Abstract
We examined the function of the conserved Val/Ile residue within the dengue virus NS5 interdomain linker (residues 263 to 272) by site-directed mutagenesis. Gly substitution or Gly/Pro insertion after the conserved residue increased the linker flexibility and created slightly attenuated viruses. In contrast, Pro substitution abolished virus replication by imposing rigidity in the linker and restricting NS5's conformational plasticity. Our biochemical and reverse genetics experiments demonstrate that NS5 utilizes conformational regulation to achieve optimum viral replication.
TEXT
Nonstructural protein 5 (NS5) is a multifunctional protein essential for dengue virus (DENV) replication. NS5 consists of an N-terminal S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MTase) domain (1) and a C-terminal RNA-dependent RNA polymerase (RdRp) domain (2–5). The MTase domain catalyzes the formation of type I RNA cap, which is critical for efficient translation and subverting the host immune surveillance (1, 6–8). The MTase domain may also be responsible for guanylyltransferase activity (9, 10). The RdRp domain (NS5 RdRp) synthesizes both negative-strand RNA from the input genomic RNA (gRNA) and the progeny plus-strand gRNA (5, 11).
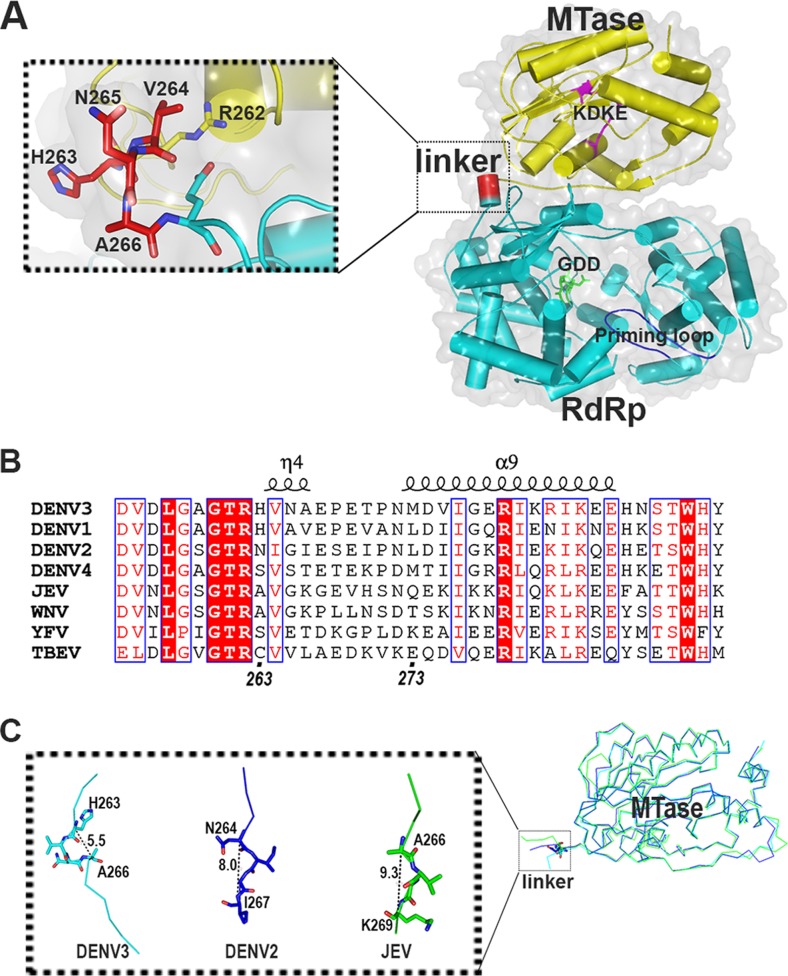
Previous structural studies on DENV NS5 individual domains revealed electron density covering residues 6 to 262 (for the MTase domain) (PDB code 3P97) (12), and residues 273 to 882 (for RdRp) (PDB code 2J7U) (5). Limited proteolysis of full-length NS5 (NS5 FL) in combination with mass determination suggested that the residues 263 to 272 (5) form the interdomain linker, and sequence comparison across various flaviviruses showed that this region is poorly conserved (Fig. 1). Small-angle X-ray scattering (SAXS) studies suggested that NS5 could adopt different conformations permitted by a flexible linker (13). Interestingly, an N-terminally extended RdRp spanning residues from 265 to 900 improved thermostability and RdRp activity compared with a shorter RdRp spanning residues 273 to 900 (14). More recently, the structures of the full-length NS5 protein from DENV3 (15) and Japanese encephalitis virus (JEV) full-length NS5 (PDB code 4K6M) (16) have been resolved. Extensive comparisons between the two structures have been reported (15). Notably, unlike JEV NS5, the DENV NS5 structure revealed a hydrophilic interdomain interface and a linker region composed of a 310-helix (amino acids [aa] 263 to 266) followed by a short loop (aa 267 to 272) (Fig. 1A and B). In DENV3 full-length NS5, the 310-helix linker adopts a compact conformation compared to JEV full-length NS5 or DENV2 MTase (1 to 296) (PDB code 1L9K) (Fig. 1C). Functionally, the MTase domain stimulates both de novo initiation and elongation activities of the RdRp domain, which implies a critical role for an interdomain linker in controlling NS5 RNA polymerization activities (17). Despite the various conformations adopted by NS5 in solution, position 264 (in DENV3 NS5) is found to be exclusively Val in flaviviruses, except for a homologous substitution of Ile (position 265) in DENV2 (Fig. 1B). In the three-dimensional (3D) structure of DENV3 NS5FL, the side chain of Val packs against side chains of R262 and E267, forming a pocket (Fig. 1A). We therefore hypothesized that this strictly conserved Val or Ile residue plays a functional role in regulating the enzymatic coupling between the NS5 MTase and NS5 RdRp domains of NS5 and also plays a role in regulating virus replication.
FIG 1.
(A) Crystal structure of DENV3 NS5 is displayed as a cartoon (PDB code 4V0R). The MTase domain is colored in yellow, the RdRp domain is in cyan, and linker residues 263 to 266 are in red. The inset shows a closeup view of the linker region. The KDKE catalytic tetrad of MTase is shown as magenta sticks and labeled; the GDD active site is in green, and the priming loop is in blue and labeled. (B) Sequence alignment of the linker regions between the MTase and RdRp domains of NS5 from DENV1 to -4 and various flaviviruses showing the secondary structures observed in DENV3 NS5. WNV, West Nile virus; YFV, yellow fever virus; TBEV, tick-borne encephalitis virus. (C) Superposition of the MTase domain of the flaviviral NS5 structures with structural information for the linker region. The distance between Cα's of the first and fourth linker residues was measured. DENV3 NS5, PDB code 4V0R; DENV2 MTase, PDB code 1L9K; JEV NS5, PDB code 4K6M.
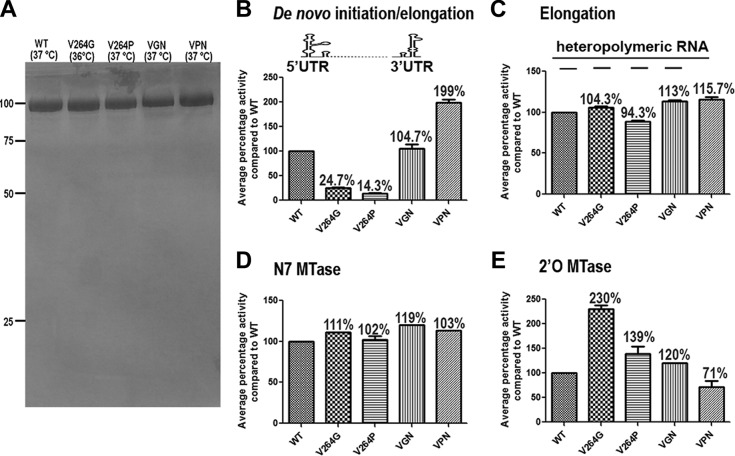
To characterize the enzymatic function of the linker mutants, the recombinant DENV3 NS5 and its V264G, V264P, VGN (G insertion after V264), and VPN (P insertion after V264) mutants were expressed, purified to >90% homogeneity (Fig. 2A), and assessed for their enzymatic activities as previously described (15). Interestingly, the four mutant NS5 proteins displayed dramatic differences in the coupled de novo initiation/elongation activity, even though their elongation activities were comparable (Fig. 2B and C). The V264G and V264P mutants showed only 25% and 15% de novo polymerase activities, respectively, compared to the wild type (WT). The VGN insertion mutant showed de novo activity similar to that of the WT, while the VPN mutant displayed a higher de novo polymerase activity. Both the WT and the four mutants displayed N7 MTase activities comparable to that of the WT protein (Fig. 2D). The V264G mutant showed a 2-fold-higher 2′-O-MTase activity than that of the WT, while the activity in the VPN mutant was 70% of that of the WT (Fig. 2E). Together, these data suggest that structural perturbation of the linker 310-helix can have differential effects on the enzymatic activities of NS5, presumably through altered conformational plasticity between the two enzymatic domains and their specific interaction with viral RNA.
FIG 2.
Protein expression and in vitro enzymatic activities of DENV3 NS5 and its mutants. (A) SDS-12% PAGE of purified WT and mutant NS5 proteins purified as described previously (19); melting temperature values measured by Thermofluor assay are indicated in parentheses. Numbers at left are molecular masses in kilodaltons. (B) A de novo initiation/elongation assay was performed using viral untranslated region (UTR) sequence as the template as described previously (19, 20). (C) Elongation assays of DENV3 WT and mutant NS5 proteins were performed with a heteropolymeric RNA template annealed with four primers as described previously (10, 13). (D) N7 MTase activities of the WT and NS5 mutant were measured with the in vitro-transcribed first 110 nucleotides (nt) of the DENV genome with unmethylated cap G (12, 21). (E) 2′-O-MTase activities of DENV3 WT and mutant NS5 proteins were measured with a 7-mer single-stranded RNA with cap 0 structure (12, 21). Two independent experiments were performed for each assay in triplicate (RdRp assays) or duplicate (MTase assays). Percent activities relative to DENV3 WT NS5 are shown above each bar.
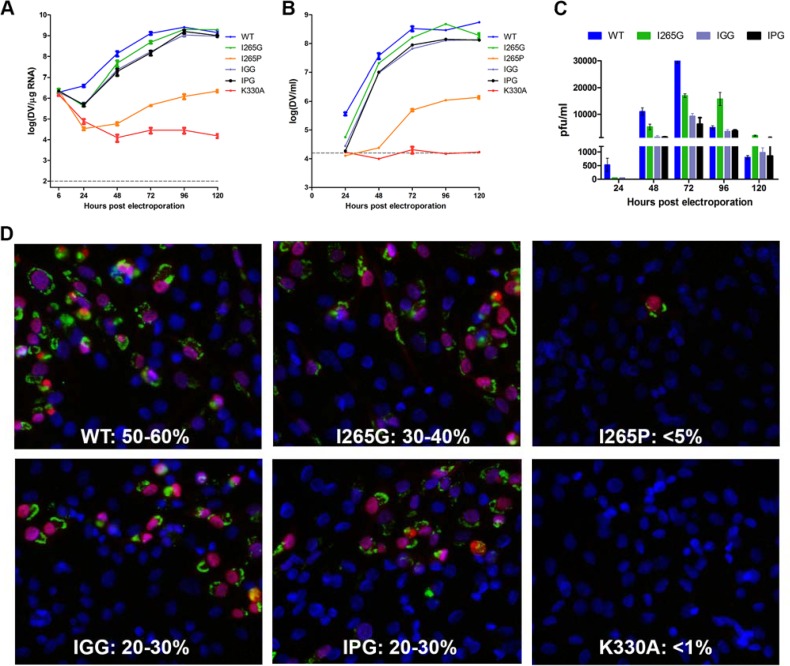
To examine the functional role of the conserved residue in the context of the virus life cycle, the same mutations as listed above were introduced into a DENV2 infectious cDNA clone (18). The viral RNA synthesis and infectious virus production capabilities of the sequence-confirmed clones and the WT were examined over the course of 5 days posttransfection by reverse transcription quantitative PCR (qPCR) and confirmed by immunofluorescence assay (IFA) staining of double-stranded RNA (dsRNA) and NS5 protein with specific antibodies (Fig. 3). Of the four analogous mutants in DENV2 (the I265G, I265P, IGG, and IPG mutants), only the I265P mutant (replacement of I265 with Pro) failed to show recovery of any viable viruses by plaque assay, similarly to the negative-control K330A mutant (18). Also, the I265P virus displayed significantly lower infectivity (<5%) than that of the WT (50 to 60%) on day 3, as indicated by the IFA staining of dsRNA and NS5. By comparison, the other three mutant viruses resulted in slightly attenuated viruses with slower growth and reduced infectivity (Fig. 3).
FIG 3.
Replication profiles of NS5 linker mutants. (A) NS5 linker mutations were introduced into an infectious DENV2 cDNA clone. Ten micrograms of in vitro-transcribed infectious clone RNA was electroporated into BHK-21 cells, and viral replication was monitored over a course of 5 days as described previously (18). Intracellular viral RNA replication was detected by reverse transcription-qPCR. The gray dashed line represents the background detection of uninfected cells. The IGG protein possesses a G insertion after I265, and the IPG protein has a P insertion after I265. (B) Extracellular viral RNA in the supernatants detected by reverse transcription-qPCR. The gray dashed line denotes background signal of uninfected supernatant. (C) Infectious virus titer measured by standard BHK-21 plaque assay. (D) IFA images showing dsRNA and NS5 costaining and percent infection of cells at 72 h postelectroporation (22).
V264G and V264P substitution mutants were defective in the de novo initiation RdRp activity—the V264G mutant showed 25% polymerase activity and the V264P mutant showed only 15% polymerase activity compared to the WT. Interestingly, in the 2′-O-MTase assay, the V264G mutant was twice as active as the WT or the other mutants. When viewed together with the reverse genetics data using the DENV2 infectious clone, the I265P mutant is severely attenuated compared to the I265G mutant. Structurally, the Pro residues in both DENV3 NS5 V264 and JEV NS5 V267 lie in allowed regions of the Ramachandran plot. However, molecular dynamics simulations of the NS5 structure revealed that this mutation in DENV causes considerable alteration in its NS5 structure and dynamics (Fig. 4A), while the same mutation in the JEV linker does not reveal significant changes in its structure compared to the wild-type protein (data not shown). The reduced de novo initiation and increased flexibility of the V264G (I265G) mutant may result in more efficient capping of newly synthesized genomic RNA compared to the rigidified V264P mutant (I265P) (Fig. 2). In the replication complex within infected cells, the intramolecular interactions between the two domains of NS5 may be regulated through interactions with RNA, NS3, and other host and viral proteins which further contribute to the stunted phenotype of the I265P mutant compared with the I265G mutant or the WT (Fig. 4B).
FIG 4.
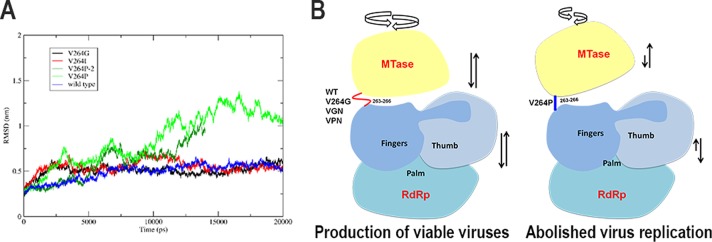
All atom root mean square deviations (RMSD) from molecular dynamics simulations of DENV NS5 structure generated using 4V0R (15) and 4HDG (23) and a schematic model depicting the dynamics of NS5 regulated by the interdomain linker. (A) Molecular dynamics simulations carried out with Gromacs-5.0.5 (www.gromacs.org) show that the V264P mutation exhibits a destabilizing effect on NS5 structure, and large conformational changes are observed in two independent simulations, while the V264G and V264I mutations have a minor effect on the structure and dynamics of NS5 compared to the wild-type protein. V264 is conserved and forms van der Waals interactions with side chains of residues R262 and E267. Molecular dynamics simulations suggest that I265 in DENV2 may adopt a similar interaction. (B) Schematic model showing that NS5 multiple enzymatic functions may require a flexible linker that permits the protein to adopt a greater variety of conformations (and thus yields viable viruses), as opposed to the V264P mutation, which abolishes 310-helix formation for the compact conformation of NS5 (suggested by molecular dynamics simulations), resulting in loss of some of the functions and abolished replication.
Our findings support the hypothesis that cross talk between MTase and RdRp domains of NS5 is essential for virus replication and that the linker region between the two domains has evolved to reach an optimal flexibility through the “greasy finger” made by residue V264/I265, which plays an important role in allowing the two domains to adopt functional orientations (17).
ACKNOWLEDGMENTS
This work was supported by NMRC grants NMRC/1315/2011 (S.G.V.) and NMRC/CBRG/0073/2014 (J.L., D.L., S.G.V.).
REFERENCES
- 1.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 3.Guyatt KJ, Westaway EG, Khromykh AA. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods 92:37–44. doi: 10.1016/S0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- 4.Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 5.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. 2008. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol 82:4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, Nonnis S, Decroly E, Selisko B, de Lamballerie X, Coutard B, Canard B, Bolognesi M. 2009. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in flavivirus. J Mol Biol 385:140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. 2009. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B. 2007. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem 282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 12.Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong HP, Liu BP, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. 2011. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem 286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bussetta C, Choi KH. 2012. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry 51:5921–5931. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim SP, Koh JH, Seh CC, Liew CW, Davidson AD, Chua LS, Chandrasekaran R, Cornvik TC, Shi PY, Lescar J. 2013. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J Biol Chem 288:31105–31114. doi: 10.1074/jbc.M113.508606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Soh S, Zheng J, Chan KW, Phoo WW, Lee CC, Tay MY, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. 2015. A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog 11(3):e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G, Gong P. 2013. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog 9:e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potisopon S, Priet S, Collet A, Decroly E, Canard B, Selisko B. 2014. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res 42:11642–11656. doi: 10.1093/nar/gku666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tay MY, Saw WG, Zhao Y, Chan KW, Singh D, Chong Y, Forwood JK, Ooi EE, Gruber G, Lescar J, Luo D, Vasudevan SG. 2015. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J Biol Chem 290:2379–2394. doi: 10.1074/jbc.M114.607341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niyomrattanakit P, Chen YL, Dong HP, Yin Z, Qing M, Glickman JF, Lin K, Mueller D, Voshol H, Lim JYH, Nilar S, Keller TH, Shi PY. 2010. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol 84:5678–5686. doi: 10.1128/JVI.02451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi PY. 2013. Ten years of dengue drug discovery: progress and prospects. Antiviral Res 100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Lim SP, Wen D, Yap TL, Yan CK, Lescar J, Vasudevan SG. 2008. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res 80:360–369. doi: 10.1016/j.antiviral.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Moreland NJ, Tay MY, Lee CC, Swaminathan K, Vasudevan SG. 2014. Identification and molecular characterization of human antibody fragments specific for dengue NS5 protein. Virus Res 179:225–230. doi: 10.1016/j.virusres.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Surana P, Satchidanandam V, Nair DT. 2014. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res 42:2758–2773. doi: 10.1093/nar/gkt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]