Abstract
The development of molecular tools to detect and report mitochondrial DNA (mtDNA) heteroplasmy will increase the discrimination potential of the testing method when applied to forensic cases. The inherent limitations of the current state-of-the-art, Sanger-based sequencing, including constrictions in speed, throughput, and resolution, have hindered progress in this area. With the advent of next-generation sequencing (NGS) approaches, it is now possible to clearly identify heteroplasmic variants, and at a much lower level than previously possible. However, in order to bring these approaches into forensic laboratories and subsequently as accepted scientific information in a court of law, validated methods will be required to produce and analyze NGS data. We report here on the development of an optimized approach to NGS analysis for the mtDNA genome (mtgenome) using the Illumina MiSeq instrument. This optimized protocol allows for the production of more than 5 gigabases of mtDNA sequence per run, sufficient for detection and reliable reporting of minor heteroplasmic variants down to approximately 0.5–1.0% when multiplexing twelve samples. Depending on sample throughput needs, sequence coverage rates can be set at various levels, but were optimized here for at least 5,000 reads. In addition, analysis parameters are provided for a commercially available software package that identify the highest quality sequencing reads and effectively filter out sequencing-based noise. With this method it will be possible to measure the rates of low-level heteroplasmy across the mtgenome, evaluate the transmission of heteroplasmy between the generations of maternal lineages, and assess the drift of variant sequences between different tissue types within an individual.
Keywords: MiSeq, mtDNA, NextGENe, heteroplasmy, next-generation sequencing, Nextera
1. Introduction
Mitochondrial (mt) DNA profiling is a well-characterized and essential tool in forensic genetics [1]. Although not a unique identifier, the high mutation rate of the mtgenome has resulted in significant variability between unrelated individuals [2]. Since mutations accumulate over the lifetime of an individual, it has been suggested that all individuals would display mtDNA heteroplasmy [3], the presence of two or more mitochondrial genotypes in a cell or individual, but at such a low level it typically cannot be detected due to limitations in available technology used to sequence mtDNA [4]. Traditionally, the detection of heteroplasmic variants in sequencing data has been limited to those with frequencies of greater than 10–20%, leaving the community to debate whether improved resolution techniques will reveal greater levels of heteroplasmy across the entire mtgenome. In recent years, it has become clear that improving the resolution of heteroplasmy detection has the potential of increasing the discrimination power of the testing results when applied to forensic cases [5].
For two decades, capillary electrophoresis (CE)-based Sanger sequencing has been the gold standard in DNA sequencing [6], but this technology is inherently hampered by limitations in speed, throughput, resolution, and associated costs. The recent introduction of next-generation sequencing (NGS) technologies has revolutionized genomic studies, providing greater throughput at a reduced cost [7–9]. NGS platforms are currently being utilized in a broad range of applications including forensic genetic investigations of STR loci [10,11], microbial community analysis [12], and cancer research [13,14].
To date, multiple reports demonstrate the potential for NGS in evaluating mtDNA using different NGS technology: Roche’s 454 [3,5,15], Illumina’s GAII [16–18], Illumina’s HiSeq 2000 [19], and Ion Torrent’s Personal Genome Machine (PGM) [20]. However, little work has been reported on the use of the MiSeq to analyze the mtgenome. The Illumina MiSeq would be an ideal candidate for mtDNA analysis as several reports have provided quality metrics showing the strength of the technology. A performance evaluation by Loman et al. [21] compared the 454 GS Junior, Ion Torrent PGM, and MiSeq platforms and determined the highest quality reads were produced by the MiSeq, with a near absence of insertion and deletion (indel) errors and a low substitution error rate of 0.1 substitutions per 100 bases. Another performance evaluation [22] assessed the Ion Torrent, Pacific Biosciences’ (Menlo Park, CA) RS, and the MiSeq platform and found the MiSeq to produce the highest number (76.45%) of error-free reads without a single mismatch or indel. This study documented low error rates for the MiSeq (<0.4%), and identified regions of DNA containing homopolymer stretches of >20 consecutive bases as being more error prone.
The current study reports on the development of an optimized protocol for sequencing the mtgenome on the Illumina MiSeq platform that can be used by the forensic community. The protocol was used to sequence 156 whole mtgenomes using a MiSeq Benchtop Sequencer and Nextera® XT library preparation.
2. Overview
2.1 MiSeq
The MiSeq sequencer uses a reversible-terminator sequencing-by-synthesis (SBS) approach capable of producing a massive parallel sequencing environment. For DNA sequencing, samples are fragmented, modified with adaptors and dual indexes, pooled (for multiplexing), and sequenced. The addition of platform specific adaptors to the fragmented DNA creates “clusterable” DNA fragments capable of binding to the flow cell of the sequencer. The addition of unique dual indexes enables the multiplexing of up to 96 samples per sequencing run. This process of sample modification is termed library preparation, and several commercially available library preparation kits are available from Illumina.
Once attached to the flow cell, the single stranded DNA fragments undergo bridge amplification, forming millions of clusters for sequencing. After cluster formation, a fluorescently-labeled terminator is imaged as each reversible ddNTP is added. The newly incorporated ddNTP is then modified to remove the fluorescent dye and the 3’-end of the DNA fragment is unblocked, allowing for the incorporation of the next base. Through this proprietary reversible terminator-based approach, each incorporated base is assessed separately, significantly reducing sequencing errors inherent to homopolymeric stretches of DNA sequence when using the dosing approaches of other NGS systems.
2.2 Library preparation
See the materials and methods section for preparation of long-range PCR products of the mtgenome. Following long-range amplification, library preparation can begin. The Nextera® XT (Illumina, Inc.) library preparation kit, designed for sequencing amplicons, small genomes, and plasmids, was selected for this study. This kit is the fastest method for sample preparation for any Illumina sequencing platform, allows multiplexing of up to 96 samples, and has the lowest DNA input requirement (1 ng).
Using the Nextera® XT library preparation, the largest available reagent cartridge (500 cycles at the time of this study) for 250 base pair (bp) paired-end reads, and having a target coverage of 20,000× per nucleotide (nt), the following Lander/Waterman equation (eq. 1) [23,24] was used to determine the theoretical total coverage for each MiSeq run.:
| (eq. 1) |
Where C is coverage; L is read length; N is number of reads; G is haploid genome length.
Project variables included a read length (L) of 250 bp (paired-end 500 cycle kit), 16 × 106 reads (N; based on Illumina v2 chemistry), and a genome length (G) of 16,569 bp, equating to each base in the mtgenome being sequenced 241,415 times on average. Dividing this estimate by our desired coverage of 20,000× allowed for multiplexing of twelve samples per run. The rationale for targeting a read count of ~20,000× was to maximize the ability to detect and report low level heteroplasmic variants. The vast majority of previously published data has involved read coverage of less than 500, which may not allow for significant improvement over Sanger sequencing in the detection of lower level variants. For example, detection of a low-level variant at a frequency of 1% would require the variant to be called by the sequencing platform 10, 50, or 200 times with a corresponding coverage of 1,000×, 5,000×, or 20,000× respectively. The lower the coverage, the less likely variants below 10% can be reported reliably.
2.3 Tagmentation
The Nextera® XT library preparation uses enzymatic fragmentation in a process termed tagmentation. During tagmentation, DNA amplicons are enzymatically fragmented by the Nextera® XT transposase and simultaneously tagged with adaptors that allow for subsequent PCR amplification to introduce the indexes. A wide variety of libraries can be sequenced, with typical libraries displaying a size distribution range of 200 bp up to 1,000–1,500 bp [28]. For a 500-cycle paired-end run, the optimal fragment size would be 250–500 bp.
2.4 Quantification with the Qubit vs. qPCR
Following the Nextera® amplification step, accurate quantification of DNA library templates is critical for achieving optimal cluster density on the flow cell and producing the highest quality data. A dsDNA-specific fluorescent dye approach, using the Qubit system or picogreen as the dye, is the suggested quantification method in the Nextera® XT protocol provided by Illumina, Inc., but quantification using qPCR yields a more accurate estimate of the “clusterable” DNA since it is specifically targeting the adaptors that allow the DNA fragments to bind to the flow cell of the MiSeq. The qPCR method also quantifies ssDNA which is viable for binding to the flow cell, which would not be detected using dsDNA-specific dyes. Of the two methods, the Qubit method is more attractive due to ease of use, lower time requirements, and cost efficiency.
2.5 Optimization
The general workflow for library preparation includes tagmentation of long-range PCR products, PCR amplification to introduce indexes, PCR clean-up, product quantification, pooling, and sequencing. Initial runs, following the manufacturer’s recommendations, produced low cluster density, averaging 575 K clusters/mm2, or an average of 50% of the maximum theoretical output. There was also an uneven distribution of reads with most areas of the genome showing ~5,000× coverage and some regions having coverage in excess of 50,000×. In an attempt to optimize cluster density and data output, as well as increase consistency and achieve more evenly distributed sequencing, we conducted a close examination of Illumina’s generalized sample preparation protocol and determined that optimization would focus on tagmentation, quantification, and PCR clean-up steps.
3 Materials and methods
The laboratory work conducted for this study was a collaborative effort by the Holland group (Forensic Science Program, Penn State University, University Park, PA) and the Makova group (Biology Department, The Pennsylvania State University, University Park, PA). All samples received identical library preparation, but were obtained, extracted, and amplified (long-range PCR) through different methods as described below.
3.1 Samples and DNA extraction
A total of 156 DNA samples were sequenced with the NGS approach on the MiSeq. DNA samples obtained by the Holland group (n=48) were from various sources including cheek swabs, previously extracted DNA, cultured cells, and extracted DNA from unknown source material. DNA was isolated using an organic extraction method. Informed, written consent was obtained from each individual supplying a DNA sample. All work for this study was conducted under a Penn State University internal review board (IRB) approved project (IRB #32047).
DNA samples obtained by the Makova group (n=108) were from cheek swabs. The isolation of genomic DNA was carried out by the Nucleic Acid Facility at Pennsylvania State University based on the method of Freeman et al. [25]. Briefly, the buccal tissues were collected using cotton swabs, placed and stored in Slagboom buffer (0.1 M NaCl, 10 mM Tris-HCl pH8, 10 mM EDTA, 0.5% SDS) with Proteinase K (0.2 mg/ml). Organic de-proteinization reagent was used to digest proteins, and genomic DNA was precipitated with isopropyl alcohol. The precipitated genomic DNA pellet was re-suspended in 250 µL of TE buffer. Cheek swab specimens were collected with informed, written consent from each individual. This study was approved by the Human Subjects Protection Office of the Pennsylvania State College of Medicine (IRB # 30432EP).
3.2 Long-range PCR
Two long-range PCR approaches were used in this study. Both long-range protocols amplified the entire mtgenome with overlapping ~8.5 kilo base (kb) fragments, but utilized different primer sets and annealing locations. The Holland laboratory amplified forty-eight samples according to the method detailed in Fendt et al. [26]. Amplification of the mtgenome was performed using the following oligonucleotide primer sets from Biosearch Technologies Inc., Novato, CA: 5'-AAATCTTACCCCGCCTGTTT-3' (forward primer A; F2480A) and 5'-AATTAGGCTGTGGGTGGTTG-3' (reverse primer A; R10858A) 5'-GCCATACTAGTCTTTGCCGC-3' (forward primer B; F10653B) and 5'-GGCAGGTCAATTTCACTGGT-3' (reverse primer B; R2688B). Two independent amplification reactions were performed, each in a total volume of 50 uL containing 5 uL of 10× TaKaRa LA PCR buffer II with 25 mM Mg2+ (Clontech, Mountain View, CA), 0.2 mM of each deoxynucleoside triphosphate (TaKaRa dNTP mixture; Clontech), 2 units TaKaRa LA Taq polymerase (Clontech), 0.0125 mg Ambion ultrapure bovine serum albumin (Life Technologies, Carlsbad, CA), 0.2 uM of each primer (Biosearch Technologies, Novato, CA), and 4 ng of template DNA. Negative controls had no added template DNA. PCR was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) thermal cycler under the following conditions: a 93°C soak for 3 min; 93°C for 15 sec, 60°C for 30 sec, 68°C for 5 min for 14 cycles; and 93°C for 15 sec, 55°C for 30 sec, 68°C for 9 min for 27 cycles. The extension time was elongated by 10 sec for each successive cycle during this last phase. PCR products (5 uL) were imaged by agarose gel electrophoresis to confirm successful amplification.
The Makova laboratory amplified 108 samples according to the method detailed in Goto et al. [16]. Amplification of the mtgenome was performed using the following oligonucleotide primer sets from Integrated DNA Technologies, Skokie, IL: 5’- GCGACCTCGGAGCAGAAC-3’ (L2817) and 5’- GTAGGCAGATGGAGCTTGTTAT-3’ (H11570) for amplicon A, and 5’-CCACTGACATGACTTTCCAA-3’ (L10796) and 5’-AGAATTTTTCGTTCGGTAAG-3’ (H3370) [27] for amplicon B. One hundred nanograms of isolated genomic DNA was used as a template in a 50-µl PCR reaction containing 2 µM of each of the two primers, 200 µM dNTP (PCR grade; Roche Applied Science, Indianapolis, IN), 3 units of Expand High Fidelity PCR Enzyme (Roche Applied Science), 1× PCR buffer with 1.5 mM Mg2+, and nuclease-free water (Teknova, Hollister, CA). The PCR parameters included a 94°C soak for 2 min; followed by 10 cycles of 94°C for 15 sec, 62.3°C for 30 sec, and 68°C for 8 min; followed by 30 cycles of 94°C for 15 sec, 62.3°C for 30 sec, and extension at 72°C for 8 min. The extension time was elongated by 5 sec for each successive cycle during this last phase. A final extension was performed at 72°C for 7 min. Amplifications were carried out in a GeneAmp PCR System 9700. PCR products (2 uL) were imaged by agarose gel electrophoresis to confirm successful amplification.
3.3 MiSeq NGS
All samples (n=156) were sequenced on Illumina’s (San Diego, CA) MiSeq benchtop sequencer, using Nextera® XT (Illumina, Inc., San Diego, CA) sample preparation, and the 500-cycle reagent kit. Specifically, samples were sequenced using paired 250 nt reads, multiplexing twelve, dual indexed samples per run. Although the Nextera® XT kit reagents were used in sample preparation, the protocol followed was a combination of the protocols available for Nextera® XT and Nextera® DNA kits. The manufacturers recommended protocol for Nextera® XT was used with the exception of the bead normalization procedure. The bead normalization step streamlines library preparation for sequencing runs containing a large number of samples (i.e., multiplexing 96 samples), but each of runs performed in this study contained only twelve samples. Therefore, quantification and dilution of individual samples was a more efficient approach to normalizing the twelve samples. Quantification and dilution is the method used in the Nextera® DNA protocol for normalization, so this protocol was followed for the remainder of the library preparation.
3.4 Tagmentation
Quality metrics generated for early MiSeq runs indicated a non-optimal fragment size range, with the production of correspondingly lower quality data. Bioanalyzer (Agilent Technologies, Palo Alto, CA) data on the size distribution of fragments indicated that our samples had an average size of approximately 1,000 bp for early runs. Theoretically, fragments larger than the optimal size range do not cluster as well due to smaller sized fragments having greater cluster efficiency. In an attempt to decrease the average fragment size of our libraries an experiment was conducted using a constant amount of Amplicon Tagment Mix (Illumina, Inc.) with varying amounts of input DNA (Table 1). The tagmentation products resulting from five different input masses from three different samples (n=15) were assessed using a Bioanalyzer to determine the fragment size distribution. Results were evaluated after MiSeq runs to determine the best ratio of long-range amplification product and tagmentation mix.
Table 1.
Tagmentation matrix with bioanalyzer results for average fragment size.
| Tagmentation Input Mass (ng) | ||||||
|---|---|---|---|---|---|---|
| Sample ID | Purified | 1.20 | 1.00 | 0.75 | 0.50 | 0.25 |
| RP438 | No | 810 | 830 | 741 | 763 | 752 |
| RP438_P | Yes | 760 | 779 | 690 | 670 | 533 |
| RP132_P | Yes | 740 | 825 | 868 | 681 | 751 |
3.5 Quantification
To assess the accuracy of our template quantification step, a subset of samples (n=92) was quantified by two methods, following the manufacture’s recommendations: 1) a dsDNA-specific fluorescent dye method (Qubit), and 2) a qPCR method (KAPA Biosystem library quantification kit). Qubit quantification targets dsDNA using an intercalating dye and the Qubit HS dsDNA kit (Invitrogen, Carlsbad, CA). Quantification using the KAPA Biosystem kit targets libraries constructed with Illumina adaptors containing the following qPCR primer sequences: P1 5’-AAT GAT ACG GCG ACC ACC GA-3’ and P2 5’-CAA GCA GAA GAC GGC ATA CGA-3’ for qPCR. Each well of a 96 well plate (VWR International, Radnor, PA) included a total reaction volume of 20 uL containing 12 uL of KAPA SYBR FAST qPCR master mix with primer premix, 3.6 uL of PCR grade water, 0.4 uL of ROX passive reference dye, and 4 uL of diluted (500×) template DNA. The qPCR was conducted using a spectrofluorimetric thermal cycler (7500 Real Time PCR System, Applied Biosystems, Foster City, CA) under the following conditions: a 95°C soak for 5 min; and 95°C for 30 sec, 60°C for 90 sec for 35 cycles. Calibration curves (arbitrarily set cycle threshold value [Ct] versus log gene copy number per uL) were created using the 452 bp DNA standards provided with the KAPA Library Quantification kit; dsDNA concentrations ranged from 0.0002 – 20 pM. Amplifications were conducted in duplicate along with negative controls (no template DNA).
For comparison of qPCR concentrations (nM) to Qubit readings (ug/uL) a conversion factor was used. Conversion factors recommended in the Nextera® DNA Sample Preparation Guide [28] are based on the average fragment size of the prepared library with 1 ng/uL being equivalent to 6 nM, 3 nM, and 1.5 nM for average fragment sizes of 250 bp, 500 bp, and 1,000–1,500 bp, respectively. Based on the average fragment size of our libraries (~650 bp), three conversion factors: 1.0 ng/uL = 1.5, 2.75, and 3.0 nM, were used to assess agreement between qPCR and Qubit quantification.
3.6 PCR clean-up
Agencourt® AMPure® XP beads (Beckman Coulter Inc., Indianapolis, IN) were used to clean-up the Nextera® XT PCR products. AMPure® XP beads are solid phase reversible immobilization (SPRI) magnetic particles used to reversibly bind DNA in the presence of polyethylene glycol (PEG) and salt. Quantification of DNA concentrations post clean-up indicated a loss of DNA during this step.
The lower than expected concentration of DNA recovered during clean-up also impacted the subsequent dilution steps and denaturation procedure. Following Illumina’s standardized protocol, after PCR clean-up, all samples are normalized to a concentration of 2 nM, pooled, denatured, initially diluted to a concentration of 20 pM, and then further diluted to give 1000 uL of the desired input concentration (15–17 pM for this study). The protocol has a standardized volume (20 uL) and normality (0.2 N NaOH) for the denaturation based on the pooled library having a starting concentration of 2 nM. Several of our samples had concentrations less than 2 nM. To accommodate these samples, the amount of buffer added to the library for the initial dilution was decreased, allowing the desired final concentration of DNA to be loaded on the flow cell. Accordingly, this increased the final normality of NaOH loaded onto the flow cell to greater than 0.001 N, which can inhibit clustering efficiency. To avoid overloading the flow cell with NaOH, the starting normality was lowered to 0.1 N, reducing the final normality on the flow cell to 0.0005 N. The pooled library was effectively denatured and the clustering efficiency was not hindered.
3.7 NGS Data analysis
All sequence data generated in this project was mapped to the revised Cambridge Reference Sequence (rCRS; GenBank ID NC_012920.1) [29,30] using the MiSeq’s integrated computer software platform, MiSeq Reporter (MSR; Illumina, Inc., San Diego, CA; v2.1.43 and v2.2.29), which uses a Burrows-Wheeler Aligner (BWA) [31] and the Genome Analysis ToolKit (GATK) for variant calling of single nucleotide polymorphisms (SNPs) and short indels. This software platform produces graphical representations of coverage and average quality scores as well as tabular output of the variant calls with respect to the rCRS. The data files generated by MSR are BAM (Binary Alignment Map), BAI (Binary Alignment Index), and FASTQ (text based format with quality scores) files. Secondary analysis was performed using Illumina’s mtDNA MSR plug-in, mtDNA Variant Analyzer (Illumina; v. 1.0.0.21), and NextGENe® (SoftGenetics, State College, PA; v2.2.3) software. The mtDNA Variant Analyzer opens data from viewer files generated through the MSR mtDNA plug-in. Within the Variant Analyzer, a question sample and known sample (i.e. rCRS) are selected for comparison, the input is interrogated, and a report is generated. The analysis in this study utilized the default settings for minimum base call quality (Q30), detection threshold (0.1 or 10%), and analysis threshold (0.25 or 25%).
Secondary analysis of the MSR generated FASTQ data was performed using the NextGENe® platform. The NextGENe® software uses a BLAST-Like Alignment Tool (BLAT) method, which employs a Smith-Waterman [32] approach with a proprietary INDEL alignment algorithm. The following filters were used in conversion of the MiSeq FASTQ files to the NextGENe® converted FASTA files: median score threshold ≥30; max number of uncalled bases ≥3; called base number of each read ≥3; and base(s) with score ≥29. This means that the quality of the reads contained in the FASTQ files was evaluated and any reads failing to surpass these thresholds were filtered (removed) and not included. The converted FASTA files were then aligned by the NextGENe® platform to the rCRS using paired reads and the following filters: matching requirement ≥12 bases and ≥85%; load paired reads library size range from 150 to 2000 bases; and ambiguous mapping, detection of large indels, and rigorous alignment were all selected. Finally, the mutation report generated by the NextGENe® software had the following filters applied: mutation percentage ≥0.5 (greater than or equal to 0.5% of the total reads); SNP allele ≥40 counts (total read counts of low level variants of at least 40); total coverage ≥200 (except for homozygous); read balance ratio ≥0.2 (read ratio of 20% or greater); and a frequency of 100% for the evaluation of variants (all observations reported). This means that for a variant sequence to be included in the report each nt position had to have a coverage of at least 200 calls, the variant had to be detected in at least forty of those calls, and low level variants had to be present in at least 0.5% of the total reads, which would require a read coverage of at least 8000. The ratio (balance) of a variant being detected in the forward versus reverse reads was evaluated for all variants (100%) and could not be less than 4:1 (0.2) in either direction.
4. Results and Discussion
In this study 156 whole mtgenomes, were sequenced using a MiSeq Benchtop Sequencer and Nextera® XT library preparation kits. Libraries consisting of 12 samples were prepared from long-range PCR products using a modification of the Nextera® XT and Nextera® DNA protocols, and then sequenced using paired-end 250 × 250 nt reads. The process of library construction was optimized through adjustment to several of the suggested protocol parameters including: input DNA for tagmentation, NaOH concentration for denaturation, and the concentration of pooled library added to the flow cell. After extensive optimization and protocol development, we were able to successfully sequence the entire mtgenome at a minimum coverage of 5K to 10K on the MiSeq. We were also able to sequence the two hypervariable regions of the genome (HV1 and HV2) at a coverage of >100K (data not shown).
Sample haplotypes are presented in supplemental table 1. Haplotype generation was accomplished through analysis of the MiSeq sequencing data using both the NextGENe® and mtDNA Variant Analyzer (Illumina) software platforms. Comparison of the haplotypes produced by the two platforms showed concordance, but due to differences in how each of the software platforms report differences from the reference sequence, manual inspection of the data (using NextGENe) was required to confirm several apparent differences. Sites requiring manual inspection were limited to calls involving length heteroplasmy in the polycytosine stretches located in HV1 (16184–16193 bp) and HV2 (303–315 bp).
The mtDNA Variant Analyzer software is very user-friendly and more of a plug-and-play approach, while the NextGENe® software allows for more detailed examination of the sequencing data. The Illumina system was more streamlined in generating haplotypes and demonstrated a higher level of accuracy in the nomenclature used to call indels. All indels that were not represented in the NextGENe® mutation report were confirmed as true mutations through manual evaluation of the NextGENe® alignment pile-up. The haplotypes produced using the NGS approach show concordance with haplotypes generated through conventional Sanger sequencing, but the MiSeq data provided more detailed information about heteroplasmy. A more comprehensive analysis of the heteroplasmy data will be forthcoming; manuscript in preparation.
4.1 Long-range PCR
The two long-range PCR approaches provided in the materials and methods section were used on independent runs and produced similar quality metrics (Table 2). The data used in this assessment was generated from mtDNA sequences only. PhiX and pUC18 control data generated during Makova runs, as well as PhiX control data generated during Holland runs, was excluded in the quality metrics. Statistical assessment of NextGENe® analyzed data did not indicate a significant difference between the two PCR approaches. Both PCR techniques showed a decrease in the quality of reads from read 1 to read 2, as indicated by the percent of reads converted by the NextGENe® software, with a decrease of 7.6% for the Holland approach and 8.3% for the Makova approach. A decrease in quality towards the ends of reads, as well as the entire sequence for read 2 has been documented by others as a general phenomenon [21,22]. Both methods had decreased coverage from approximately the first nt of the reference genome through nt 600, especially after position 300, where a homopolymeric stretch of cytosines is located. In addition, coverage decreased from nt 16,480 to the last nt of the genome sequence, which did not affect coverage in HV1. Initially this was thought to be due to alignment challenges when attempting to align a circular genome using a linear reference template. However, reorganization of the reference sequence (i.e., placing the mtgenome origin in the middle of the linearized reference genome) continued to produce reduced coverage. Overall, both methods produced high-quality sequencing data spanning the entire length of the mtgenome.
Table 2.
Comparison of the two long-range PCR approaches based on quality metrics generated by the NextGENe® and MiSeq Reporter software platforms.
| NextGENe Analysis | |||
|---|---|---|---|
| Long-Range PCR Protocol | Input file Read 1 Converted Reads (% of total) |
Input file Read 2 Converted Reads (% of total) |
Aligned to rCRS Read 1 & 2 (% of total) |
| Holland average±one stnd dev. | 97.8±1.1 | 90.4±3.8 | 96.2±4.9 |
| Makova average±one stnd dev. | 96.2±3.6 | 88.3±7.3 | 87.8±21.9 |
| Welch t-test p-value* | 0.0000 | 0.0000 | 0.0002 |
Two-tailed Welch t-test with 95% Confidence Level
4.2 Tagmentation
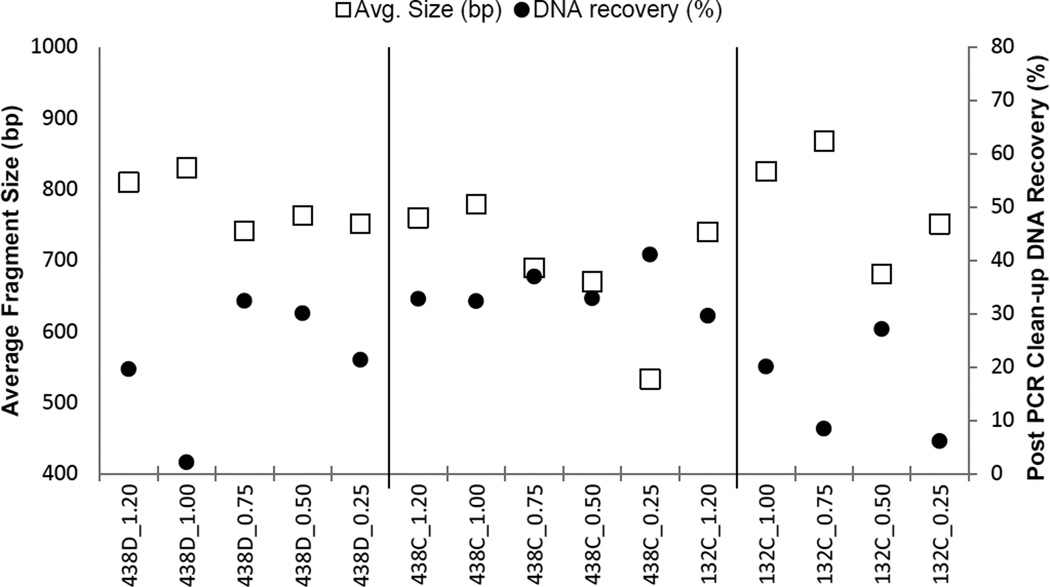
The average fragment size following tagmentation decreased with decreasing amounts of input DNA template. Considering that adaptors and indexes can account for 130–165 bp of the resulting DNA fragment length after tagmentation, and to achieve complete sequencing of the fragments with paired 250 nt reads, an input mass of 0.25–0.50 ng of long-range amplicon produced the best average fragment size (~650 bp). Another consideration was how much DNA was being recovered after AMPure® bead clean-up of the Nextera® PCR products. In the Nextera® DNA protocol, the samples are normalized to a concentration of 2 nM before pooling the library. To date, approximately 25% of our post-PCR clean-up products have had a concentration less than 2 nM. Although samples less than 2 nM are not precluded from successful sequencing, these samples required modification to the sample preparation protocol and presented a challenge to maintain NaOH concentrations that would not hinder cluster formation. Figure 1 shows the average fragment size, as well as the percent of DNA recovered after PCR clean-up. From this data it was determined that a tagmentation input mass of 0.5 ng produced the best balance between the average fragment size and recovering as much DNA as possible following clean-up of Nextera® PCR products.
Figure 1.
Average fragment size and DNA recovery observed during tagmentation optimization.
4.3 Quantification
A total of 92 samples were quantified using both fluorometric (Qubit) and qPCR methods. Using Illumina’s suggested conversion factor of 1.0 ng/uL = 1.5 nM, 91% of the DNA concentrations measured by qPCR were greater than the Qubit measurement. The increased concentration could be due to ssDNA that is not quantifiable using the Qubit’s fluorescent intercalating dye chemistry. A statistical assessment of the agreement between the two quantification methods was performed, with the Qubit readings converted to nM concentrations using three conversion factors: 1.5 ng/uL = 1.5, 2.75, and 3.0 nM. Agreement between the direct qPCR and converted Qubit measurements was assessed using the statistical method of Bland and Altman [33,34]. Evaluating the data by this method gives a more accurate assessment of agreement than the typical approach of comparing two methods through the calculation of a correlation coefficient (R). This is because a high R value does not necessarily correspond to a high degree of agreement between the methods. Perfect agreement requires all data points to lie on the line of equality, which would have a high R value, but any straight line would also produce a high R value. The Bland and Altman approach evaluates the degree of agreement between the two methods using basic graphing and statistical calculations providing insight into how much the two measurement techniques are likely to differ from each other. The degree to which the two methods can differ without a negative effect is project specific. The Bland and Altman approach uses a plot of the difference between the methods versus their mean and calculated “limits of agreement” (eq. 2) to determine if the two methods can be used interchangeably.
| (eq. 2) |
Where đ is the mean difference and s is the standard deviation of the differences.
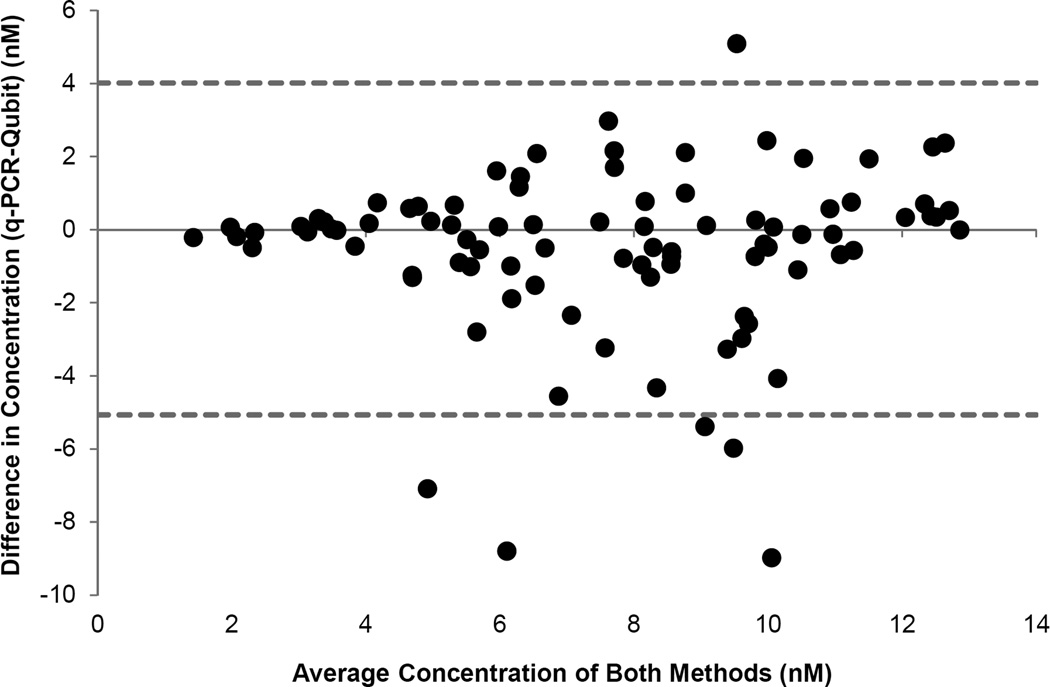
If the two methods produce the exact same reading for all measurements, all data points would lie on a line of equality. Although plotting the data with a line of equality helps to visualize the degree of agreement between the two methods of quantification, a plot of the difference between the two methods versus their mean (Figure 2) can be more informative. Since we do not know the true value, the mean of the two methods of quantification is the best available estimate. By plotting this number against the difference, an evaluation of any relationship between the true value and measurement error can be made. Basic statistical calculations are presented in Table 3. It was determined that using a conversion factor of 2.75 for the Qubit data would produce the closest approximation of the concentrations determined through qPCR. The Bland and Altman plot (Figure 1) for data using the 2.75 conversion factor shows 86 of the 92 data points (93.5%) falling within the calculated limits of agreement. Interestingly, five of the six data points falling outside the limits of agreement represent data points in which the concentration measured by qPCR was less than the Qubit measurements, with two samples falling below detection by qPCR. The two samples that were below detection were not diluted (i.e., not normalized to 2 nM), yet still processed and pooled with the other samples. Even though these samples were below detection through qPCR analysis, and calculated to be 7.1 nM and 8.8 nM as measured by the Qubit, they still accounted for 12.4% and 13.3% of the clusters on the flow cell for their respective runs. It is unclear why these samples, which clearly contained clusterable DNA, were below qPCR detection. Therefore this outcome may have been due to primer binding issues with the qPCR assay, or human error.
Figure 2.
Bland and Altman plot of differences between quantification methods (Qubit and qPCR) versus the mean of those measurements. Dashed lines represent the calculated limits of agreement (95%). In this plot, Qubit measurements were converted using a factor of 2.75 nM = 1 ng/uL.
Table 3.
Basic statistics for Bland and Altman assessment of Qubit and q-PCR quantification methods for conversion factors 1.5 nM, 2.75 nM, and 3.0 nM = 1 ng/uL.
| Basic Statistic | q-PCR vs. Qubit1.5 | q-PCR vs. Qubit2.75 | q-PCR vs. Qubit3.0 | |
|---|---|---|---|---|
| avg diff (đ) | 2.48 | −0.52 | −1.13 | |
| stnd dev (s) | 2.23 | 2.27 | 2.36 | |
| limit of agreement |
(đ−2s) (đ+2s) |
−1.97 | −5.07 | −5.84 |
| 6.94 | 4.02 | 3.59 | ||
| stnd error đ | 0.23 | 0.24 | 0.25 | |
| stnd error of the limit | 0.40 | 0.41 | 0.43 | |
| 95%CI for bias | 2.02 | −0.99 | −1.61 | |
| 2.95 | −0.05 | −0.64 | ||
| 95% CI for lower limit | −2.77 | −5.88 | −6.68 | |
| −1.17 | −4.25 | −4.99 | ||
| 95% CI for upper limit | 6.14 | 3.20 | 2.74 | |
| 7.74 | 4.83 | 4.43 | ||
Using a conversion factor of 2.75 nM = 1 ng/uL, the mean difference (đ) within the data set was −0.52, with a standard deviation of 2.27. This was determined to be acceptable for library quantification, and the last two MiSeq runs were prepared using only Qubit quantification. Based on a 12 sample run, an equal distribution of flow cell clusters (or reads) would equate to 100% divided by 12, or an average of 8.33% per sample. The standard deviation of the percent of reads identified passing filter per run was 3.21 and 2.54, respectively, for each of the last two runs using Qubit alone for quantification. For the eight runs using both methods of quantification, the standard deviation ranged between 0.92–5.67. Other quality metrics for all ten runs can be found in Table 4.
Table 4.
Quality metrics generated by MiSeq Reporter for sequencing runs that used both Qubit and q-PCR methods for quantification and sequencing runs using only Qubit quantification after optimization.
| Quantification Method |
Sample Count |
Cluster (K/m2) |
Yield (Gb) |
>Q3 (%) |
Clusters (K) |
Passing % |
Unaligned (K) |
Unindexed (K) |
Duplicate (K) |
|---|---|---|---|---|---|---|---|---|---|
| Qubit + q− | 12 | 697 | 6.0 | 84.8 | 11,76 | 91.2 | 2,18 | 242 | 3,98 |
| 12 | 307 | 2.9 | 87.6 | 5,54 | 91.5 | 1,77 | 147 | 1,06 | |
| 11 | 634 | 5.8 | 89.7 | 11,23 | 91.6 | 4,09 | 180 | 2,19 | |
| 11 | 666 | 6.2 | 87.0 | 11,90 | 91.9 | 3,46 | 222 | 2,91 | |
| 11 | 1107 | 8.1 | 80.4 | 15,73 | 79.1 | 4,82 | 505 | 4,12 | |
| 11 | 991 | 7.7 | 81.1 | 15,03 | 82.0 | 5,30 | 355 | 3,53 | |
| 11 | 1323 | 10.5 | 82.8 | 20,47 | 83.4 | 7,21 | 377 | 5,58 | |
| 11 | 1181 | 9.7 | 82.5 | 18,95 | 85.7 | 5,41 | 376 | 5,18 | |
| 12 | 550 | 5.0 | 89.6 | 9,77 | 91.8 | 173 | 463 | 3,20 | |
| 12 | 863 | 7.7 | 88.3 | 14,89 | 89.9 | 336 | 691 | 5,72 | |
| avg. | 854. | 7.0 | 84.5 | 13633. | 87.0 | 3549. | 353. | 3454. | |
| stnd. | 317. | 2.3 | 3.5 | 4416. | 4.8 | 2323. | 167. | 1498. | |
| Qubit | 12 | 992 | 8.7 | 86.6 | 16,90 | 89.3 | 421 | 761 | 6,51 |
| 11 | 986 | 8.5 | 84.9 | 16,60 | 88.6 | 5,73 | 319 | 4,44 | |
| avg. | 989. | 8.6 | 85.8 | 16754. | 89.0 | 3077. | 540. | 5482. | |
| stnd. | 4.2 | 0.1 | 1.2 | 212. | 0.5 | 3756. | 312. | 1465. |
4.4 PCR Clean-up
Quantification of DNA concentrations following PCR clean-up with AMPure® beads indicated a loss of DNA during clean-up and impacted the subsequent denaturation process and dilution steps. To increase DNA recovery, the amount of AMPure® beads was increased from a ratio of 0.5× to 1.0× per reaction, the ethanol concentration was decreased from 80% (Illumina protocol) to 70% (AMPure® protocol), and the final dry time before eluting DNA into buffer solution was decreased to 10 minutes. None of these alterations to the protocol resulted in substantial increases in the amount of DNA recovered, and therefore, were not adopted. The apparent loss of DNA was ultimately attributed to the exclusion of DNA fragments less than 200 bp during the clean-up process. Bioanalyzer data (not shown) generated from fifteen samples clearly showed peaks (65 –80% of the total area) representing fragment sizes below 200 bp in samples collected pre-PCR clean-up that were not detected in samples collected post-PCR clean-up.
4.5 Alignment
All high quality sequencing data (i.e., not removed by quality filters) was analyzed using the MSR and NextGENe® software platforms. The MSR software analyzed a total of 202 million paired end reads with an average of 78.7±15.6% of the forward and 70.7±14.2% of the reverse reads mapping to the rCRS. The NextGENe® software analyzed a total of 200 million paired reads, with an average of 95.5±7.2% of the forward and 87.5±10.3% of the reverse reads successfully converted (Q-score filter ≥30) from MiSeq-generated FASTQ files to NextGENe®-generated FASTA files. Of the successfully generated FASTA files, 84.3±15.4% of the combined forward and reverse reads mapped to the rCRS, with an average per nt coverage of 17,027 reads and a range of coverage averaging 1,035 to 48,655 reads, respectively. It should be noted that these numbers are artificially low due to the NextGENe® software having a coverage cut-off of 65,535 reads, and that some of our samples exceeded this coverage at various nucleotide locations across the mtgenome.
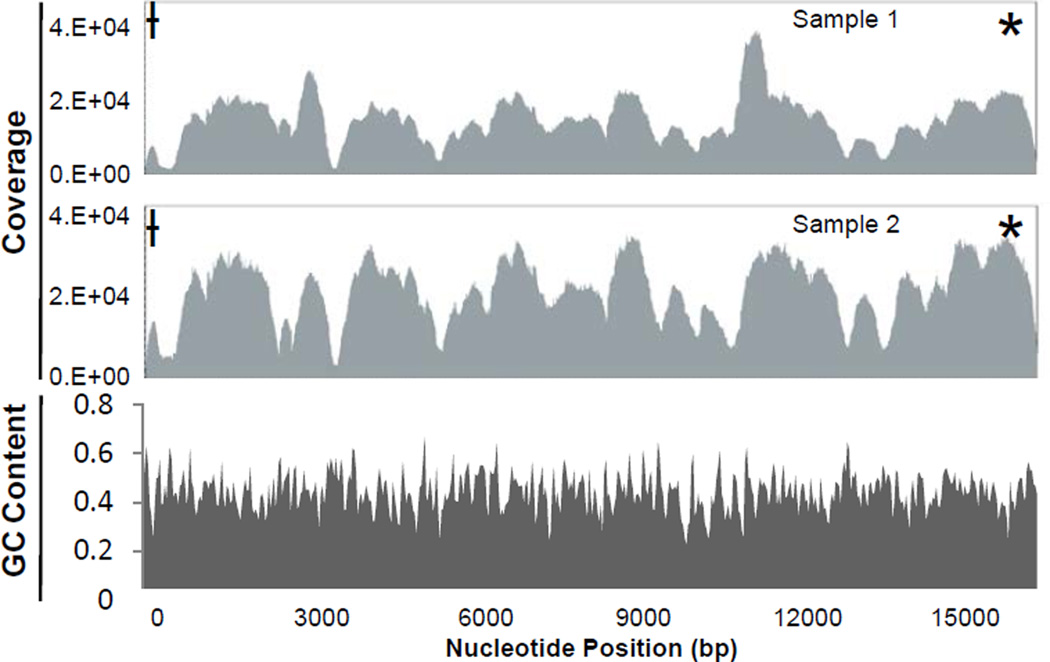
A coverage curve, representing the number of times each nt in the mtgenome was sequenced, is graphically represented in the analysis by both software platforms (NextGENe® coverage curves for two representative samples presented in Figure 3). For the majority of samples, both alignment methods produced a characteristic distribution of reads across the entire mtgenome, with specific areas of the genome consistently producing higher or lower coverage. This type of reproducible coverage bias has been reported previously for several NGS technologies. An evaluation of the Roche 454, Illumina GA, and ABI SOLiD sequencing platforms showed that each technology generated a distinctive yet reproducible characteristic pattern of non-uniform coverage that was theorized to be due largely to local sequencing characteristics [35]. Sequencing technologies are known to be susceptible to various forms of bias, with several studies reporting a correlation between coverage bias and GC content. Dohm et al. [36] and Teytelman et al. [37] found a positive correlation between greater coverage and higher GC content (GC content of genomes sequenced was 35–38%). Bentley et al. [38] also found a decrease in sequencing coverage for areas of the human genome with GC contents <4% and >76%.
Figure 3.
Representative coverage curves and GC content plot (NextGENe® software). GC content calculated using the rCRS, based on a rolling window of 39 base pairs.
Hypervariable region 2 (HV2)
* Hypervariable region 1 (HV1)
HV2 D-loop extension
The GC content of the mtgenome was evaluated manually and using the NextGENe® software. Manual calculation in the areas of low coverage spanning bp 300–600 (50%) and 3350–3650 (52%) indicated the GC content in these areas fell outside the range of GC content found by other studies to correlate to coverage bias. A plot of GC content across the entire rCRS was generated by the NextGENe® software based on a rolling window of 39 bp, and showed fluctuations in GC content ranging from approximately 25% to 60% with an average content of 44.4% across the entire genome (Figure 3). Neither of these evaluations offered a satisfactory explanation for the uneven distribution of coverage.
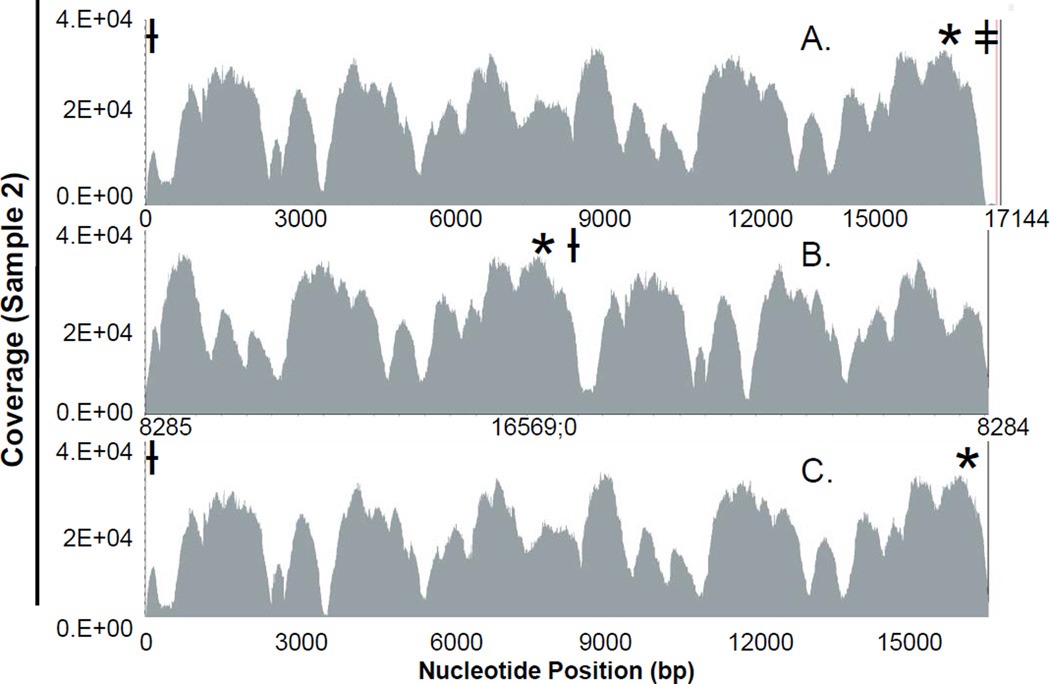
Although the specific reason for the uneven distribution of coverage in this study is unknown, attempts to assess the apparent coverage bias were made including using a rearranged rCRS as the reference sequence and inclusion of lower quality data originally excluded from alignment due to quality filtering. Low coverage due to difficulties in aligning the data to a linearized circular mtgenome was assessed by aligning the sequencing data to two different rearrangements of the rCRS (Figure 4). One rearrangement extended the rCRS by adding the D-loop (bp 0–575) sequence to the end of the sequence (i.e., after bp 16,569; Figure 4A), while the other rearrangement located the origin of the mtgenome in the middle of the linearized sequence (Figure 4B). Neither approach resulted in a significant increase in coverage in the regions of interest.
Figure 4.
- Alignment to an extended version of the rCRS with D-loop addition after nucleotide 16,569.
- Alignment to a rearranged rCRS with the origin located in the middle of the linearized genome.
- Alignment to the rCRS (nt 0 thru 16,569).
Raw data not included in alignment analyses (i.e., removed reads) due to quality filtering was also aligned to the rCRS to assess whether specific areas of the mtgenome are prone to producing lower quality data (results not shown). On average, the removed reads represented 6.4%±3.0% of the total number of high quality reads, and 4.3%±1.2% of the average coverage of the high quality reads. With an average coverage of 18,794.0±7897.7 reads for the high quality data, this translates to an insignificant average increase in coverage of 810±225.5 reads, distributed across the entire genome. Coverage profiles generated from the removed reads generally mimicked the read distribution of the coverage profiles of the corresponding high quality reads, and did not indicate that specific regions of the mtgenome are prone to producing lower quality data.
4.6 Quality Filtering
Initial assessment of the MiSeq data indicated that the most common sequencing errors are apparent A>C transversions. This type of error has been reported in other sequencing studies using Illumina’s Genome Analyzer [36,39,40], which employs the same SBS chemistry as the MiSeq. One way to decrease the number of these reported errors is to apply a quality filter evaluating the balance ratio of forward to reverse reads. The NextGENe® software allows a balance ratio to be set that effectively rejects identified variants with an imbalance of frequency in the forward or reverse read. In the initial assessment, applying a balance ratio of 0.33 (2:1 ratio) to a randomly selected sample decreased the number of reported A>C transversions across the entire genome with the ratio of transitions to transversions (ti:tv) increasing from 0.67 to 28. A ratio of 15:1 has been reported in previous analyses of the hypervariable regions of the mtgenome [41–46]. Ultimately it was determined that a lower balance ratio of 0.2 (4:1 ratio) further reduced the number of A>C transversions (for all samples the average ti:tv = 21.2) while not excluding potential SNPs.
5. Conclusions
High quality MiSeq data was generated using 0.5 ng of long range PCR product for tagmentation, a concentration of 0.1 N NaOH for denaturation of the prepared sequencing library, and loading of the indexed library at a concentration of 17 pM. These parameters consistently produced flow cell densities averaging 845 K/mm2, data outputs averaging 7.0 Gb of data with an average of 85% of the reads having Q scores ≥30, and routinely generated sequence data for the entire mtgenome at a coverage of 5K to 10K reads, with coverage in the HV1 and HV2 hypervariable segments of the control region at >100K reads. Using this optimized method, we now have the ability to expand studies on heteroplasmy detection and rates [5,16], evaluate transmission of heteroplasmic variants within maternal lineages and between different tissues types, and conduct a variety of other studies that will need to be addressed before an NGS approach using the MiSeq system can be fully validated and brought online in a forensic crime laboratory.
The MiSeq produced reliable and reproducible data using our optimized approach. As expected, NGS results were consistent with Sanger data generated on the same samples. In general, NGS techniques have proven to be a reliable way to generate sequence data for the mtgenome [20], and our results were consistent with these previous findings. Dose dependent NGS sequencing chemistries do not perform as well with long homopolymeric stretches when compared to the reverse terminator chemistry of the MiSeq [21]. In our hands, the MiSeq produced reliable results for the typical homopolymeric stretches found in the HV1 and HV2 regions of the mtgenome. However, analysis software packages with effective alignment algorithms will be essential to ensure that sequencing errors are not introduced at the analysis level. The mtDNA Variant Analyzer software from Illumina is a user-friendly system that had the most accurate nomenclature for indels, but this platform does not allow for detailed evaluation of the sequencing pile-up, and in comparison to other platforms, the reanalysis of data using different filter settings is a more complicated and time consuming process. The NextGENe® software from SoftGenetics, Inc., is an excellent tool for this purpose, but did lack some of the tools that will be needed to analyze mtDNA data in forensic laboratories; for example, proper nomenclature for indels, and flexible/user driven filtering parameters to remove noise in the data, while retaining information for true mixtures, including heteroplasmy. Therefore, further development of the available software tools will be required for the comprehensive analysis of mtDNA sequence data for forensic purposes.
The deep coverage rates of data generated on the MiSeq allowed for the routine detection of low level heteroplasmy across the entire mtgenome during our study. While minor variants were easily detected above 1%, variants less than 1% were readily and reproducibly observed. Development of methods for accurately assessing error rates will be needed so that thresholds of detection can be set. In addition, assuming that heteroplasmy can be reliably detected, forensic laboratories would be able to report heteroplasmy on a routine basis. If this is the case, statistical models will be needed to determine the relative frequency of not only observing an mtDNA haplotype, but also the haplotype in the presence of heteroplasmy. The likelihood ratio approach used in the identification case of Nicholas Romanov serves as an existing model [47], however, a better understanding of the rates of heteroplasmy, preferably on a per nucleotide basis, are desirable in order to more effectively apply the approach. In the meantime, conservative models should be further developed to bridge the gap while those studies are being completed. Identifying a robust statistical approach will be a key feature of applying NGS techniques for the analysis of mtDNA sequence in forensic cases.
Highlights.
The MiSeq produced reliable and reproducible data using our optimized approach
Consistently averaged 7.0 Gb output of high quality data (85+% of the reads Q≥30)
NGS results were consistent with Sanger data generated on the same samples
Deep coverage on the MiSeq allowed for detection of low-level heteroplasmy
Minor variants ≥1% easily detected with variants <1% readily/reproducibly observed
Acknowledgements
Funding for this study was provided in part by Battelle of Columbus, OH, and The Eberly College of Science, Forensic Science Program, Penn State University, University Park, PA. Additional funding was provided by the Penn State Clinical and Translational Science Institute and the Pennsylvania Department of Health using Tobacco Settlement Funds (the Department specifically disclaims responsibility for any analyses, interpretations or conclusions). The authors wish to thank Anton Nekrutenko and Boris Rebolledo-Jaramillo (Biochemistry & Molecular Biology Department, The Pennsylvania State University, University Park, PA, USA) for bioinformatics support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Melton T, Holland C, Holland M. Forensic mitochondrial DNA analysis: current practice and future potential. Forensic Science Review. 2012;24:101–122. [PubMed] [Google Scholar]
- 2.Just RS, Leney MD, Barritt SM, Los CW, Smith BC, Holland TD, et al. The use of mitochondrial DNA single nucleotide polymorphisms to assist in the resolution of three challenging forensic cases. Journal of Forensic Science. 2009;54:887–891. doi: 10.1111/j.1556-4029.2009.01069.x. [DOI] [PubMed] [Google Scholar]
- 3.Payne BAI, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horevath R, et al. Universal heteroplasmy of human mitochondrial DNA. Human Molecular Genetics. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salas A, Lareu MV, Carracedo A. Heteroplasmy in mtDNA and the weight of evidence in forensic mtDNA analysis: a case report. International Journal of Legal Medicine. 2001;114:186–190. doi: 10.1007/s004140000164. [DOI] [PubMed] [Google Scholar]
- 5.Holland MM, McQuillan MR, O’Hanlon KA. Second generation sequencing allows for mtDNA mixture deconvolution and high resolution detection of heteroplasmy. Croat Med Journal. 2011;52:299–313. doi: 10.3325/cmj.2011.52.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berglund EC, Kiialainen A, Syvänen A-C. Next-generation sequencing technologies and applications for human genetic history and forensics. Investigative Genetics. 2011;2:23–37. doi: 10.1186/2041-2223-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biology. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Research. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, et al. A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454. Genome Biology. 2010;11:R15. doi: 10.1186/gb-2010-11-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bornman DM, Hester ME, Schuetter JM, Kasoji MD, Minard-Smith A, Barden CA, et al. Short-read, high-throughput sequencing technology for STR genotyping. Biotechniques. 2012:1–6. [PMC free article] [PubMed] [Google Scholar]
- 11.Fordyce SL, Ávila-Acros MC, Rockenbauer E, Børsting C, Frank-Hansen R, Petersen FT, et al. High-throughput sequencing of core STR loci for forensic genetic investigations using the Roche Genome Sequencer FLX platform. BioTechniques. 2011;51:127–133. doi: 10.2144/000113721. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirsch S, Klein CA. Sequence error storms and the landscape of mutations in cancer. PNAS. 2012;109:14289–14290. doi: 10.1073/pnas.1212246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. PNAS. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaragoza MV, Fass J, Diegoli M, Lin D, Arbustini E. Mitochondrial DNA variant discovery and evaluation in human cardiomyopathies through next-generation sequencing. PLoS ONE. 2010;5:e12295. doi: 10.1371/journal.pone.0012295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto H, Dickins B, Afgan E, Paul IM, Taylor J, Makova KD, et al. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biology. 2011;12:R59. doi: 10.1186/gb-2011-12-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Wu J, Dressman DC, lacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal turmour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. The American Journal of Human Genetics. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S, Wang J, Zhang VW, Li F-Y, Landsverk M, Cui H, et al. Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Human Mutation. 2013;34:882–893. doi: 10.1002/humu.22307. [DOI] [PubMed] [Google Scholar]
- 20.Parson W, Strobl C, Huber G, Zimmermann B, Gomes SM, Souto L, et al. Evaluation of next generation mtGenome sequencing using the Ion Torrent Personal Genome Machine (PGM) Forensic Science International: Genetics. 2013;7:543–549. doi: 10.1016/j.fsigen.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nature Biotechnology. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 22.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341–353. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Illumina Inc. Illumina technical note: sequencing. Estimating sequence coverage. 2011 http://res.illumina.com/documents/products/technotes/technote_coverage_calculation.pdf. [Google Scholar]
- 24.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 25.Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behavior Genetics. 2003;33:67–72. doi: 10.1023/a:1021055617738. [DOI] [PubMed] [Google Scholar]
- 26.Fendt L, Zimmermann B, Daniaux M, Parson W. Sequencing strategy for the whole mitochondrial genome resulting in high quality sequences. BMC Genomics. 2009;10:139–149. doi: 10.1186/1471-2164-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka M, Hayakawa M, Ozawa T. Automated sequencing of mitochondrial DNA. Methods in Enzymology. 1996:264. doi: 10.1016/s0076-6879(96)64037-3. [DOI] [PubMed] [Google Scholar]
- 28.Illumina Inc. Nextera® DNA sample preparation guide. (Part #15027987 Rev. B.) 2012 http://supportres.illumina.com/documents/myillumina/2a3297c5-8a34-4fc5-a148-3e16666fd65e/nextera_dna_sample_prep_guide_15027987_b.pdf. [Google Scholar]
- 29.Anderson S, Bankier AT, Barrell BG, MH dB, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 30.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genetics. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith TF, Waterman MS. Identification of common molecular subsequences. Journal of Molecular Biology. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 33.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. Measuring agreement in method comparison studies. STatistical Methods in Medical Research. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 35.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biology. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Research. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teytelman L, Özaydin B, Zill O, Lefrançois P, Snyder M, Rine J, et al. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biology. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu W, Hashimoto S-I, Morishita S. Efficient frequency-based de novo short-read clustering for error trimming in next-generation sequencing. Genome Research. 2009;19:1309–1315. doi: 10.1101/gr.089151.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forster L, Forster P, Gurney SMR, Spencer M, Huang C, Rohl A, et al. Evaluating length heteroplasmy in human mitochondrial DNA control region. International Journal of Legal Medicine. 2010;124:133–142. doi: 10.1007/s00414-009-0385-0. [DOI] [PubMed] [Google Scholar]
- 42.Irwin JA, Saunier JL, Niederstätter H, Strouss KM, Sturk KA, Diegoli TM, et al. Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples. Journal of Molecular Evolution. 2009;68:516–527. doi: 10.1007/s00239-009-9227-4. [DOI] [PubMed] [Google Scholar]
- 43.Strouss K. Master's thesis. Washington, DC.: George Washington University; 2006. Relative evolutionary rate estimation for sites in the mtDNA control region. [Google Scholar]
- 44.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humas and chimpanzees. Molecular Biology and Evolution. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humas and chimpanzees. 10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 45.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 46.Wakeley J. Substitution rate variation among sites in hypervariable region 1 of human mitochondrial DNA. Journal of Molecular Evolution. 1993;37:613–623. doi: 10.1007/BF00182747. [DOI] [PubMed] [Google Scholar]
- 47.Pavel II, Wadhams MJ, Roby RK, Holland MM, Weedn VW, Parsons TJ. Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nature Genetics. 1996;12:417–420. doi: 10.1038/ng0496-417. [DOI] [PubMed] [Google Scholar]