ABSTRACT
More than 500,000 people die each year from the liver diseases that result from chronic hepatitis B virus (HBV) infection. Therapeutic vaccines, which aim to elicit an immune response capable of controlling the virus, offer a potential new treatment strategy for chronic hepatitis B. Recently, an evolved, high-titer vaccine platform consisting of Semliki Forest virus RNA replicons that express the vesicular stomatitis virus glycoprotein (VSV G) has been described. This platform generates virus-like vesicles (VLVs) that contain VSV G but no other viral structural proteins. We report here that the evolved VLV vector engineered to additionally express the HBV middle surface envelope glycoprotein (MHBs) induces functional CD8 T cell responses in mice. These responses were greater in magnitude and broader in specificity than those obtained with other immunization strategies, including recombinant protein and DNA. Additionally, a single immunization with VLV-MHBs protected mice from HBV hydrodynamic challenge, and this protection correlated with the elicitation of a CD8 T cell recall response. In contrast to MHBs, a VLV expressing HBV core protein (HBcAg) neither induced a CD8 T cell response in mice nor protected against challenge. Finally, combining DNA and VLV-MHBs immunization led to induction of HBV-specific CD8 T cell responses in a transgenic mouse model of chronic HBV infection. The ability of VLV-MHBs to induce a multispecific T cell response capable of controlling HBV replication, and to generate immune responses in a tolerogenic model of chronic infection, indicates that VLV vaccine platforms may offer a unique strategy for HBV therapeutic vaccination.
IMPORTANCE HBV infection is associated with significant morbidity and mortality. Furthermore, treatments for chronic infection are suboptimal and rarely result in complete elimination of the virus. Therapeutic vaccines represent a unique approach to HBV treatment and have the potential to induce long-term control of infection. Recently, a virus-based vector system that combines the nonstructural proteins of Semliki Forest virus with the VSV glycoprotein has been described. In this study, we used this system to construct a novel HBV vaccine and demonstrated that the vaccine is capable of inducing virus-specific immune responses in mouse models of acute and chronic HBV replication. These findings highlight the potential of this new vaccine system and support the idea that highly immunogenic vaccines, such as viral vectors, may be useful in the treatment of chronic hepatitis B.
INTRODUCTION
Despite the availability of an effective prophylactic vaccine, over 240 million people remain chronically infected with hepatitis B virus (HBV) (1). Although HBV infection can often be asymptomatic, chronic infection can lead to long-term consequences, such as liver cirrhosis and hepatocellular carcinoma. Currently, the main options for HBV therapy include nucleos(t)ide analogues and alpha interferon (IFN-α), but these treatments have several limitations. Nucleos(t)ide analogues effectively suppress virus replication but do not eliminate the infection, and once treatment is stopped, the virus rapidly rebounds. Furthermore, long-term treatment with these antivirals can result in the generation of drug-resistant mutants. In contrast to nucleos(t)ide analogues, IFN-α, which has both antiviral and immunomodulatory activities, can produce more durable results in some patients. However, IFN-α treatment is often associated with a high incidence of side effects, which makes it a suboptimal treatment option. Therefore, the design of new effective treatments for HBV-associated infection and disease is essential.
One potential treatment strategy in development is the use of therapeutic vaccines. Though DNA and protein vaccines have been unable to elicit responses capable of controlling HBV infection (2), more immunogenic strategies have shown promising results in animal models. For instance, in a chimpanzee model, a retroviral vector expressing HBcAg induces HBV-specific immune responses and, in some animals, clearance of HBV (3). Adenovirus vectors have also shown efficacy in woodchuck (4) and mouse (5) models of chronic HBV infection. Finally, we have previously shown that a highly immunogenic vesicular stomatitis virus (VSV)-based vector can elicit HBV-specific immune responses in a transgenic mouse model that exhibits a severely tolerized immune response to HBV (6).
Despite the success of these vaccine approaches in experimental animal models, the translational use of virus-based vectors in humans is often hindered by safety concerns. For this reason, in this study, we investigated a novel vaccine vector that lacks detectable pathogenicity in mouse and monkey models (7, 8). This hybrid vector consists of a Semliki Forest virus (SFV) RNA replicon, which expresses the nonstructural proteins (replicase) of SFV and the VSV glycoprotein (G) in place of the normal SFV structural proteins (capsid and glycoproteins). During SFV replication, the nonstructural proteins mediate the formation of light bulb-shaped replication complexes (spherules), which initiate at the plasma membrane and are then trafficked into perinuclear vacuoles (9). When VSV G protein is expressed from an SFV replicon in place of the SFV viral structural proteins, vesicles containing SFV RNA and VSV G bud from the cells (10) and are likely derived from spherules containing VSV G (11). These virus-like vesicles (VLVs) can be further engineered to express additional foreign genes with no strict size limit to the insert (12). Furthermore, VLVs demonstrate an ability to induce potent immune responses while lacking pathogenicity in mouse and nonhuman primate models (7, 8). Recently, this VLV system has been evolved to reach high titers, further improving its potential as a vaccine vector while still lacking detectable pathogenicity (11). In this study, we used this high-titer evolved vector system to explore how VLVs expressing either the middle version of the HBV envelope protein (MHBs) or the HBV core protein (HBcAg) may be developed as a therapeutic vaccine for chronic HBV infection.
MATERIALS AND METHODS
Cell lines, plasmids, and recombinant viruses.
Hamster BHK-21 (BHK) epithelial cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml of penicillin, and 2 mM l-glutamine.
The open reading frame of the HBV (ayw serotype; GenBank accession no. V01460.1) core antigen (HBcAg) and the middle version of the HBV envelope protein (MHBs) were PCR amplified, adding on cloning sites for PacI and Sbf1 on the 5′ and 3′ ends, respectively. Primers used include the following: for MHBs, 5′-GAT CGA TCT TAA TTA AAA TGC AGT GGA ATT CCA CAA CCT TC-3′ (forward) and 5′-GAT CGA TCC CTG CAG GAA TGT ATA CCC AAA GAC AAA AGA AAA TTG-3′ (reverse), and for HBcAg, 5′-GAT CGA TCT TAA TTA AAA TGG ACA TCG ACC CTT ATA AAG AAT TTG (forward) and 5′-GAT CGA TCC CTG CAG GAC ATT GAG ATT CCC GAG ATT GAG ATC-3′ (reverse). The genes were then cloned into plasmid pCMV-SFVT2AG to create pCMV-SFVHBcT2AG and pCMV-SFVmST2AG, respectively. pCMV-SFVT2AG is a plasmid derived from pCMV-SFVG-P50R (11) that has been engineered with a PacI to Sbf1 cloning site downstream of the SFV subgenomic RNA promoter. This plasmid also encodes a ribosomal T2A skipping site (13) downstream of the cloning site but upstream of the Indiana serotype VSV glycoprotein. A similar plasmid encoding the New Jersey serotype VSV glycoprotein (pCMV-SFVT2AGNJ) was also used for cloning in order to produce VLVs that express the HBV proteins and the alternate glycoprotein. VLV-MHBs (Indiana serotype), VLV-HBcAg, or VLVNJ-MHBs was recovered by transfecting BHK cells with plasmid pCMV-SFVmST2AG, pCMV-SFVHBcT2AG, or pCMV-SFVmST2AGNJ, respectively. Cells were observed for cytopathic effects, and supernatants were collected when nearly all cells appeared rounded and/or detached. Supernatants were centrifuged briefly to remove cell debris. In some cases, VLVs were concentrated by centrifugation through a 100-kDa Amicon Ultra filter (EMD Millipore Corporation). Aliquots were stored at −80°C.
VSV-MHBs that encodes the same HBV MHBs sequence as VLV-MHBs was previous described (14). VSV-HBcAg that encodes the same HBV HBcAg sequence as VLV-HBcAg was generated using methods previously described (14).
Quantification of VLV titers was performed by serial dilution on BHK cells and indirect immunofluorescence microscopy to detect VSV G protein expression in infectious centers, and titers were quantified as immunofluorescence units (IFU) per ml. VSV titer determination was completed by serial dilution and plaque assay on BHK cells and quantified as PFU per ml.
Mice.
Six- to 8-week-old C57BL/6 × BALB/c F1 (CB6F1J) mice were obtained from Jackson Laboratory (Farmington, CT). For transgenic (Tg) mouse experiments, 1.3.32 HBV Tg mice (15) and BALB/c mice (Charles River, Wilmington, MA) were crossed for one generation to obtain HBV.CB6F1 mice. Mice were screened at 5 to 6 weeks after birth for serum HBeAg levels as a measure of transgene expression. All mice were housed at the Yale University School of Medicine animal facilities, and all experiments were performed in accordance with Yale Institutional Animal Care and Use Committee-approved procedures.
ELISPOT.
T cell responses were determined using an IFN-γ enzyme-linked immunospot (ELISPOT) set (BD Biosciences) according to the manufacturer's protocol. Briefly, 96-well plates were coated overnight with purified anti-mouse IFN-γ antibody (1:200). Plates were rinsed and then blocked for 2 h using DMEM supplemented with 10% fetal bovine serum, 100 μg/ml of penicillin, and 2 mM l-glutamine. Splenocytes were purified from mice by passaging spleens through 70-μm strainers (BD Falcon) and treating the cell suspension with ACK lysing buffer (Lonza). After washing with Hanks' balanced salt solution (HBSS; Invitrogen), cells were suspended in DMEM and seeded at 2 × 105/well. Peptides representing previously described epitopes present in MHBs and HBcAg (Table 1) were used to stimulate cells overnight at 37°C. Cells were washed from plates using phosphate-buffered saline (PBS)-Tween (0.05% [vol/vol]), and biotinylated anti-mouse IFN-γ antibody (1:250) was added for 2 h at 25°C. After washing, streptavidin-horseradish peroxidase (HRP) (1:100) was added to wells and incubated for 1 h at 25°C. Following the final washes, 3-amino-9-ethyl-carbazole (AEC) chromogen substrate (BD Biosciences) was added to the wells and allowed to develop at 25°C for 20 to 40 min. Distilled water was added to stop the reaction, and the plates were allowed to air dry before spot-forming cells (SFC) were enumerated.
TABLE 1.
HBV surface envelope and HBcAg CD8 T cell epitopesa
| Epitope | Protein | Position | Sequence | MHC | Reference(s) |
|---|---|---|---|---|---|
| 191 | HBV S | 191–202 | IPQSLDSWWTSL | Ld | 30 |
| 353 | HBV S | 353–360 | VWLSVIWM | Kb | 31 |
| 364 | HBV S | 364–372 | WGPSLYSIL | Dd | 32 |
| 371 | HBV S | 371–378 | ILSPFLPL | Kb | 32, 33 |
| 87 | HBcAg | 87–95 | SYVNTNMGL | Kd | 34 |
| 93 | HBcAg | 93–100 | MGLKFRQL | Kb | 35 |
| 131 | HBcAg | 131–139 | AYRPPNAPI | Kd | 36 |
MHC, major histocompatibility complex; S, surface.
ELISAs.
In order to detect antibody responses to MHBs (HBsAb) and HBcAg (HBcAb) and serum HBeAg levels, either direct (HBsAb and HBeAg) or competitive (HBcAb) enzyme-linked immunosorbent assays (ELISAs; International Immunodiagnostics) were performed by following the manufacturer's protocol. Serum samples were diluted in FBS at 1:5 for antibody detection or 1:50 for HBeAg detection.
Immunization and challenge protocols.
Mice were immunized intramuscularly with 107 IFU of VLV-MHBs or VLV-HBcAg, 107 PFU of VSV-MHBs, or 50 μg of pCMV-MHBs. In some cases, mice were boosted 4 weeks postimmunization with 107 IFU/PFU of either VSV-MHBs, VLV-MHBs, or VLVNJ-MHBs.
For HBV challenge experiments, mice were immunized as described above and 6 weeks later challenged using an HBV hydrodynamic challenge protocol (16). Briefly, 10 μg of a plasmid encoding a 1.3-mer overlength copy of the HBV genome (pHBV1.3) was injected into the tail vein of the mouse in a volume approximately equal to 9% of the animal's body weight, resulting in volumetric overload of the circulatory system and uptake of plasmid DNA into hepatocytes. HBV mRNA expression from the plasmid results in production of HBV DNA replication intermediates and proteins.
Isolation of intrahepatic leukocytes.
Intrahepatic leukocytes were isolated as described previously (14). Briefly, liver pieces were passed through a 70-μm-pore strainer, and the resulting suspension was treated for 30 min at 37°C with 0.5 mg/ml of collagenase D (Roche). Cells were then washed with HBSS, resuspended in 44% (vol/vol) Percoll in HBSS, and layered over 56% Percoll in PBS (vol/vol). After centrifugation of the gradient for 30 min at 850 × g, the cells at the interphase were collected. The cells were then washed with HBSS and resuspended in DMEM for further analysis.
Immunohistochemistry.
Liver tissue was collected and fixed in 10% buffered formalin phosphate (Fisher Scientific). After paraffin embedding, HBV core protein was detected by immunohistochemical staining performed by Yale University Research Histology using anti-HBcAg polyclonal rabbit antibody (Dako).
Data analysis.
Data are shown as means of the indicated number of replicates, and error bars indicate standard errors of the means. A 2-tailed Student t test was used to determine significant differences in CD8 T cell responses in normal and transgenic mice. P values of <0.05 were considered statistically significant.
RESULTS
VLV construction and expression.
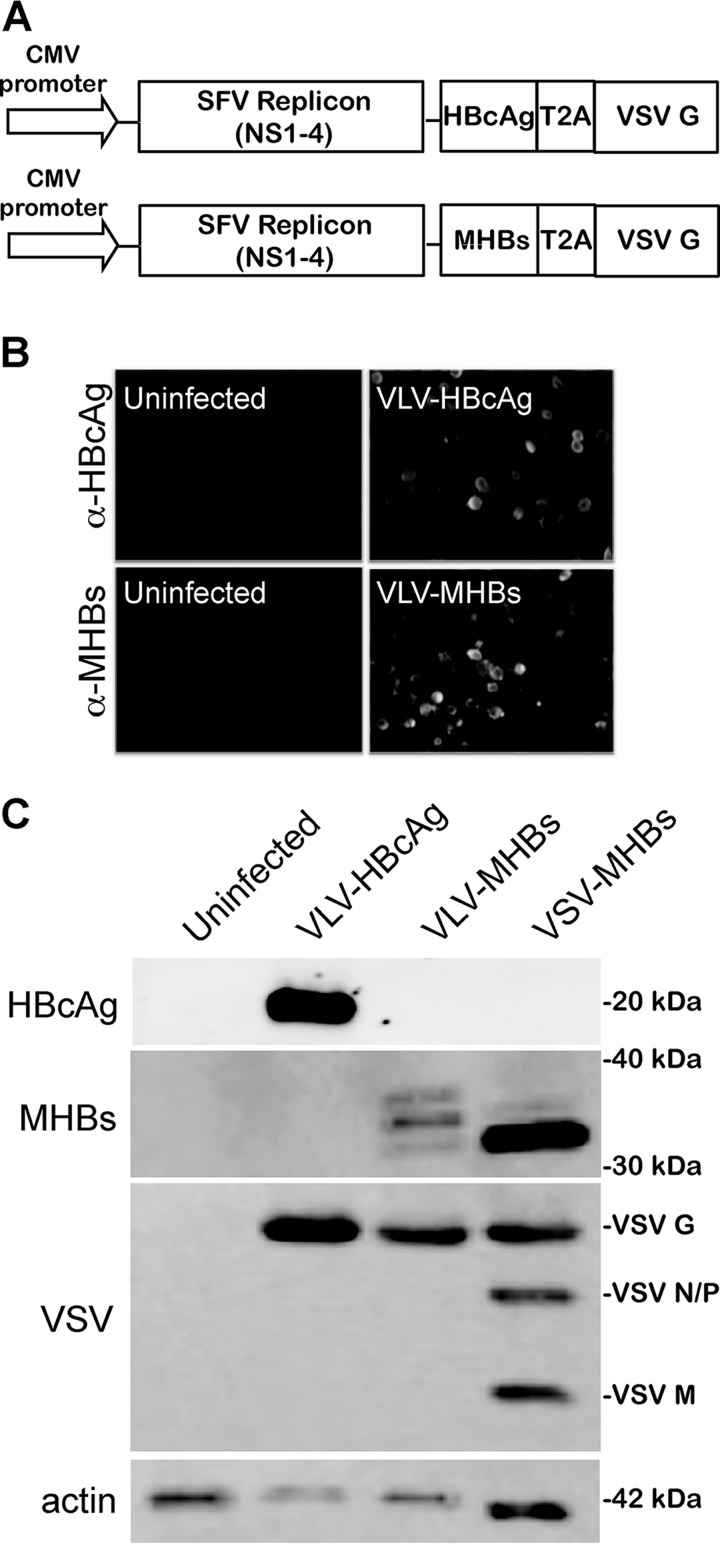
In order to construct VLV vectors that express HBV proteins, we PCR amplified MHBs and HBcAg coding sequences, adding restriction sites for PacI and Sbf1 on the 5′ and 3′ ends, respectively. The fragments were then cloned into a DNA plasmid encoding the SFV nonstructural proteins and the Indiana serotype VSV G. This plasmid has been engineered so that the gene of interest is inserted upstream of a ribosomal T2A skipping site (13) and the VSV G protein (Fig. 1A). The T2A site allows for expression of both the HBV protein and the VSV G protein from the same promoter and, importantly, ensures readthrough of the HBV protein prior to VLV production. This expression strategy gives a more stable platform than a dual-promoter platform (data not shown), which has been found to rapidly lose expression of the foreign protein after passage (12).
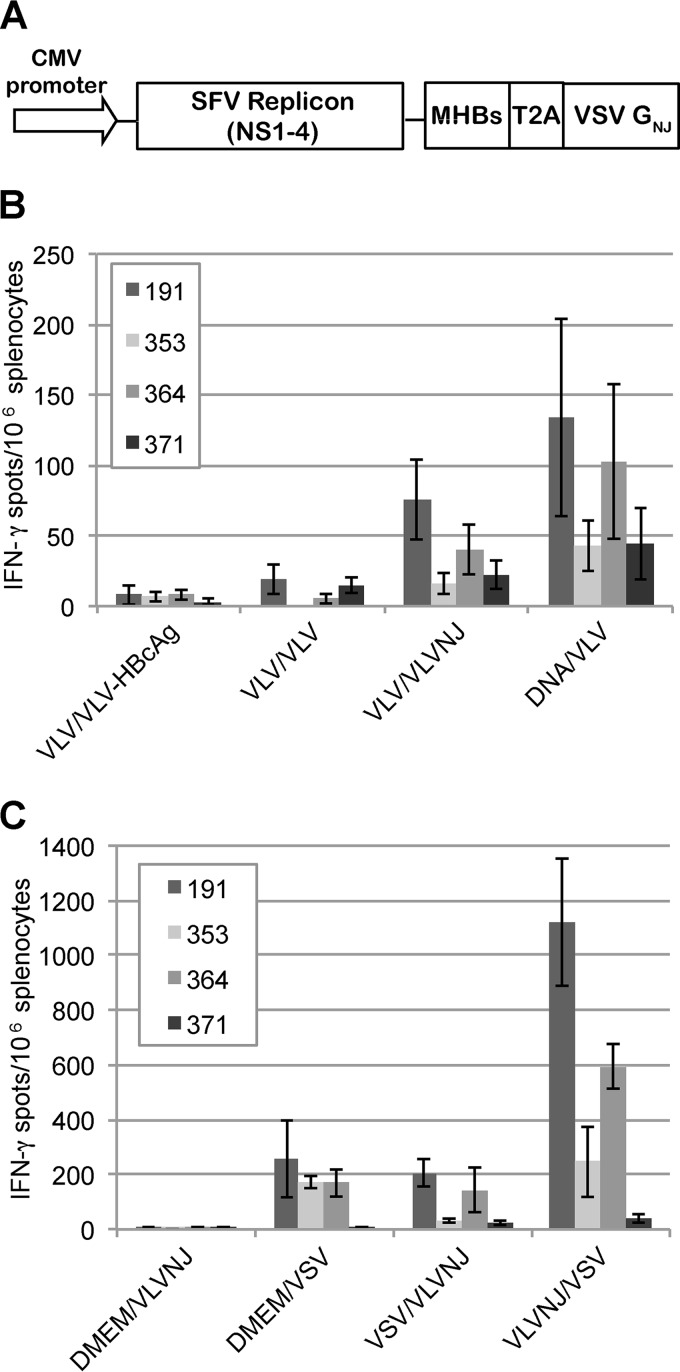
FIG 1.
VLV constructs that express HBV proteins. (A) Diagram of VLV DNA constructs used to generate the evolved VLVs expressing MHBs or HBcAg. Sequences encoding the HBV proteins are inserted under the viral subgenomic RNA promoter upstream of a T2A ribosomal skipping site and the VSV G protein. CMV, cytomegalovirus. (B) Immunofluorescence images of BHK cells stained with anti-HBcAg or anti-MHBs (preS2) antibody 24 h after transfection with VLV DNA constructs. (C) Western blot analysis of protein expression using anti-HBcAg or anti-MHBs (preS2) antibody 24 h after infection of BHK cells with VLVs or 5 h after infection with VSV-MHBs. The multiple MHBs bands in the VLV-MHBs-infected cells likely represent the p30/gp33/gp36 forms of the protein.
After cloning into the VLV vector backbone, the VLVs were recovered, and expression of the HBV proteins was confirmed. After an initial transfection of the VLV plasmid, indirect immunofluorescence using antibody specific for HBcAg (Dako) or the preS2 region of MHBs (S26; Santa Cruz Biotechnology) was performed to determine if the proteins were expressed. Indeed, transfection of the VLV plasmids leads to expression of MHBs or HBcAg (Fig. 1B). Next, it was determined if the recovered VLVs could express the HBV proteins after infection. Consistent with the transfection data, both HBcAg and MHBs were readily detected in lysates of infected BHK cells by Western blotting using anti-HBcAg or the MHBs-specific anti-preS2 antibody (Fig. 1C). The multiple MHBs bands observed in the VLV-MHBs-infected cells likely represent the p30 unglycosylated, gp33 singly glycosylated, and gp36 doubly glycosylated forms of the protein. Additionally, expression of the VSV G protein after infection with the generated VLVs was also observed (Fig. 1C). As a control, VSV-MHBs infection of BHK cells resulted in expression of MHBs as well as all other VSV structural proteins (Fig. 1C). VLVs for immunization experiments were typically recovered at titers above 108 IFU/ml (data not shown).
Immunization with VLVs induces HBV-specific immune responses.
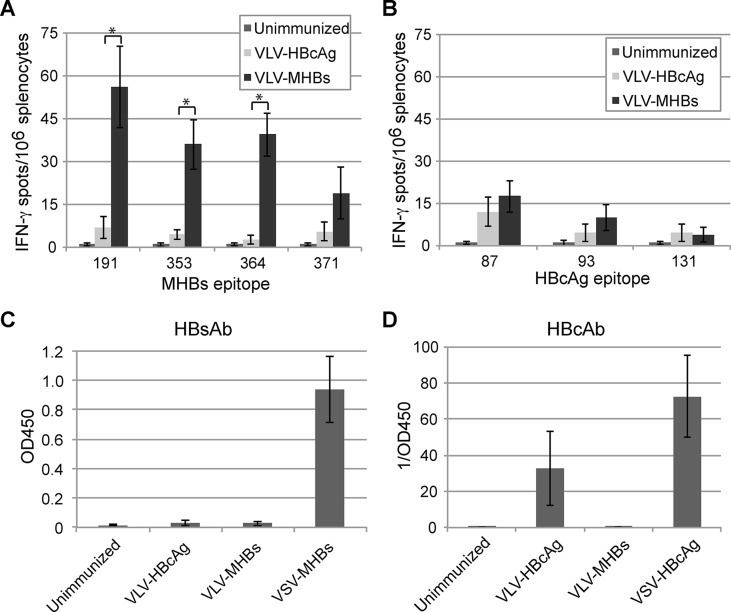
To determine if the VLVs expressing HBV proteins would elicit immune responses in naive mice, CB6F1 mice were intramuscularly immunized with 107 IFU of either VLV-MHBs or VLV-HBcAg. Because CD8 T cell responses would be important in a therapeutic vaccine, we first performed IFN-γ ELISPOT assays 7 days postimmunization, stimulating cells with 1 of 4 CD8 T cell peptide epitopes present in MHBs or 1 of 3 epitopes present in HBcAg (Table 1). Immunization with VLV-MHBs elicited detectable responses to at least 3 of the 4 epitopes tested (Fig. 2A). In contrast, no HBcAg responses were detected above background in mice immunized with VLV-HBcAg (Fig. 2B).
FIG 2.
VLVs expressing HBV proteins induce HBV-specific immune responses. (A and B) Mice were immunized intramuscularly with 1 × 107 IFU of VLVs expressing either MHBs or HBcAg. CD8 T cell responses to MHBs or HBcAg were measured by IFN-γ ELISPOT assays at day 7 postimmunization (n = 10). (C and D) Mice were immunized intramuscularly with 1 × 107 IFU of VLVs or 1 × 107 PFU of VSV expressing either MHBs or HBcAg, and serum antibody responses were determined by ELISA at day 30 postimmunization (n = 5). *, P < 0.01.
Although it is unclear if antibody responses to HBV would be necessary for an effective therapeutic vaccine, anti-HBs antibody could potentially play a role in neutralizing free virus and therefore preventing infection or reinfection of host hepatocytes. To determine if the VLV vaccines were able to elicit HBV-specific antibody responses, mice were immunized as described above. Thirty days postimmunization, serum HBsAb or HBcAb levels were examined by ELISA. In contrast to the T cell responses, VLV-MHBs immunization did not elicit detectable HBsAb responses (Fig. 2C). In contrast, immunization with VLV-HBcAg did elicit antibodies specific for HBcAg, though these responses were not as high as a nonattenuated recombinant VSV vector expressing HBcAg (VSV-HBcAg; Fig. 2D). Taken together, these data indicate that the VLV vaccine platform is capable of expressing HBV proteins that can elicit HBV-specific immune responses.
Vaccination with VLV-MHBs elicits a better CD8 T cell response than other immunization strategies.
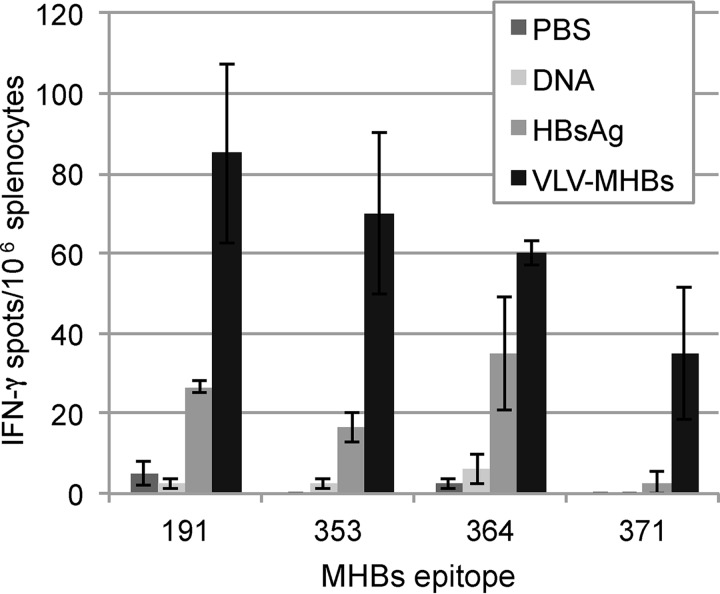
Platforms tested therapeutically for HBV infection, including recombinant DNA and protein vaccination, do not elicit immune responses capable of controlling chronic HBV infection (2). Since CD8 T cell responses would likely play a major role in viral clearance, we directly compared T cell responses generated by VLV-MHBs immunization to those induced with recombinant DNA or recombinant protein immunization. Interestingly, responses to VLV-MHBs were more consistently detectable and generally stronger than the other immunization strategies (Fig. 3). Furthermore, in mice in which responses were detected, the responses were directed toward more epitopes in the mice immunized with VLV-MHBs. These data indicate that VLV-MHBs immunization elicits better CD8 T cell responses than other tested immunization strategies.
FIG 3.
VLV-MHBs induces superior CD8 T cell responses compared to DNA or protein. Mice were intramuscularly immunized with 50 μg of recombinant DNA expressing MHBs (pCMV-MHBs), 10 μg of recombinant HBsAg, or 107 IFU of VLV-MHBs, and CD8 T cell responses were determined 7 days postimmunization by IFN-γ ELISPOT assay (n ≥ 4).
CD8 T cells elicited by VLV-MHBs are protective in a model of acute HBV infection.
To determine if the CD8 T cells induced by VLV-MHBs are capable of recognizing HBV antigen in the liver and controlling virus replication, an HBV hydrodynamic injection protocol (16), which models an acute HBV infection, was used. This model involves injection of an HBV DNA expression plasmid (pHBV1.3) into the tail vein of the mouse in a volume approximately equal to 9% of the mouse's body weight. This volumetric overload forces uptake of the DNA by hepatocytes and results in expression of HBV proteins and replication intermediates. Because there is no viral entry step in this protocol, protection is dependent on T cell-mediated mechanisms to clear HBV infection (14). Mice were immunized with DMEM, 107 IFU of VLV-MHBs or VLV-HBcAg, or 107 PFU of VSV-MHBs or VSV-HBcAg. Six weeks postimmunization, the hydrodynamic injection was performed, and the mice were monitored for HBV replication.
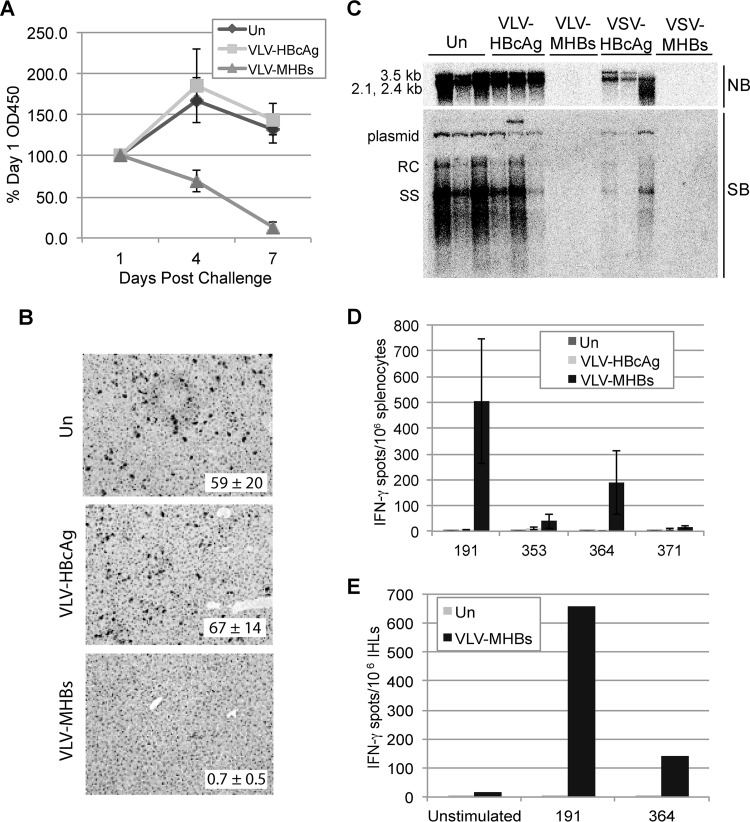
To examine HBV protein expression after challenge, HBeAg expression in the serum and HBcAg expression in the liver were measured. In all mice, serum HBeAg levels significantly increased by day 1 postchallenge (data not shown). In mice left unimmunized or immunized with VLV-HBcAg, HBeAg continued to increase at day 4 and then plateaued at day 7 postchallenge. However, in mice immunized with VLV-MHBs, serum HBeAg decreased at day 4 and became almost undetectable by day 7 postchallenge (Fig. 4A). HBcAg expression in the liver was also determined at day 7 postchallenge by immunohistochemistry. Consistent with the detected HBeAg levels, the number of HBcAg-expressing hepatocytes was significantly reduced in mice immunized with VLV-MHBs compared to unimmunized or VLV-HBcAg-immunized mice (Fig. 4B).
FIG 4.
VLV-MHBs-immunized mice are protected from HBV hydrodynamic challenge. Mice were immunized with 107 IFU of VLV-HBcAg or VLV-MHBs or with 107 PFU of VSV-MHBs or VSV-HBcAg. HBV replication was then induced by hydrodynamic injection of pHBV1.3 plasmid (10 μg) at 6 weeks postimmunization. (A) Serum HBeAg levels were determined by ELISA at days 1, 4, and 7 after challenge (n ≥ 4). Percentages of day 1 levels are shown. (B) HBcAg expression in the liver was determined by immunohistochemistry at day 7 postchallenge. Quantifications represent average numbers of HBcAg-positive cells per 100× field (n = 3 with 5 fields counted per sample). Representative images are shown. (C) At day 7 postchallenge, Northern (NB) and Southern blot (SB) analyses were used to determine liver-associated HBV RNA and DNA levels, respectively (n ≥ 3). Representative samples are shown. RC, relaxed circular DNA; SS, single-stranded DNA. (D and E) The CD8 T cell recall response was measured at day 7 postchallenge by IFN-γ ELISPOT assay in either splenocytes (n ≥ 4) (D) or intrahepatic leukocytes (pooled samples from 2 or more mice) (E). Un, unimmunized.
Next, HBV mRNA and DNA in the liver were examined by Northern and Southern blotting, respectively. These analyses demonstrated that VLV-MHBs immunization completely protected mice from HBV replication (Fig. 4C). Interestingly, the level of protection demonstrated by VLV-MHBs immunization was similar to that observed for VSV-MHBs, a vector that induces very strong HBV-specific immune responses. In agreement with their inability to induce T cell responses, the VLV- and VSV-HBcAg vectors did not protect mice from challenge (Fig. 4C).
Finally, since CD8 T cells are responsible for clearance in this model, the T cell recall response was measured by performing IFN-γ ELISPOT assays 7 days postchallenge on both splenocytes (Fig. 4D) and intrahepatic leukocytes (Fig. 4E). The results demonstrate a strong CD8 T cell response to at least 2 of the 4 epitopes tested. Importantly, these responses are approximately 5-fold greater than the response induced after primary immunization (compare Fig. 4D to Fig. 2A). These results suggest that VLV-MHBs particles induce memory T cells that can be recalled and expanded upon HBV challenge, and that the T cells induced by VLV-MHBs are capable of controlling HBV infection in this model in the absence of detectable antibody.
Combining immunization strategies in a prime-boost protocol improves HBV-specific immune responses.
Although VLV-MHBs particles elicit protective T cell responses in a single dose, overcoming immunotolerance during chronic infection may require multiple immunizations, but antibodies to the VSV G protein would likely neutralize a VLV boosting vector of the same serotype. Therefore, in order to investigate prime-boost strategies using the VLV vaccine platform, we first constructed an additional VLV construct. This construct also expresses MHBs, but the VSV G protein (Indiana serotype) was replaced with the New Jersey serotype VSV G (VLVNJ-MHBs) (Fig. 5A). After constructing VLVNJ-MHBs, a prime-boost experiment was performed, intramuscularly priming with VLV-MHBs or 50 μg of a DNA plasmid expressing MHBs (pCMV-MHBs). Then, mice were boosted 4 weeks later with VLV-HBcAg, VLV-MHBs, or VLVNJ-MHBs. Seven days postboost, CD8 T cell responses were analyzed by performing IFN-γ ELISPOT assays. Boosting with VLV-HBcAg or VLV-MHBs did not induce a measurable response, as expected (Fig. 5B). Boosting with VLVNJ-MHBs did yield a measurable HBV-specific response; however, the response observed was no greater than the primary response observed with VLV-MHBs immunization, suggesting that the boost was suboptimal. Priming with DNA and then boosting with VLV-MHBs tended to yield a response greater than that observed for primary immunization alone (Fig. 5B). These results suggest that combining immunization platforms may induce stronger T cell responses than using VLVs alone.
FIG 5.
Prime-boost protocols using VLVs improve HBV-specific CD8 T cell responses. (A) Diagram of VLVNJ-MHBs construct. The VSV glycoprotein expressed is derived from the New Jersey serotype VSV. (B and C) IFN-γ ELISPOT assays conducted 1 week postboost in mice primed and boosted with the designated MHBs-expressing vectors (n ≥ 5). VLV, VLV-MHBs; VLVNJ, VLVNJ-MHBs; VSV, VSV-MHBs; DNA, pCMV-MHBs.
To further investigate heterologous prime-boost strategies, a prime-boost experiment combining VLV and VSV immunization was performed. In this experiment, mice were primed and boosted with various combinations of VLV-MHBs, VLVNJ-MHBs, or VSV-MHBs (Fig. 5C). Combining VLVNJ-MHBs immunization with VSV-MHBs immunization led to significant increases in HBV-specific T cells. Importantly, performing a VLVNJ-MHBs prime followed by a VSV-MHBs boost yielded T cell responses more than 3 times greater than any of the other immunization protocols. Together, these data indicate that combining heterologous immunization strategies in prime-boost protocols can lead to significant improvements in HBV-specific CD8 T cell responses.
VLV-MHBs immunization induces CD8 T cell responses in a model of chronic infection.
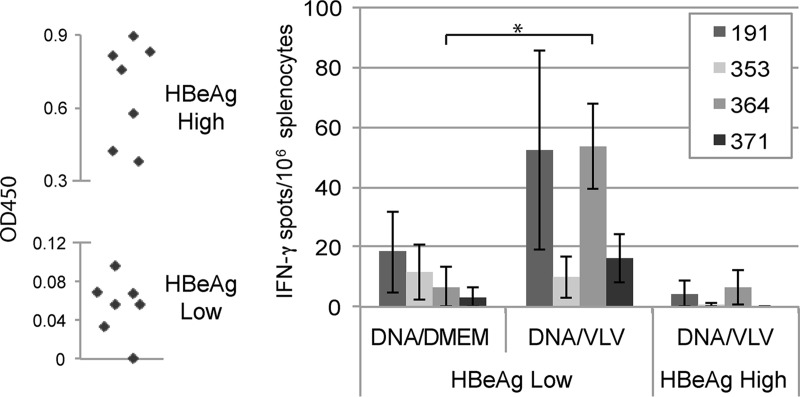
In order to test the therapeutic potential of VLV-MHBs immunization, we utilized 1.3.32 HBV transgenic mice as a model of chronic infection. These mice express variable levels of the HBV transgene, which can be measured by serum HBeAg level (15). HBV therapeutic vaccination would likely be used in combination with other treatment modalities such as antiviral or immunomodulatory drugs. To model the lower antigen load that might be present in a person receiving long-term antiviral therapy, HBeAglow mice were immunized. A single immunization with VLV-MHBs did not induce any HBV-specific immune responses (data not shown). However, priming with DNA and then boosting with VLV-MHBs did induce T cell responses to at least one epitope (Fig. 6). Though immune responses were detected in HBeAglow mice, VLV-MHBs vaccination did not elicit T cell responses in HBeAghigh mice. These results suggest that combination of VLV immunization with other treatment strategies to lower antigenemia may represent a novel approach for HBV therapy.
FIG 6.
Prime-boost protocol using VLV-MHBs induces CD8 T cell responses in HBV transgenic mice. HBV 1.3 transgenic mice were screened for HBeAg expression and separated into HBeAglow (optical density at 450 nm [OD450] < 0.1) or HBeAghigh (OD450 > 0.3) groups. All mice were primed with 50 μg of pCMV-MHBs and boosted 4 weeks later with DMEM (n = 3) or 107 IFU of VLV-MHBs (n ≥ 4). IFN-γ ELISPOT assays were performed 1 week postboost. *, P < 0.05.
DISCUSSION
Virus-based vectors offer unique vaccine platforms due to their ability to induce potent immune responses even, in some cases, despite immunotolerance. However, many current viral vectors have extensive safety concerns that must be overcome before widespread use in humans. For our studies using the nonpathogenic VLV system, we chose to make vaccines expressing either MHBs or HBcAg since these proteins are known to elicit T cell responses in humans (17–19), and responses to the viral structural proteins, in comparison to the nonstructural proteins, may be more likely to contribute to HBV clearance in humans (20). We chose to use MHBs, rather than small HBs, because of the possible potential to generate T cell responses to preS2 sequences (although no known mouse preS2 CD8 T cell epitopes have been defined). We did not use large HBs, as this protein contains the HBV receptor-binding domain and thus could alter VLV tropism if it were to become incorporated into the vesicles. However, we recognize that the different secretion patterns of small, middle, and large HBs may affect the balance between the humoral and cellular immune responses generated to these proteins, and further studies would be needed to determine which proteins (or combination of proteins) elicit the optimal response.
In contrast to VLV-MHBs, which generated an MHBs-specific CD8 T cell response after a single immunization, we were unable to generate an HBcAg-specific CD8 T cell response with a single VLV-HBcAg administration. This result was not completely unexpected since we previously observed that HBcAg delivery by VSV or lentiviral vectors also did not generate a measurable response in CB6F1 mice (data not shown). It is possible that inherent properties of HBcAg, such as its high degree of stability or resistance to proteasome-mediated degradation (21), make it difficult for the host to generate CD8 T cell responses when the antigen is delivered by certain viral vaccine systems. Indeed, expression of HBeAg, the secreted form of HBcAg, results in suppression of HBcAg responses (22). Although the precore region necessary for secretion of HBeAg is not present in our construct, precore is not absolutely necessary for cytoplasmic HBeAg production (23), and alternative folding or localization of HBcAg could potentially suppress the anti-HBcAg response.
Although HBV clearance is usually associated with HBsAg seroconversion (24), the exact mechanisms and contributions of serconversion are unknown. HBsAb would clear circulating free virus and prevent infection or reinfection of host hepatocytes; however, whether this is absolutely required in the presence of a strong cellular immune response is unknown. Regardless, the VLV-MHBs vaccine examined here did not elicit HBsAg-specific antibodies in naive mice. One possibility for this result may lie in the design of the VLV construct and the topology of MHBs. During normal production of MHBs, the C terminus of the protein must be translocated across the endoplasmic reticulum (ER) membrane (25). The T2A ribosomal skipping site used to express MHBs and VSV G from the same promoter adds 18 additional amino acids onto the C terminus of the protein. These amino acids may alter processing, glycosylation, and secretion of MHBs, which could affect antibody production. Indeed, we did not observe secretion of MHBs after in vitro VLV-MHBs infection (data not shown), and the somewhat different pattern of MHBs expression in VLV- and VSV-infected cells (Fig. 1C) may also be a consequence of this additional sequence. However, differences in infectivity, replication kinetics, and cytopathic effects between VSV and VLV make it difficult to precisely compare protein expression by the two systems, and future studies will be needed to analyze alternative modes of MHBs expression from the VLV system to determine if optimization would allow for enhanced immune responses to MHBs.
In the HBV hydrodynamic challenge model, we observed a marked reduction in HBV replication in mice immunized with VLV-MHBs. However, unlike for mice immunized with VSV-MHBs (14), we did not observe a significant elevation in serum alanine aminotransferase (ALT) levels beyond that induced by the injection (data not shown). This result combined with the lack of histopathology (data not shown) suggests that noncytolytic mechanisms may play a significant role in clearance. Given that noncytolytic methods are an important mode of HBV clearance (26) and that the cytolytic function of T cells can often mediate disease pathogenesis (26, 27), the lack of observable liver disease in VLV-MHBs-immunized mice suggests that this vaccine may not induce an overly pathogenic immune response.
The aim of boosting strategies is to expand the memory T cell pool to a particular antigen. Generally during boosting, a secondary response greater than the primary response can be observed. Unexpectedly, however, boosting with a serotype switch VLV construct resulted in a CD8 T cell response of a magnitude similar to that of the primary response to VLV-MHBs (Fig. 6). One possibility for this result is that a single immunization with VLV-MHBs does not induce a memory response capable of being boosted. However, since we observed T cell expansion associated with protection from hydrodynamic challenge at 6 weeks postimmunization, VLV-MHBs likely does induce a durable memory response. Nevertheless, it is possible that the kinetics of memory development may not allow for optimum boosting at 4 weeks postimmunization. Indeed, it is unknown how long antigen persists after VLV inoculation, a factor that can influence memory T cell development (28). Alternatively, the kinetics of the secondary expansion may be such that analysis at 1 week postboost did not accurately reflect the peak response. Finally, priming with the weaker VLV (VLVNJ-MHBs) and boosting with the more immunogenic VLV-MHBs may improve responses, as delivering a less inflammatory vaccine for the prime may strengthen prime boost vaccination approaches by favoring memory rather than effector CD8 T cell differentiation (29). Further experiments analyzing these factors may improve the CD8 T cell response after boost.
Interestingly, immunization of the HBV transgenic mice with a DNA prime and VLV boost elicited a response most consistently to the 364 epitope in MHBs. Conversely, this epitope was subdominant to the 191 epitope in experiments performed with normal mice. Furthermore, the 191 epitope, to which stronger responses were induced in the nontransgenic mice, yielded a relatively variable response in the transgenic mice. It is possible that given its immunodominance, T cells reactive to the 191 epitope are under a more severe state of tolerance in the transgenic mice. We previously reported that in low-level antigen-expressing transgenic mice, responses can be elicited to the 191 epitope by a VSV vector (6), suggesting that the 191-specific T cells are not completely deleted in these animals. However, they are likely rare, have extremely low affinity or avidity for the peptide, express high levels of inhibitory receptors, and/or are impacted by other tolerance mechanisms, which may all contribute to the inability of VLV-MHBs to elicit consistent responses to the 191 epitope. This result highlights the likelihood that T cell responses to subdominant epitopes may be important in therapeutic vaccination strategies.
Overall, the VLV vector expressing MHBs demonstrates that a viral vaccine system, likely in combination with other therapeutic strategies, may lead to an effective chronic HBV treatment. Furthermore, the ability of the VLV system to induce responses even in the tolerogenic environment of HBV transgenic mice suggests that the system may also be useful for designing vaccines for other chronic infections. Indeed, VLVs have already shown promise in the HIV field (7, 8). Further investigation into vaccines for other pathogens and diseases may highlight the versatility and utility of the VLV vaccine system.
ACKNOWLEDGMENTS
This work was supported by sponsored research grants from Connecticut Innovations (CI) and the Connecticut Bioscience Innovation Fund (CBIF) through CaroGen Corporation (Hamden, CT) to Yale University. Additional support came from NIH grants R01AI045510 and R56AI105409 (to J.K.R.). T.D.R. was also supported by NIH predoctoral training grant T32AI055403.
We thank Bijan Almassian, Valerian Nakaar, and Joseph Rininger (CaroGen) for useful discussions.
T.D.R., N.F.R., J.K.R., and M.D.R. are inventors on patents related to the VLV technology that is being commercialized by CaroGen.
This paper is dedicated to the memory of the first author, Tracy D. Reynolds.
REFERENCES
- 1.World Health Organization. July 2015, posting date Hepatitis B fact sheet. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs204/en/. [Google Scholar]
- 2.Liu J, Kosinska A, Lu M, Roggendorf M. 2014. New therapeutic vaccination strategies for the treatment of chronic hepatitis B. Virol Sin 29:10–16. doi: 10.1007/s12250-014-3410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sällberg M, Hughes J, Javadian A, Ronlov G, Hultgren C, Townsend K, Anderson CG, O'Dea J, Alfonso J, Eason R, Murthy KK, Jolly DJ, Chang SM, Mento SJ, Milich D, Lee WT. 1998. Genetic immunization of chimpanzees chronically infected with the hepatitis B virus, using a recombinant retroviral vector encoding the hepatitis B virus core antigen. Hum Gene Ther 9:1719–1729. doi: 10.1089/hum.1998.9.12-1719. [DOI] [PubMed] [Google Scholar]
- 4.Kosinska AD, Zhang E, Johrden L, Liu J, Seiz PL, Zhang X, Ma Z, Kemper T, Fiedler M, Glebe D, Wildner O, Dittmer U, Lu M, Roggendorf M. 2013. Combination of DNA prime-adenovirus boost immunization with entecavir elicits sustained control of chronic hepatitis B in the woodchuck model. PLoS Pathog 9:e1003391. doi: 10.1371/journal.ppat.1003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin P, Dubois C, Jacquier E, Dion S, Mancini-Bourgine M, Godon O, Kratzer R, Lelu-Santolaria K, Evlachev A, Meritet JF, Schlesinger Y, Villeval D, Strub JM, Van Dorsselaer A, Marchand JB, Geist M, Brandely R, Findeli A, Boukhebza H, Menguy T, Silvestre N, Michel ML, Inchauspe G. 26 November 2014. TG1050, an immunotherapeutic to treat chronic hepatitis B, induces robust T cells and exerts an antiviral effect in HBV-persistent mice. Gut doi: 10.1136/gutjnl-2014-308041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobleigh MA, Wei X, Robek MD. 2013. A vesicular stomatitis virus-based therapeutic vaccine generates a functional CD8 T cell response to hepatitis B virus in transgenic mice. J Virol 87:2969–2973. doi: 10.1128/JVI.02111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose NF, Publicover J, Chattopadhyay A, Rose JK. 2008. Hybrid alphavirus-rhabdovirus propagating replicon particles are versatile and potent vaccine vectors. Proc Natl Acad Sci U S A 105:5839–5843. doi: 10.1073/pnas.0800280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schell JB, Rose NF, Bahl K, Diller K, Buonocore L, Hunter M, Marx PA, Gambhira R, Tang H, Montefiori DC, Johnson WE, Rose JK. 2011. Significant protection against high-dose simian immunodeficiency virus challenge conferred by a new prime-boost vaccine regimen. J Virol 85:5764–5772. doi: 10.1128/JVI.00342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spuul P, Balistreri G, Kaariainen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J Virol 84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rolls MM, Webster P, Balba NH, Rose JK. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 11.Rose NF, Buonocore L, Schell JB, Chattopadhyay A, Bahl K, Liu X, Rose JK. 2014. In vitro evolution of high-titer, virus-like vesicles containing a single structural protein. Proc Natl Acad Sci U S A 111:16866–16871. doi: 10.1073/pnas.1414991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolls MM, Haglund K, Rose JK. 1996. Expression of additional genes in a vector derived from a minimal RNA virus. Virology 218:406–411. doi: 10.1006/viro.1996.0211. [DOI] [PubMed] [Google Scholar]
- 13.Doronina VA, Wu C, de Felipe P, Sachs MS, Ryan MD, Brown JD. 2008. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol 28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobleigh MA, Buonocore L, Uprichard SL, Rose JK, Robek MD. 2010. A vesicular stomatitis virus-based hepatitis B virus vaccine vector provides protection against challenge in a single dose. J Virol 84:7513–7522. doi: 10.1128/JVI.00200-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69:6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang PL, Althage A, Chung J, Chisari FV. 2002. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci U S A 99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penna A, Chisari FV, Bertoletti A, Missale G, Fowler P, Giuberti T, Fiaccadori F, Ferrari C. 1991. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J Exp Med 174:1565–1570. doi: 10.1084/jem.174.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayersina R, Fowler P, Guilhot S, Missale G, Cerny A, Schlicht HJ, Vitiello A, Chesnut R, Person JL, Redeker AG, Chisari FV. 1993. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol 150:4659–4671. [PubMed] [Google Scholar]
- 19.Sobao Y, Sugi K, Tomiyama H, Saito S, Fujiyama S, Morimoto M, Hasuike S, Tsubouchi H, Tanaka K, Takiguch M. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J Hepatol 34:922–929. doi: 10.1016/S0168-8278(01)00048-4. [DOI] [PubMed] [Google Scholar]
- 20.Kakimi K, Isogawa M, Chung J, Sette A, Chisari FV. 2002. Immunogenicity and tolerogenicity of hepatitis B virus structural and nonstructural proteins: implications for immunotherapy of persistent viral infections. J Virol 76:8609–8620. doi: 10.1128/JVI.76.17.8609-8620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nath N, Hickman K, Nowlan S, Shah D, Phillips J, Babler S. 1992. Stability of the recombinant hepatitis B core antigen. J Clin Microbiol 30:1617–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milich DR, Chen MK, Hughes JL, Jones JE. 1998. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol 160:2013–2021. [PubMed] [Google Scholar]
- 23.Ou JH, Laub O, Rutter WJ. 1986. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci U S A 83:1578–1582. doi: 10.1073/pnas.83.6.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters M, Vierling J, Gershwin ME, Milich D, Chisari FV, Hoofnagle JH. 1991. Immunology and the liver. Hepatology 13:977–994. doi: 10.1002/hep.1840130529. [DOI] [PubMed] [Google Scholar]
- 25.Bruss V. 2007. Hepatitis B virus morphogenesis. World J Gastroenterol 13:65–73. doi: 10.3748/wjg.v13.i1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25–36. doi: 10.1016/S1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 27.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. 2003. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol 77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaech SM, Wherry EJ, Ahmed R. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 29.Nolz JC, Harty JT. 2011. Strategies and implications for prime-boost vaccination to generate memory CD8 T cells. Adv Exp Med Biol 780:69–83. doi: 10.1007/978-1-4419-5632-3_7. [DOI] [PubMed] [Google Scholar]
- 30.Ando K, Moriyama T, Guidotti LG, Wirth S, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. 1993. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J Exp Med 178:1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schirmbeck R, Bohm W, Fissolo N, Melber K, Reimann J. 2003. Different immunogenicity of H-2 Kb-restricted epitopes in natural variants of the hepatitis B surface antigen. Eur J Immunol 33:2429–2438. doi: 10.1002/eji.200324125. [DOI] [PubMed] [Google Scholar]
- 32.Sette AD, Oseroff C, Sidney J, Alexander J, Chesnut RW, Kakimi K, Guidotti LG, Chisari FV. 2001. Overcoming T cell tolerance to the hepatitis B virus surface antigen in hepatitis B virus-transgenic mice. J Immunol 166:1389–1397. doi: 10.4049/jimmunol.166.2.1389. [DOI] [PubMed] [Google Scholar]
- 33.Schirmbeck R, Dikopoulos N, Kwissa M, Leithauser F, Lamberth K, Buus S, Melber K, Reimann J. 2003. Breaking tolerance in hepatitis B surface antigen (HBsAg) transgenic mice by vaccination with cross-reactive, natural HBsAg variants. Eur J Immunol 33:3342–3352. doi: 10.1002/eji.200324403. [DOI] [PubMed] [Google Scholar]
- 34.Kuhröber A, Wild J, Pudollek HP, Chisari FV, Reimann J. 1997. DNA vaccination with plasmids encoding the intracellular (HBcAg) or secreted (HBeAg) form of the core protein of hepatitis B virus primes T cell responses to two overlapping Kb- and Kd-restricted epitopes. Int Immunol 9:1203–1212. doi: 10.1093/intimm/9.8.1203. [DOI] [PubMed] [Google Scholar]
- 35.Kuhöber A, Pudollek HP, Reifenberg K, Chisari FV, Schlicht HJ, Reimann J, Schirmbeck R. 1996. DNA immunization induces antibody and cytotoxic T cell responses to hepatitis B core antigen in H-2b mice. J Immunol 156:3687–3695. [PubMed] [Google Scholar]
- 36.Chen A, Wang L, Zhang J, Zou L, Jia Z, Zhou W, Wan Y, Wu Y. 2005. H-2 Kd-restricted hepatitis B virus-derived epitope whose specific CD8+ T lymphocytes can produce gamma interferon without cytotoxicity. J Virol 79:5568–5576. doi: 10.1128/JVI.79.9.5568-5576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]