed] [Google Scholar]
- 12.Ong CK, Chan SY, Campo MS, Fujinaga K, Mavromara-Nazos P, Labropoulou V, Pfister H, Tay SK, ter Meulen J, Villa LL. 1993. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol 67:6424–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Schiffman M, Herrero R, DeSalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2013. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One 8:e72565. doi: 10.1371/journal.pone.0072565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burk RD, Harari A, Chen Z. 2013. Human papillomavirus genome variants. Virology 445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alibegashvili T, Clifford GM, Vaccarella S, Baidoshvili A, Gogiashvili L, Tsagareli Z, Kureli I, Snijders PJ, Heideman DA, van Kemenade FJ, Meijer CJ, Kordzaia D, Franceschi S. 2011. Human papillomavirus infection in women with and without cervical cancer in Tbilisi, Georgia. Cancer Epidemiol 35:465–470. doi: 10.1016/j.canep.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Aruhuri B, Tarivonda L, Tenet V, Sinha R, Snijders PJ, Clifford G, Pang J, McAdam M, Meijer CJ, Frazer IH, Franceschi S. 2012. Prevalence of cervical human papillomavirus (HPV) infection in Vanuatu. Cancer Prev Res 5:746–753. doi: 10.1158/1940-6207.CAPR-11-0515. [DOI] [PubMed] [Google Scholar]
- 17.Bardin A, Vaccarella S, Clifford GM, Lissowska J, Rekosz M, Bobkiewicz P, Kupryjanczyk J, Krynicki R, Jonska-Gmyrek J, Danska-Bidzinska A, Snijders PJ, Meijer CJ, Zatonski W, Franceschi S. 2008. Human papillomavirus infection in women with and without cervical cancer in Warsaw, Poland. Eur J Cancer 44:557–564. doi: 10.1016/j.ejca.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst 87:796–802. [DOI] [PubMed] [Google Scholar]
- 19.Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, Anh PT, Ferreccio C, Hieu NT, Matos E, Molano M, Rajkumar R, Ronco G, de Sanjose S, Shin HR, Sukvirach S, Thomas JO, Tunsakul S, Meijer CJ, Franceschi S. 2005. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet 366:991–998. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- 20.Dai M, Bao YP, Li N, Clifford GM, Vaccarella S, Snijders PJ, Huang RD, Sun LX, Meijer CJ, Qiao YL, Franceschi S. 2006. Human papillomavirus infection in Shanxi Province, People's Republic of China: a population-based study. Br J Cancer 95:96–101. doi: 10.1038/sj.bjc.6603208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Vuyst H, Ndirangu G, Moodley M, Tenet V, Estambale B, Meijer CJ, Snijders PJ, Clifford G, Franceschi S. 2012. Prevalence of human papillomavirus in women with invasive cervical carcinoma by HIV status in Kenya and South Africa. Int J Cancer 131:949–955. doi: 10.1002/ijc.26470. [DOI] [PubMed] [Google Scholar]
- 22.Dondog B, Clifford GM, Vaccarella S, Waterboer T, Unurjargal D, Avirmed D, Enkhtuya S, Kommoss F, Wentzensen N, Snijders PJ, Meijer CJ, Franceschi S, Pawlita M. 2008. Human papillomavirus infection in Ulaanbaatar, Mongolia: a population-based study. Cancer Epidemiol Biomarkers Prev 17:1731–1738. doi: 10.1158/1055-9965.EPI-07-2796. [DOI] [PubMed] [Google Scholar]
- 23.Foliaki S, Brewer N, Pearce N, Snijders PJ, Meijer CJ, Waqatakirewa L, Clifford GM, Franceschi S. 2014. Prevalence of HPV infection and other risk factors in a Fijian population. Infect Agents Cancer 9:14. doi: 10.1186/1750-9378-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschi S, Rajkumar T, Vaccarella S, Gajalakshmi V, Sharmila A, Snijders PJ, Munoz N, Meijer CJ, Herrero R. 2003. Human papillomavirus and risk factors for cervical cancer in Chennai, India: a case-control study. Int J Cancer 107:127–133. doi: 10.1002/ijc.11350. [DOI] [PubMed] [Google Scholar]
- 25.Halec G, Schmitt M, Dondog B, Sharkhuu E, Wentzensen N, Gheit T, Tommasino M, Kommoss F, Bosch FX, Franceschi S, Clifford G, Gissmann L, Pawlita M. 2013. Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. Int J Cancer 132:63–71. doi: 10.1002/ijc.27605. [DOI] [PubMed] [Google Scholar]
- 26.Hammouda D, Munoz N, Herrero R, Arslan A, Bouhadef A, Oublil M, Djedeat B, Fontaniere B, Snijders P, Meijer C, Franceschi S. 2005. Cervical carcinoma in Algiers, Algeria: human papillomavirus and lifestyle risk factors. Int J Cancer 113:483–489. doi: 10.1002/ijc.20600. [DOI] [PubMed] [Google Scholar]
- 27.Keita N, Clifford GM, Koulibaly M, Douno K, Kabba I, Haba M, Sylla BS, van Kemenade FJ, Snijders PJ, Meijer CJ, Franceschi S. 2009. HPV infection in women with and without cervical cancer in Conakry, Guinea. Br J Cancer 101:202–208. doi: 10.1038/sj.bjc.6605140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodakarami N, Clifford GM, Yavari P, Farzaneh F, Salehpour S, Broutet N, Bathija H, Heideman DA, van Kemenade FJ, Meijer CJ, Hosseini SJ, Franceschi S. 2012. Human papillomavirus infection in women with and without cervical cancer in Tehran, Iran. Int J Cancer 131:E156–E161. doi: 10.1002/ijc.26488. [DOI] [PubMed] [Google Scholar]
- 29.Li LK, Dai M, Clifford GM, Yao WQ, Arslan A, Li N, Shi JF, Snijders PJ, Meijer CJ, Qiao YL, Franceschi S. 2006. Human papillomavirus infection in Shenyang City, People's Republic of China: a population-based study. Br J Cancer 95:1593–1597. doi: 10.1038/sj.bjc.6603450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 31.Okolo C, Franceschi S, Adewole I, Thomas JO, Follen M, Snijders PJ, Meijer CJ, Clifford GM. 2010. Human papillomavirus infection in women with and without cervical cancer in Ibadan, Nigeria. Infect Agents Cancer 5:24. doi: 10.1186/1750-9378-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raza SA, Franceschi S, Pallardy S, Malik FR, Avan BI, Zafar A, Ali SH, Pervez S, Serajuddaula S, Snijders PJ, van Kemenade FJ, Meijer CJ, Shershah S, Clifford GM. 2010. Human papillomavirus infection in women with and without cervical cancer in Karachi, Pakistan. Br J Cancer 102:1657–1660. doi: 10.1038/sj.bjc.6605664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherpa AT, Clifford GM, Vaccarella S, Shrestha S, Nygard M, Karki BS, Snijders PJ, Meijer CJ, Franceschi S. 2010. Human papillomavirus infection in women with and without cervical cancer in Nepal. Cancer Causes Control 21:323–330. doi: 10.1007/s10552-009-9467-z. [DOI] [PubMed] [Google Scholar]
- 34.Tshomo U, Franceschi S, Dorji D, Baussano I, Tenet V, Snijders PJ, Meijer CJ, Bleeker MC, Gheit T, Tommasino M, Clifford GM. 2014. Human papillomavirus infection in Bhutan at the moment of implementation of a national HPV vaccination programme. BMC Infect Dis 14:408. doi: 10.1186/1471-2334-14-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu RF, Dai M, Qiao YL, Clifford GM, Liu ZH, Arslan A, Li N, Shi JF, Snijders PJ, Meijer CJ, Franceschi S. 2007. Human papillomavirus infection in women in Shenzhen City, People's Republic of China, a population typical of recent Chinese urbanisation. Int J Cancer 121:1306–1311. doi: 10.1002/ijc.22726. [DOI] [PubMed] [Google Scholar]
- 36.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J Clin Microbiol 40:779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole ST, Danos O. 1987. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. Phylogeny of papillomaviruses and repeated structure of the E6 and E7 gene products. J Mol Biol 193:599–608. [DOI] [PubMed] [Google Scholar]
- 38.Meissner J. 1997. Sequencing errors in reference HPV clones, p III-110–III- 123 In Myers G, Sverdrup F, Baker C, McBride A, Munger K, Bernard H-U, Meissner J (ed), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 39.Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. 2013. The papillomavirus episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res 41:D571–D578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arias-Pulido H, Peyton CL, Torrez-Martinez N, Anderson DN, Wheeler CM. 2005. Human papillomavirus type 18 variant lineages in United States populations characterized by sequence analysis of LCR-E6, E2, and L1 regions. Virology 338:22–34. doi: 10.1016/j.virol.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 41.Vrtacnik Bokal E, Kocjan BJ, Poljak M, Bogovac Z, Jancar N. 2010. Genomic variants of human papillomavirus genotypes 16, 18, and 33 in women with cervical cancer in Slovenia. J Obstet Gynaecol Res 36:1204–1213. doi: 10.1111/j.1447-0756.2010.01316.x. [DOI] [PubMed] [Google Scholar]
- 42.Arroyo SL, Basaras M, Arrese E, Hernaez S, Andia D, Esteban V, Garcia-Etxebarria K, Jugo BM, Cisterna R. 2012. Human papillomavirus (HPV) genotype 18 variants in patients with clinical manifestations of HPV related infections in Bilbao, Spain. Virol J 9:258. doi: 10.1186/1743-422X-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerqueira DM, Raiol T, Veras NM, von Gal Milanezi N, Amaral FA, de Macedo Brigido M, Martins CR. 2008. New variants of human papillomavirus type 18 identified in central Brazil. Virus Genes 37:282–287. doi: 10.1007/s11262-008-0263-8. [DOI] [PubMed] [Google Scholar]
- 44.Ong CK, Nee S, Rambaut A, Bernard HU, Harvey PH. 1997. Elucidating the population histories and transmission dynamics of papillomaviruses using phylogenetic trees. J Mol Evol 44:199–206. doi: 10.1007/PL00006136. [DOI] [PubMed] [Google Scholar]
- 45.Perez S, Cid A, Inarrea A, Pato M, Lamas MJ, Couso B, Gil M, Alvarez MJ, Rey S, Lopez-Miragaya I, Melon S, Ona M. 2014. Prevalence of HPV 16 and HPV 18 lineages in Galicia, Spain. PLoS One 9:e104678. doi: 10.1371/journal.pone.0104678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pista A, Oliveira A, Barateiro A, Costa H, Verdasca N, Paixao MT. 2007. Molecular variants of human papillomavirus type 16 and 18 and risk for cervical neoplasia in Portugal. J Med Virol 79:1889–1897. doi: 10.1002/jmv.21002. [DOI] [PubMed] [Google Scholar]
- 47.Schlecht NF, Burk RD, Palefsky JM, Minkoff H, Xue X, Massad LS, Bacon M, Levine AM, Anastos K, Gange SJ, Watts DH, Da Costa MM, Chen Z, Bang JY, Fazzari M, Hall C, Strickler HD. 2005. Variants of human papillomaviruses 16 and 18 and their natural history in human immunodeficiency virus-positive women. J Gen Virol 86:2709–2720. doi: 10.1099/vir.0.81060-0. [DOI] [PubMed] [Google Scholar]
- 48.Sun Z, Liu J, Wang G, Zhou W, Liu C, Ruan Q. 2012. Variant lineages of human papillomavirus type 18 in northeast China populations characterized by sequence analysis of E6, E7, and L1 regions. Int J Gynecol Cancer 22:930–936. doi: 10.1097/IGC.0b013e318253a994. [DOI] [PubMed] [Google Scholar]
- 49.Villa LL, Sichero L, Rahal P, Caballero O, Ferenczy A, Rohan T, Franco EL. 2000. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J Gen Virol 81:2959–2968. [DOI] [PubMed] [Google Scholar]
- 50.Yang L, Yang H, Wu K, Shi X, Ma S, Sun Q. 2014. Prevalence of HPV and variation of HPV 16/HPV 18 E6/E7 genes in cervical cancer in women in south west China. J Med Virol 86:1926–1936. doi: 10.1002/jmv.24043. [DOI] [PubMed] [Google Scholar]
- 51.Cornet I, Gheit T, Franceschi S, Vignat J, Burk RD, Sylla BS, Tommasino M, Clifford GM, IARC HPV Variant Study Group . 2012. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J Virol 86:6855–6861. doi: 10.1128/JVI.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen AA, Heideman DA, Boon D, Chen Z, Burk RD, De Vuyst H, Gheit T, Snijders PJ, Tommasino M, Franceschi S, Clifford GM. 2014. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology 448:356–362. doi: 10.1016/j.virol.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen AA, Heideman DA, Boon D, Gheit T, Snijders PJ, Tommasino M, Franceschi S, Clifford GM, IARC HPV Variant Study Group. 2014. Human papillomavirus 45 genetic variation and cervical cancer risk worldwide. J Virol 88:4514–4521. doi: 10.1128/JVI.03534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lurchachaiwong W, Junyangdikul P, Termrungruanglert W, Payungporn S, Sampatanukul P, Tresukosol D, Niruthisard S, Trivijitsilp P, Karalak A, Swangvaree S, Poovorawan Y. 2010. Whole-genome sequence analysis of human papillomavirus type 18 from infected Thai women. Intervirology 53:161–166. doi: 10.1159/000274977. [DOI] [PubMed] [Google Scholar]
- 55.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Solomon D, Castle PE, Burk RD. 2010. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res 70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xi LF, Kiviat NB, Hildesheim A, Galloway DA, Wheeler CM, Ho J, Koutsky LA. 2006. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst 98:1045–1052. doi: 10.1093/jnci/djj297. [DOI] [PubMed] [Google Scholar]
- 57.Sichero L, Ferreira S, Trottier H, Duarte-Franco E, Ferenczy A, Franco EL, Villa LL. 2007. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int J Cancer 120:1763–1768. doi: 10.1002/ijc.22481. [DOI] [PubMed] [Google Scholar]
- 58.Sotlar K, Stubner A, Diemer D, Menton S, Menton M, Dietz K, Wallwiener D, Kandolf R, Bultmann B. 2004. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J Med Virol 74:107–116. doi: 10.1002/jmv.20153. [DOI] [PubMed] [Google Scholar]
- 59.Pim D, Bergant M, Boon SS, Ganti K, Kranjec C, Massimi P, Subbaiah VK, Thomas M, Tomaic V, Banks L. 2012. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J 279:3530–3537. doi: 10.1111/j.1742-4658.2012.08709.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.