All life processes are governed at the chemical level. Knowledge of how single molecules interact, provides a fundamental understanding of Nature. An aspect of molecular interactions, is the self-assembly of supramolecular structures. For example, membrane fusion requires the assembly of a supramolecular complex formed when proteins in opposing bilayers interact. Membrane fusion is essential for numerous cellular processes, including hormone secretion, enzyme release, or neurotransmission. In living cells, membrane fusion is mediated via a specialized set of proteins present in opposing bilayers.1,2 Target membrane proteins, SNAP-25 and syntaxin (t-SNAREs) and secretory vesicle-associated protein (v-SNARE), are part of the conserved protein complex involved in fusion of opposing lipid membranes.1,2 The structure and arrangement of membrane-associated full length SNARE complex, was first examined using atomic force microscopy (AFM).3 Results from the study demonstrate that t-SNAREs and v-SNARE, when present in opposing bilayers, interact in a circular array to form supramolecular ring complexes each measuring a few nanometers.3 The ring-complex helps in establishing continuity between the opposing bilayers. 3 In contrast in the absence of membrane, soluble v- and t-SNAREs fail to assemble in any specific pattern, or form such conducting pore structures.3
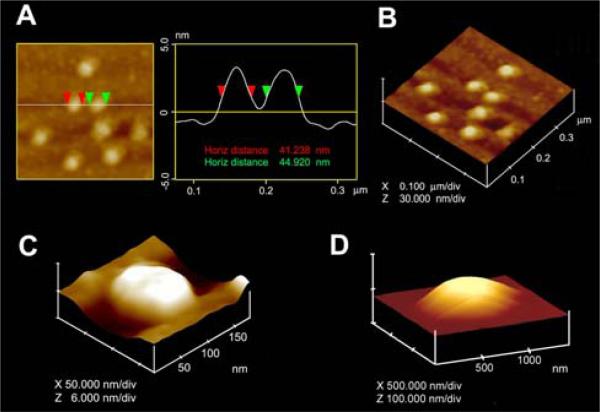
SNARE-ring complexes ranging in size from approximately 15 nm to 300 nm in diameter are formed when t-SNARE-reconstituted and v-SNARE-reconstituted lipid vesicles meet. Since vesicle curvature would dictate the contact area between opposing vesicles, this broad spectrum of SNARE complexes observed, may be due to the interaction between SNARE-reconstituted vesicles of different size. To test this hypothesis, t-SNARE- and v-SNARE-reconstituted liposomes (proteoliposomes) of distinct diameters were used. Lipid vesicles of different sizes used in the study were isolated using published extrusion method.4,5 The size of each vesicle population was further assessed using the AFM (Fig. 1). AFM section analysis demonstrates the presence of small 40-50 nm-in diameter vesicles isolated using a 50 nm extruder filter (Fig. 1A, B). Similarly, representative samples of large vesicles measuring 150-200 nm and 800-1000 nm were obtained using different size filters in the extruder. Such large vesicles are shown in the AFM micrograph (Fig. 1C, D). Analysis of vesicle size using photon correlation spectroscopy, further confirmed the uniformity in the size of vesicles within each vesicle population (data not shown).
Figure 1.
AFM micrograph of t-SNARE and v-SNARE reconstituted liposomes of different sizes. Note the ~40-50 nm vesicles (A,B), the ~150 nm (C) and ~800 nm vesicle (D).
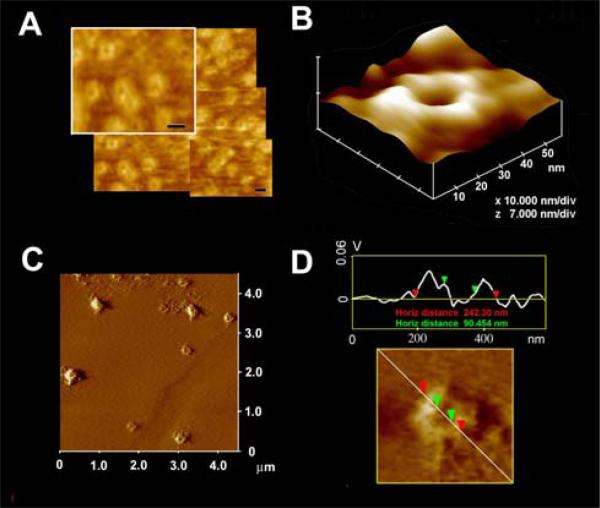
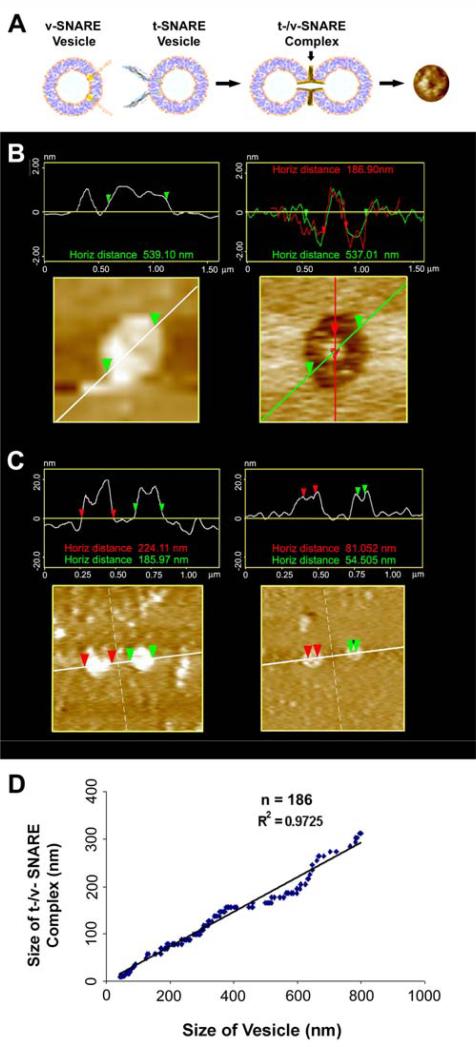
The morphology and size of the SNARE complex formed by the interaction of t-SNARE- and v-SNARE-reconstituted vesicles of different diameter were examined using the AFM (Fig. 2). In each case, the t-SNARE and v-SNARE proteins in opposing proteoliposomes, interact and self-assemble in a circular pattern, forming pore-like structures. The interaction and arrangement of SNAREs in a characteristic ring pattern were observed for all populations of proteoliposomes examined (Fig. 2A-D). However, the size of the SNARE complex was found to be dictated by the diameter of the proteoliposomes used (Fig. 2). When small (~50 nm) t-SNARE- and v-SNARE-reconstituted vesicles were allowed to interact, SNARE-ring complexes of ~20 nm in diameter were generated (Fig. 2 A, B). With increase in the diameter of proteoliposomes, larger t-/v-SNARE complexes were formed (Fig. 2 C, D). A strong linear relationship between size of the SNARE complex and vesicle diameter is demonstrated from these studies (Fig. 3). The experimental data fit well with the high correlation coefficient, R2=0.9725 between vesicle diameter and SNARE-complex size (Fig. 3).
Figure 2.
Representative AFM micrograph of t-/v-SNARE complexes formed when small (A, B) or large (C, D) t-SNARE and v-SNARE reconstituted vesicles interact with each other. Note the formation of different size SNARE complexes, which are arranged in a ring pattern. Bar= 20nm. AFM section analysis (D) shows the size of a large SNARE complex.
Figure 3.
SNARE complex is directly proportional to vesicle diameter. Schematic diagram depicting the interaction of t-SNARE and v-SNARE reconstituted vesicles. At the extreme right, is a single t-/v-SNARE complex imaged by AFM (A). AFM images of vesicles before and after their removal by the AFM cantilever tip, exposing the t-/v-SNARE complex (B). Interacting t-SNARE and v-SNARE vesicles imaged by AFM at low (<200 pN) and high forces (300-500 pN). Note, at low imaging forces, only the vesicle profile is imaged (left C). However at higher forces, the soft vesicle is flattened, allowing the SNARE complex to be imaged (right C). Plot of vesicle diameter vs. size of the SNARE complex. Note the high correlation coefficient (R2=0.9725) between vesicle diameter and the size of the SNARE complex (D).
Unlike calculated values, which fundamentally assume vesicles to be non-deformable and hard spheres, our experimental data suggest that these artificial lipid vesicles, similar to secretory vesicles, are soft. Hence, once v-SNARE and t-SNAREs from opposing vesicles meet, the initial SNARE complex formed, pulls the opposing bilayers closer to each other. As a consequence, vesicles become flattened, which then leads to an increase in contact area between the opposing vesicles. The result is a further increase in t-/v-SNARE contacts, allowing the formation of larger SNARE-ring complexes. In case of hard vesicles, flattening would be unlikely, and therefore result in forming smaller SNARE-ring complexes. On the other hand, deformation of soft vesicles leads to an increase in contact area between the opposing bilayers and a resultant increase in Gibbs free energy. In an elastic membrane, the surface free energy is given by the equation: (1/2)ka(ΔA)2/A0, where ka is the bending modulus, ΔA, the increase in surface area, and A0, the initial unstressed area.6 Therefore, an increase in surface area results in an increase in the Gibbs free energy, and the spontaneous fusion between opposing bilayers become less probable.6-8 Hence, large vesicles are less fusogenic than smaller vesicles. This would explain why neurons being fast secretory cells, possess small 40-50 nm in diameter vesicles, for rapid and efficient fusion and release of neurotransmitters at the nerve terminal,9,10 compared to a slow secretory cell like the exocrine pancreas, with larger (200-1,200 nm in diameter) secretory vesicles, for the slow and prolonged release of digestive enzymes.11,12
In summary, we have demonstrated that the size of a self-assembled supramolecular SNARE protein complex can be controlled. This has implications in the regulated fusion of artificial lipid membranes, which may find use in the controlled delivery of lipid-encapsulated drugs and the transport of molecules.
Supplementary Material
Acknowledgment
Supported by NIH grants (BPJ).
Footnotes
Supporting Information Available: Experimental procedures is available via the internet at http://pubs.acs.org
References
- 1.Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, Zemelman BV, McNew JA, Westerman B, Gmachi M, Parlati F, Söllner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 3.Cho SJ, Kelly M, Rognlien KT, Cho J, Hoerber JKH, Jena BP. Biophys. J. 2002;83:2522–2527. doi: 10.1016/s0006-3495(02)75263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR. Biochim. Biophys. Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- 5.Jeremic A, Kelly M, Cho JA, Cho SJ, Horber JK, Jena BP. Cell Biol. Int. 2004;28:19–31. doi: 10.1016/j.cellbi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Israelachvili J. Intermolecular and surface forces. 2nd Ed. Academic Press; San Diego, CA: 1992. [Google Scholar]
- 7.Ohki SJ. Memb. Biol. 1984;77:265–275. doi: 10.1007/BF01870574. [DOI] [PubMed] [Google Scholar]
- 8.Wilschut J, Duzgunes N, Papahadjopoulos D. Biochemistry. 1981;20:3126–3133. doi: 10.1021/bi00514a022. [DOI] [PubMed] [Google Scholar]
- 9.Cho WJ, Jeremic A, Rognlien KT, Zhvania MG, Lazrishvili I, Tamar B, Jena BP. Cell Biol. Int. 2004;28:699–708. doi: 10.1016/j.cellbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Kelly M, Cho WJ, Jeremic A, Abu-Hamdah R, Jena BP. Cell Biol. Int. 2004;28:709–716. doi: 10.1016/j.cellbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Jena BP, Schneider SW, Geibel JP, Webster P, Oberleithner H, Sritharan KC. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13317–13322. doi: 10.1073/pnas.94.24.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider SW, Sritharan KC, Geibel JP, Oberleithner H, Jena BP. Proc. Natl. Acad. Sci. U.S.A. 1997;94:316–321. doi: 10.1073/pnas.94.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.