Abstract
Significance: The redox code is a set of principles that defines the positioning of the nicotinamide adenine dinucleotide (NAD, NADP) and thiol/disulfide and other redox systems as well as the thiol redox proteome in space and time in biological systems. The code is richly elaborated in an oxygen-dependent life, where activation/deactivation cycles involving O2 and H2O2 contribute to spatiotemporal organization for differentiation, development, and adaptation to the environment. Disruption of this organizational structure during oxidative stress represents a fundamental mechanism in system failure and disease. Recent Advances: Methodology in assessing components of the redox code under physiological conditions has progressed, permitting insight into spatiotemporal organization and allowing for identification of redox partners in redox proteomics and redox metabolomics. Critical Issues: Complexity of redox networks and redox regulation is being revealed step by step, yet much still needs to be learned. Future Directions: Detailed knowledge of the molecular patterns generated from the principles of the redox code under defined physiological or pathological conditions in cells and organs will contribute to understanding the redox component in health and disease. Ultimately, there will be a scientific basis to a modern redox medicine. Antioxid. Redox Signal. 23, 734–746.
Introduction
In a seminal article in 1958, Bücher and Klingenberg (15) examined pathways of hydrogen in the living organization, addressing the interactions within the network of redox reactions extending over essential functions of living cells. In considering the distinct cytological spaces and their relationship with biosynthesis, bioenergetics, and cellular respiration, the ground was laid for fruitful development of these research areas in dynamic biochemistry. Since then, the enormous advances in techniques of analysis and in noninvasive molecular imaging of cells, tissues, and intact organisms, as well as genomics and the discoveries of important redox enzyme systems have led to novel insights in redox biology (13, 38).
Early research clarified a fundamental issue in the assessment of kinetic versus thermodynamic properties in the analysis of redox processes in biology. Specifically, the steady states of deviation from thermodynamic equilibrium provide a source of order (85). The specific location of an enzymatic activity in subcellular structures or in distinct organs, that is, a kinetic potential, is used to influence the reduction–oxidation state of a given redox couple, defined by its thermodynamic redox potential as expressed by the Nernst equation. This enables multiple chemical and associated physical processes to occur simultaneously within the physical space, obeying the same thermodynamic forces, having distinct spatial and temporal response characteristics, and supporting differentiated functions. The two major systems addressed here, the nicotinamide nucleotide (NAD and NADP) systems and thiol systems (protein thiols, glutathione [GSH], and associated disulfides), substantially differ in organization and exemplify the central operational structures of redox biology.
The redox code of these metabolic and protein components provides a critical complement to the genetic code, the epigenetic code (80), and the histone code in the molecular logic of life. Unlike the genetic code and histone code, which underlie information storage and transfer for use, the epigenetic code and the redox code define the operations of the genetic and histone codes in the organizational structure, differentiation, and adaptation of an organism to the environment. Like the genetic and histone codes, the redox code has a fundamental simplicity, yet it is elaborated with considerable complexity in multicellular organisms. In particular, redox signaling and redox control in multicellular organisms evolved and diversified with the increase in atmospheric O2 about 600 million years ago (54, 93). The present review highlights key principles of the redox code derived from the extensive literature on redox sensing and signaling in diverse processes of circadian variation, cell cycle, apoptosis, and organismic defense (51). The term “Redox Code” applies to the redox organization of cells, tissues, and organisms. It is not confined to mammalian systems, and it extends ultimately to all living matter. While focusing mainly on eukaryotic mammalian systems in this review, it is understood that other eukaryotic, for example, plant and fungal systems, as well as prokaryotic, for example, bacterial, systems have elaborated redox codes describing their respective metabolic organization.
Principles of the Redox Code
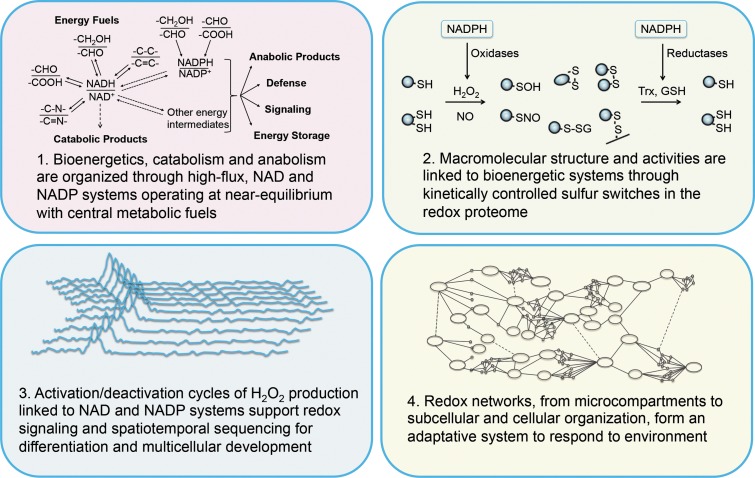
The redox code consists of four principles by which biological systems are organized (Fig. 1). The first is the use of the reversible electron accepting and donating properties of nicotinamide in NAD and NADP to provide organization of metabolism, operating at near equilibrium. Substrate oxidations are linked to reduction of NAD+ and NADP+, which in turn are linked to ATP production, catabolism, and anabolism, respectively. The second principle is that metabolism is linked to protein structure through kinetically controlled redox switches in the proteome, which determine tertiary structure, macromolecular interactions and trafficking, activity, and function. The abundance of proteins and reactivity of sulfur switches with oxidants vary over several orders of magnitude to determine specificity in biological processes. The third principle is that of redox sensing, in that activation/deactivation cycles of redox metabolism, especially involving H2O2, support spatiotemporal sequencing in differentiation and life cycles of cells and organisms. The fourth principle is that redox networks form an adaptive system to respond to the environment from microcompartments through subcellular systems to the levels of cell and tissue organization. This adaptive redox network structure is required to maintain health in a changing environment and, if functionally impaired, contributes to disease and organism failure.
FIG. 1.
The redox code. The four principles of the redox code by which biological systems are organized (see text).
Redox Players
Redox biology occurs in a multidimensional space, in which the fundamental basis set, that is, the full spectrum of redox elements and dimensionality of this space, remains incompletely understood. In developing the redox code, we focused on a limited set of combinations to outline central principles for bioenergetics, metabolic and structural organization, and interaction of an organism with its environment. As indicated below, it does not incorporate the full spectrum of redundant and unique uses of specific redox systems [see also Buettner et al. (17)]. Consequently, the redox code should be viewed as a central framework rather than specific vectors leading to specific redox states and associated biological states as observed in the life cycles of diverse life forms. Future elaboration will be needed to fully encompass the selenoproteins, hemoproteins, cuproproteins, iron–sulfur proteins, flavoproteins, pteroproteins, molybdoproteins, manganoproteins, and other redox elements and interacting systems.
The NAD and NADP systems along with thiol/disulfide systems are central to the redox code. These are outlined in Table 1 and addressed in more detail below. The NAD system uses powerful dehydrogenases, active in reversible two-electron transfer catalysis, in the metabolism of biomolecules with covalent bonds of C, O, S, N, and H. [NADH]/[NAD+], the ratio of concentrations of free NADH and NAD+, is in equilibrium with the reduced and oxidized substrates of major cellular dehydrogenases (11). The [NADH]/[NAD+] couple is central to catabolism and energy supply. The [NADPH]/[NADP+] system is set apart from the [NADH]/[NAD+] and is central to anabolism, defense, and control of thiol/disulfide systems by controlling the redox state, for example, in the GSH and the thioredoxin (Trx) systems. The [NADPH]/[NADP+] and [NADH]/[NAD+] systems are linked by activity of mitochondrial energy-linked transhydrogenase (40, 127). One-electron transfer is biologically less relevant in the NAD and NADP systems, but important in associated components, such as FAD and FMN systems, the flavin-linked reactions (30), and in iron–sulfur clusters (7). Redox chemistry of thiols involves both one-electron and two-electron transfer reactions of cysteine (Cys)/cystine (CySS), GSH/glutathione disulfide (GSSG), protein thiols and oxidized forms, and protein-bound cysteines/disulfides. The selenium analog of cysteine, selenocysteine, also plays an important role in controlling the redox function of proteins [see Jacob et al. (47)]. The role of selenium in selenoprotein biology [see Refs. (44, 84)] and the chemical properties of selenocysteine versus cysteine (76, 94) have been studied intensively. Protein thiols are subject to other modifications (e.g., nitrosylation, sulfhydration, metal ion binding) impacting redox activities (51). These and other redox players provide an enriched diversity to the central redox organizational structure described here.
Table 1.
Properties and Comparison of the Nicotinamide Adenine Dinucleotide and Thiol/Disulfide Systems in Redox Biology
| Parameter | NAD+, NADP+ | References | Thiol/disulfide | References |
|---|---|---|---|---|
| Type of control | Near-equilibrium system; thermodynamic control | (15) | Nonequilibrium system; kinetic control | (26, 31) |
| Capacity/rates | High flux | (85) | Low fluxa | (85) |
| Stoichiometry | 2 e− | (15) | 1 e− or 2 e− | (16, 55) |
| Nature of redox relationships | Redox coupling | (43, 122) | Redox switch; redox sensing | (37, 51, 90) |
| Biological role | Chemical, metabolic, and energetic organization | (47, 100) | Structural, spatial, and temporal organization | (21, 47, 100) |
| Subcellular distribution | Compartment specific | (22, 42, 46, 86) | Compartment specific; microcompartments | (71) |
| Example | Oxidative phosphorylation; sirtuins | (73) | OxyR; NF-κB; Nrf2; AP-1, HIF-1α | (23, 56, 92, 99, 110, 119) |
For discussion, see text.
There are exceptions, for example, the lipoamide in pyruvate dehydrogenase is high flux, likewise ribonucleotide reductase in an S-phase T cell.
HIF-1α, hypoxia-inducible factor-1α; NAD, NADP, nicotinamide adenine dinucleotide; NF-κB, nuclear factor-κB; Nrf2, nuclear factor-E2-related factor 2.
First Principle: Metabolic Organization
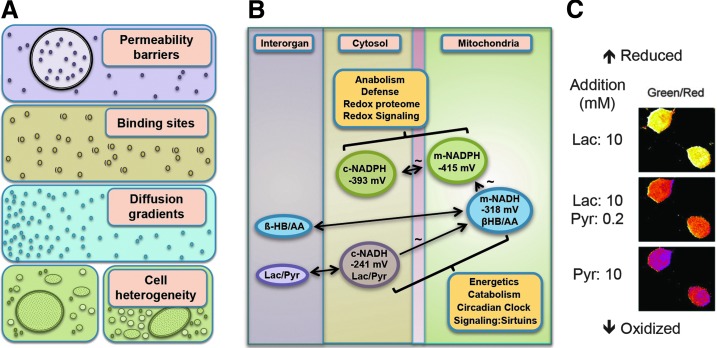
Metabolism is organized through high-flux thermodynamically controlled NAD and NADP systems. The relevant property for redox systems is the activity of the redox partners, which is closely dependent upon their free concentrations within cellular and subcellular compartments. This concept is emphasized by the nonequivalence of tissue levels (nmol/g wet weight) with the free concentrations (nM) (101). Differences in free concentrations among compartments may result from permeability barriers, binding to macromolecules, nonhomogeneous distribution, and also heterogeneity of cell types in organs and tissues (Fig. 2A). Recent technological progress allows the resolution of redox partners within microcompartments (37) and such resolution can be expected to enable refined investigation in redox biology.
FIG. 2.
Metabolism is organized through high-flux NAD and NADP systems. (A) Multiple types of compartmentation affect free concentration of redox partners: microheterogeneity (permeability barriers, binding sites, diffusion gradients) and macroheterogeneity. (B) The NAD and NADP systems are maintained at steady states near thermodynamic equilibrium in mitochondria, cytosol, and other subcellular redox spaces to control energetics, catabolism, and anabolism. The cytoplasmic and mitochondrial NAD systems are respectively equilibrated throughout the body through lactate (Lac)/pyruvate (Pyr) and β-hydroxybutyrate (HB)/acetoacetate (AA). Mitochondrial NADH and NADPH systems are connected through energy-linked transhydrogenase. The midpoint potentials (two-electron half-cell reduction potentials) are given in mV. (C) Cytosolic NADH/NAD+ redox state imaged in cells with a genetically encoded fluorescent biosensor, Peredox, calibrated using exogenous lactate (Lac) and pyruvate (Pyr). (A, B) Modified from Sies (100, 101); (C) from Hung et al. (46) with permission of the publisher.
Detailed studies of the three major NAD-linked dehydrogenases showed that the systems operate near equilibrium (15, 42, 43). The cytosolic redox poise of [NADH]/[NAD+] is at a set point of −241 mV (Fig. 2B), substantially more oxidized than expected from the total cytosolic NADH and NAD+ content (14, 100). This difference is attributable to powerful protein-binding sites for NADH; the redox buffer capacity of these titratable NADH-binding sites provides a cytosolic set point at about 1 μM free NADH, despite much higher total cytosolic NADH (pNADH=6). Operation of the NAD system at near equilibrium allows cells to support catabolic needs while maintaining capability to supply ATP energy according to cellular needs. Studies of subcellular compartments showed that the redox potential of the cytosolic and the mitochondrial [NADH]/[NAD+] are vastly different by about two orders of magnitude or ca. 60 mV (15, 122). The mitochondrial matrix NAD operates at a more negative redox potential (86), −318 mV, providing a greater reductive force for mitochondrial ATP production. Noninvasive redox titrations have been extensively applied to study dynamic steady states using redox indicator metabolites in combination with organ spectrophotometry (14); newer methods use genetically encoded fluorescent biosensors (Fig. 2C) (46).
The NADP system provides an important counterpart to the NAD system, supporting anabolic reactions of metabolism, organismic defense, and maintenance of the thiol/disulfide systems. The [NADPH]/[NADP+] system is equilibrated with NADP-linked dehydrogenases and operates at a more negative redox potential than the NAD system, that is, −393 mV in the cytosol and −415 mV in the mitochondrial matrix (Fig. 2B) (100). As mentioned above, the mitochondrial NAD and NADP systems are connected by energy-linked transhydrogenase. This enzyme may act as a sensor that determines the positioning of either NAD or NADP at the near-equilibrium steady state [see Yin et al. (127)]. The reducing force provided by these potentials supports biosynthesis concurrently with NAD-dependent catabolism and energy production; the specificity of the NAD- or NADP-linked systems provides a central logic to these divergent needs in metabolism. This molecular logic is further illustrated by the use of the NADP system for defensive purposes, especially activation of O2 for otherwise unfavorable oxidation of xenobiotics and reduction of O2 for the generation of superoxide anion used in phagocytic killing of pathogens.
Second Principle: Linkage of Metabolism to Structure
The linkage of redox metabolism to structure involves the redox proteome. The redox proteome is a subset of the epiproteome, which includes phosphorylation, acylation, methylation, and other modifications of the translated proteome. Although functionally integrated with the systems controlling these other covalent modifications, the redox proteome provides a distinct basis to link metabolism to structure through kinetically controlled sulfur switches. These molecular redox switches determine tertiary structure, molecular activity, macromolecular interactions, and trafficking principally through changes of thiol and disulfide structures.
The nature of the thiol systems differs from the nicotinamide nucleotide systems, in that most elements operate at low flux rather than near equilibrium (Table 1). This characteristic is exemplified by steady-state redox potentials of extracellular low-molecular-weight thiol/disulfide couples, including Cys/CySS and GSH/GSSG pools (55). Using the Nernst equation, the calculated redox potentials in human plasma are −75 and −150 mV, respectively. While compartmentation and other factors (Fig. 2A) complicate interpretation of thiol/disulfide systems in cells and tissues, lack of equilibration in this cell-free aqueous space emphasizes the kinetic limitation. Reaction rates calculated from rate constants and concentrations for thiol/disulfide couples are too slow for most redox elements to equilibrate in living organisms. This has recently been emphasized in a discussion of the GSH redox system (8).
Cellular Cys/CySS and GSH/GSSG couples are maintained at a substantially more reduced state than the extracellular counterparts. Like the NAD and NADP systems, redox potentials differ in subcellular compartments, with the mitochondria more negative (reducing) and endoplasmic reticulum more positive (oxidizing) than cytosol. The thiol/disulfide set point for specific Cys residues in proteins is analogous to the NADH-binding sites mentioned above, with the thiol reactivity of individual protein Cys being tuned by location within the protein structure. Unlike the NADH sites, most of which have catalytic functions supporting energetics and metabolism, most protein thiols have noncatalytic functions, for example, protein folding, regulation of activity, control of macromolecular interactions, and directional trafficking (47). The low-molecular-weight thiols interact both enzymatically and nonenzymatically with proteins, presenting challenges to identify corresponding thiol/disulfide systems connected to them (26). Additionally, protein thiols undergo other chemical reactions and interact with metal ions, either of which can impact biological function.
Despite the extensive number of specific Cys (>200,000 encoded in the human genome) (50) and variation in reactivity (>6 orders of magnitude) and abundance (>6 orders of magnitude), progress is being made in understanding the redox organizational structure. Current evidence points to evolution of the Cys proteome (70) during the same period as evolution of multicellular organisms, advanced O2 sensing, and H2O2 signaling systems (54, 93). The Cys proteome increased with organismic complexity, from 0.5% of the proteome in prokaryotes to 2.2% in mammals (70). The expected Cys content in proteins based upon the random use of tRNAs is (3.1%), indicating that increase in Cys in the proteome occurred, despite selection pressure against use (51). This coevolution of the Cys proteome with the O2 sensing and H2O2 signaling in multicellular evolution is consistent with known structure–function relationships involving protein thiols in complex organisms.
Measurements of cells and tissues show that Cys occurs with different steady-state oxidation in proteins (33, 60). Use of thiol-trapping agents and mass spectrometry in the absence of added oxidants shows that median percentage oxidation of protein Cys is 6%–14%. These findings are consistent with measures of steady-state redox potentials of the central thiol antioxidants, Trx and GSH, which show that these couples are not in thermodynamic equilibrium with the reductant couple, NADPH/NADP+. Inhibitors of GSH and Trx antioxidant systems increase fractional oxidation (6), and extensive studies with nutrient deprivation, mitochondrial respiratory inhibitors, and oxidant challenges show that oxidation of the thiol proteome is dynamic and reversible. Thus, the redox proteome is characterized by widespread occurrence of protein Cys with partial oxidation.
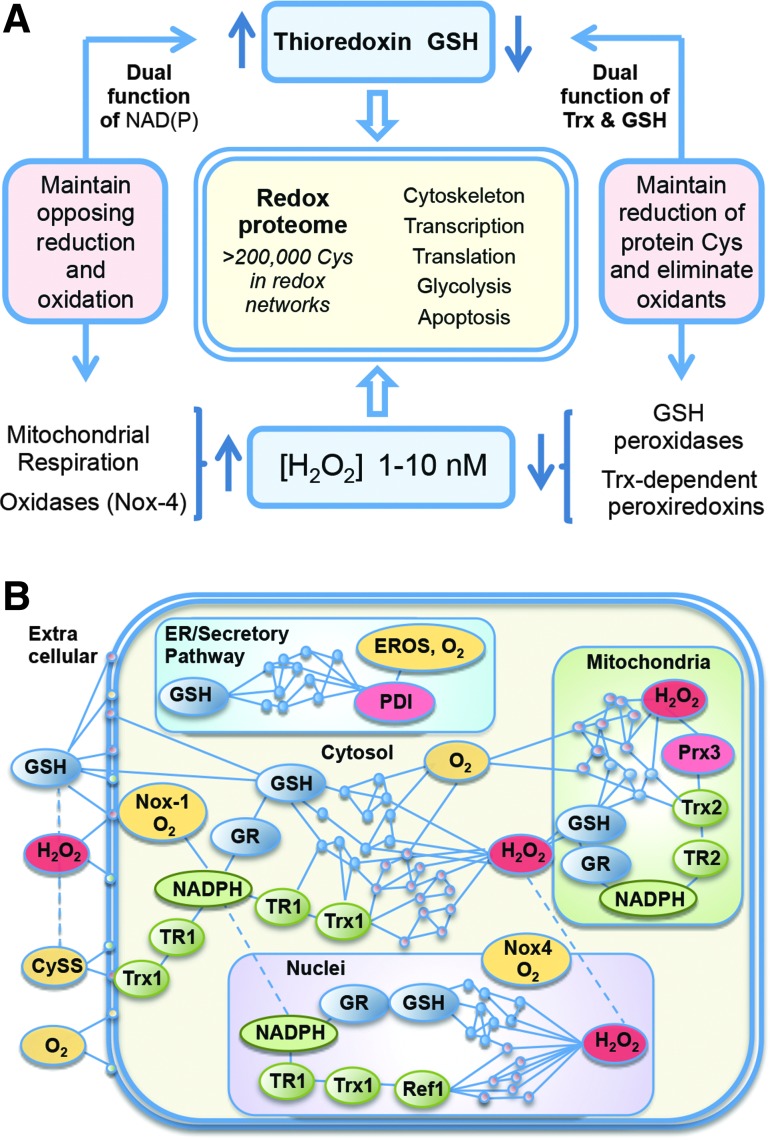
A common operational structure governs the redox-sensitive Cys through metabolism dependent upon the NAD and NADP systems. In this structure, H2O2 is the most central oxidant, and Trx and GSH are the most central reductants (Fig. 3A). Measurements of steady-state concentration show that H2O2 in the mammalian liver is 1–10 nM (18, 82, 103, 105). Constitutive H2O2 is derived from mitochondrial respiration dependent upon NADH (10, 34, 87) and activities of NADPH oxidase (Nox) enzymes dependent upon NADPH (2, 13, 87). As a consequence, H2O2 production depends upon activity levels in cells. H2O2 production is countered by Trx and GSH/glutaredoxin (Grx) systems, both functioning to reduce disulfides, but with differing specificities for protein Cys (62, 65). Importantly, both Trx and GSH systems depend upon NADPH-linked reductases, creating a system in which protein oxidation and reduction are inherently balanced yet responsive to cellular activity through NADPH availability. This links energetics and metabolism to redox structure, which in turn uses oxidant signals through nuclear factor-E2-related factor 2 (Nrf2), peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α), nuclear factor-κB (NF-κB), and other transcription factor systems to more broadly integrate energy efficiency, resistance of cells to injury, and cell repair (11, 23, 25, 28, 56, 61, 86, 92, 98, 118). The stability of this redox structure is further assured by the use of Trx and GSH (with GSH peroxidases) as reductants for hydroperoxides, thereby providing two levels of opposing reactions to assure stable steady-state operation of this redox network structure (Fig. 3A).
FIG. 3.
The redox proteome is organized through kinetically controlled sulfur switches linked to NAD and NADP systems. (A) Redox states of Cys in functional networks are maintained by opposing oxidation and reduction reactions. NADPH is the principal reductant used to maintain the redox states of the Trx and GSH systems. These systems maintain the redox proteome, with GSH functioning through glutaredoxins (not shown). Trx and GSH also regulate H2O2 concentrations through Prx and GSH peroxidases, respectively. H2O2 is generated metabolically by mitochondria and about 25 different oxidases. Nox-4 is included because of constitutive activity with NADPH and O2; different Nox systems support a range of signaling activities, while other H2O2-producing enzymes support a range of biosynthetic and detoxification functions. (B) Protein Cys in subcellular compartments is regulated by oxidation (red) and reduction (blue/green) to maintain organization structure through control of distribution, interaction, and activity. Cys, cysteine; CySS, cystine; EROS, endoplasmic reticulum oxidase system; GR, glutathione disulfide reductase; GSH, glutathione; Nox, NADPH oxidase; PDI, protein disulfide isomerase; Prx, peroxiredoxin; Ref-1, redox factor-1 (AP endonuclease-1); TR, thioredoxin reductase; Trx, thioredoxin.
The fraction of the Cys proteome that undergoes reversible oxidation is unknown. Redox proteomics data provide evidence for thousands of redox-sensitive protein thiols (Fig. 3B), but a much lesser number have descriptions of associated function. Some have detailed understanding, such as those involved in transcription control via OxyR in prokaryotes (91), Nrf-2 (56), NF-κB (115), P53 (88), AP-1 (1), glucocorticoid receptor (78), and other transcriptional systems. In NF-κB, for instance, oxidation of Cys-38 in p65 or Cys-62 in p50 serves to control DNA binding. The redox pathway controlling reduction uses NADPH, thioredoxin reductase-1 (TR1), Trx1, and the nuclear redox factor-1 (Ref-1) to maintain the DNA-binding structure (115, 126). Nuclear peroxiredoxin-1 enhances activity, suggesting that endogenous H2O2 supports oxidative inactivation. Measurement of steady-state oxidation of each component in the pathway and sensitivity to changes in NADPH supply further show that the redox pathway is kinetically limited (50). Other examples of specific redox-controlled proteins occur in the respiratory chain NADH dehydrogenase (6), mitochondrial thioredoxin reductase-2 (TR2), and Trx2 control activation of apoptosis (113) and actin-associated proteins (33). The GSH systems provide alternative reductases that not only have some redundancy to Trx systems but also have some specific functions in parallel with Trx. By itself, GSH is a slow reductant. However, Grx uses GSH to catalyze reduction of disulfides. Grx1 in the cytosol/nucleus and Grx2 in mitochondria remove S-glutathionyl groups formed during cell signaling, regulation, or oxidative stress. Functional modules for translation, glycolysis, lipid metabolism, and mRNA and miRNA processing and trafficking Cys are associated within functional networks in subcellular compartments according to steady-state oxidation (Fig. 3B) (33). The central logic to the dynamic operational structure of these diverse systems lies within the stable redox proteome linked to NADH/NAD+ and NADPH/NADP+ metabolism.
Third Principle: Redox Signaling and Spatiotemporal Differentiation
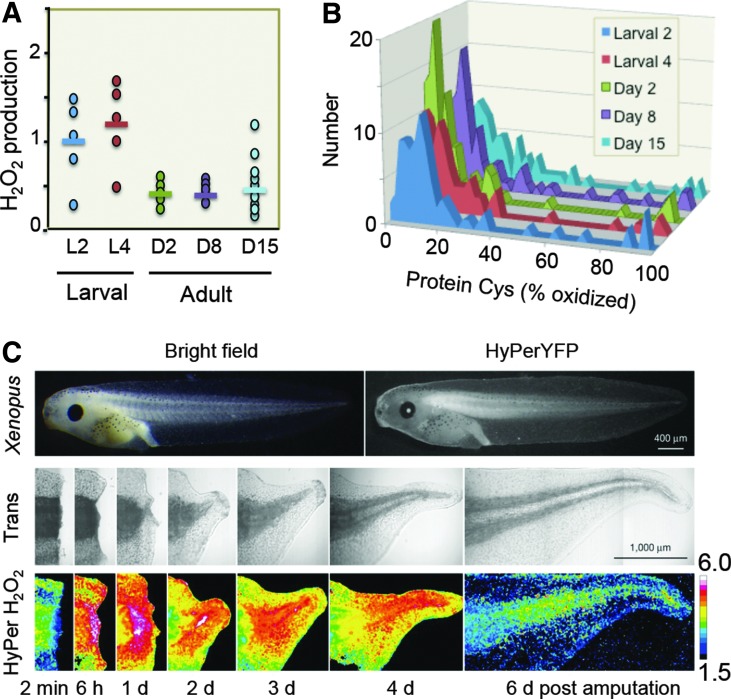
The evolution of O2 sensing, H2O2 signaling, and the Cys proteome following appearance of an O2 atmosphere naturally suggests a relationship between these processes; the significance of such a relationship is crystallized by studies with modern redox probe technologies. Since the discovery of nonphagocytic Nox enzymes in mammalian systems, considerable research has focused on the role of H2O2 and Nox in cell signaling (35, 59). Redox probe studies now show that during Caenorhabditis elegans development, a temporal wave of H2O2 production occurs in the larval stage, followed by a decrease in H2O2 upon emergence of the adult (Fig. 4A) (58). Selective oxidation of protein thiols accompanies this H2O2 production (Fig. 4B). The data highlight the dynamic redox organizational structure that occurs within the temporal framework of multicellular development.
FIG. 4.
Activation/deactivation cycles of H2O2 production support redox signaling and spatiotemporal organization of complex multicellular systems. (A) Redox imaging using HyPer reveals temporal sequence of H2O2 production in Caenorhabditis elegans development. Endogenous hydroperoxide levels during the life span of C. elegans were measured with HyPer ratios in the body wall muscle cells at different stages during their life span. Each symbol represents the HyPer ratio of an individual animal; the bar illustrates the average HyPer ratio per day. Experiments were performed a minimum of three times, and a representative graph shown here is redrawn from the original. One-way ANOVA, followed by the Tukey multiple comparison test, was performed on the log-transformed HyPer ratio and showed that the larval levels were different from the adult levels (p<0.05). Results were independently confirmed by Amplex UltraRed measurement of H2O2, which were also significantly different. (B) Protein Cys oxidation occurs in association with H2O2 production in C. elegans development. A complete list of proteins and standard deviations are given as an online supplement (http://dx.doi.org/10.1016/j.molcel.2012.06.016) to the original publication (58). (C) Redox imaging reveals the spatiotemporal sequence of H2O2 production in Xenopus laevis wound healing. Sequence of H2O2 production in tadpole tail regeneration is shown for days postamputation in the X. laevis line expressing HyPerYFP. The numbers give the excitation ratio of HyPerYFP490nm/HyPerYFP402nm, and the color scale reflects this ratio. (A, B) Modified from Knoefler et al. (58) and (C) from Love et al. (64), with permission from the publisher.
Redox probe studies of tadpole tail regeneration extend this to a spatiotemporal framework (Fig. 4C) (64). Assuming that the indicator responses measure H2O2 and not pH or other effects, the results show that enhanced production of H2O2 occurs at the wound site following tail amputation and progresses as a wave associated with regeneration. A tissue-scale gradient of hydrogen peroxide also mediates rapid wound detection in zebrafish (77). While molecular and pharmacologic manipulations have effectively demonstrated cellular redox mechanisms and roles in disease, the advanced bioimaging methods have enabled recognition of the third principle of the redox code, that is, activation/deactivation cycles of H2O2 metabolism support complex time-dependent processes in the life cycle of development of cells and organisms. Indeed, this echoes O2 sensing via hypoxia-inducible factor-1α (HIF-1α), a signaling system that requires O2 rather than H2O2 and provides signaling in multicellular organisms (99).
In spatiotemporal redox signaling, relatively high rates of local H2O2 production (2) provide an important contrast to the slow reaction rates that govern the bulk of the redox proteome described above (second principle). Localized H2O2 production may represent only a small fraction of total cellular O2 utilization, but results in locally high H2O2 concentrations (2). Diffusion of H2O2 and its rapid metabolism confines the redox signal to the vicinity of production. Specific proteins, such as peroxiredoxins, will react with hydroperoxides with substantial velocity. In principle, specificity could be obtained by the rate constants of redox elements for reaction with H2O2. An alternative is that peroxiredoxins or other proteins with high reactivity with H2O2 could serve as oxidative catalysts to provide specific oxidation of target proteins. This could occur in linear pathways or with a network reaction structure, with scaling of reactivities and abundance of nodes within the network. In such a network structure, one oxidative catalyst could have multiple protein targets, each with relatively high reactivity toward subsets of the cysteine or methionine proteome. Such a hierarchical structure could coordinate functional subsets of the proteome broadly over space and time.
Multiple other factors could also provide specificity. For instance, cationic and other proximal amino acids have a considerable effect on the reactivity of thiols with H2O2. pH is regionally controlled, especially in the vicinity of membranes, and thiol redox reactions are highly pH dependent. Metal ions are also important to stimulate (e.g., Cu2+) or inhibit (e.g., Zn2+) oxidation and these could impart specificity through protein–protein or other domain-specific structural interactions. The detailed mechanism and specificity of redox signaling by hydrogen peroxide needs to be further elaborated (57, 123).
Redox organization is well established in association with organellar function. For instance, in the endoplasmic reticulum, an endoplasmic reticulum oxidase system uses O2 and protein disulfide isomerase to introduce disulfides into proteins for the cell surface and secretion (116). This vectorial processing with successive oxidation results in relatively oxidized steady states of thiol/disulfide couples in extracellular compartments. Import into mitochondria uses similar redox processing, and multiple redox processes control transcription factor translocation into nuclei (32). One can extend these concepts to the cross talk between signaling mechanisms, for example, the widespread use of redox switches in kinase signaling pathways, such as oxidation of active site 215Cys in protein-tyrosine phosphatase 1B (PTP1B), underscores the spatiotemporal organizational structure. Subsets of protein Cys provide additional functional diversity through selective nitrosylation, sulfhydration, and binding to metal ions (51, 125) and irreversible modification of Cys by reaction with electrophilic metabolites (39). Reversible oxidation of Met to MetSO at the protein surface also occurs (29) and may provide a parallel redox network. Refined redox imaging tools will enable detailed mapping to enhance understanding of specialized redox functions.
O2 sensing allows maintaining NAD and NADP systems for energetics and metabolism, as defined above (first principle), while activation/deactivation cycles of H2O2 signaling direct targeted activities within a multicellular structure. Complementary NAD functions have recently been reviewed and highlight the importance of the subcellular location of NAD synthesis and the diverse roles of NAD in chromatin stability, DNA repair, circadian rhythms, and longevity (22). As an example, NAD+ activates sirtuins, which catalyze deacetylation of histones (68). Sirtuins are regulated by the [NADH]/[NAD+] ratio (27), and the [NADH]/[NAD+] ratio activates SIRT1-mediated autophagy (48). NAD plays a role in mitonuclear protein imbalance, which has been described as a conserved mechanism in the regulation of energy metabolism (45).
Thus, the molecular logic of differentiation and development in multicellular systems is fundamentally linked to three simple principles of redox biology defining the organization of energetics and metabolism, the linkage of metabolism to macromolecular structure and function, and the activation/deactivation cycles of H2O2 production that direct spatiotemporal organization. Although governed at all levels by the structural design of the NAD system to support energetics and metabolism, detailed knowledge of activation/deactivation cycles of H2O2 production and responsive redox protein systems will enhance the understanding of the differentiated functions and functional limitations in redox biology.
Fourth Principle: Adaptation to the Environment
The intricate relationships of NAD metabolism and circadian rhythms (74, 89, 96) underscore the central logic of redox systems in supporting organismic adaptation to environmental changes, including availability of nutrient resources, exposure to hazardous agents, and vulnerability to predators. This highlights the fourth principle of the redox code that redox networks, from microcompartments through subcellular systems to the levels of cell and tissue organization, form an adaptive system to respond to the environment. This adaptive redox network structure is required to maintain health in a changing environment and, if functionally impaired, contributes to disease and organism failure.
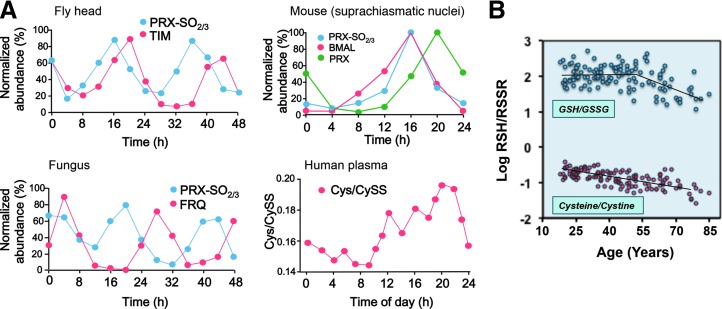
The fundamental linkage of diurnal changes to the light cycle is evident for photosynthetic organisms, but day–night cycles also determine the timing for food access in animals, and associated activities entrain susceptibility to environmental hazards and predation. Recently elucidated circadian redox clocks emphasize the conserved nature of diurnal variation of redox control in eukaryotes (Fig. 5A) (24). Coordination of NAD control of metabolism with NADPH/H2O2 control of spatiotemporal organization of the redox proteome supports key cellular activities, including mitosis and meiosis, differentiation, activation, senescence, and apoptosis, and complex multicellular processes, including fertilization, development, wound healing, and repair (95). The central positioning of H2O2 and thiol redox states in space and time is emphasized by the circadian variation in peroxiredoxins, abundant proteins that metabolize H2O2 and are central to redox signaling, and control of the thiol proteome (Fig. 5A).
FIG. 5.
Redox network structure supports adaptation to the environment. (A) Oxidation cycles for Prx are linked to clock protein expression. The oxidation cycle for peroxiredoxin-6 (Prx-SO2/3) is shown for fly (Drosophila melanogaster) with clock protein timeless (TIM), fungus (Neurospora crassa) with clock protein (FRQ), and mouse (Mus musculus) Prx1 with circadian protein BMAL. Similar variation in Cys/CySS is shown for human plasma. Figures were redrawn and simplified from original publications; originals should be consulted for statistical measurements and additional details and validations. (B) Progressive oxidation with age, shown by human plasma Cys/CySS and GSH/GSSG, contributes to decline in adaptability. (A) Composite modified from Edgar et al. (24) and Blanco et al. (9); (B), modified from Jones et al. (49). BMAL, transcriptional activator, core component of circadian clock; GSSG, glutathione disulfide; Prx1, peroxiredoxin-1.
Mechanistic studies also provide knowledge of GSH/GSSG oxidation during the life cycle of mammalian cells (32), reversible glutathionylation of actin (69), mitochondrial NADH dehydrogenase (6) to integrate structure and metabolism, and genetic control to modulate GSH redox systems. A yeast model system studied with a fluorescence imaging probe (71) showed a substantially more negative GSH redox potential (very low free GSSG); the relevance for mammalian cell physiology will need to be addressed. Diurnal variation in the GSH system occurs in humans and is linked to the timing of food intake and the precursor amino acid, Cys (Fig. 5A).
Activity cycles are connected to nutritional needs and environmental threats, linking all specialized adaptive mechanisms ultimately to central homeostatic control of energetics, metabolism, and proteomic structure. Thus, redox integration is fundamental to the logic of these adaptive mechanisms, evident in the activity and response characteristics of the immune system, redox gradients within physical barriers, and adaptive responses, such as thermogenesis. GSH-dependent detoxification of reactive chemicals, protection from oxidants, and control of toxic metals, each depend upon this integrated redox network to tolerate and thrive in the presence of environmental challenges. NAD+ and the redox proteome networks are linked to life span with an incipient oxidation of steady-state thiol redox systems with age, providing a redox indicator for aging (Fig. 5B).
Together, the four principles of the redox code approach the central theory for the redox organizational structure of life. Along with the evolved genetic code to define an organism and epigenetic and histone codes to guide the use of the genetic code, the redox code supports organized implementation of the genetic code within energetic, metabolic, and environmental constraints of the organism. Knowledge of the network structure embodied in this code provides new avenues for redox medicine to understand and treat diseases.
Implications for Redox Medicine
Health and disease are distinguished, among other parameters, by maintenance or disruption of the network of redox steady states. The concept of oxidative stress describes deviation from physiological redox steady state toward oxidation [see Sies (104) for recent discussion]. As outlined above, the very functioning of cell metabolism is governed by redox processes. Fine-tuning of reaction cascades and/or direct involvement in signaling pathways involves molecular redox switches and participation of redox-active metabolites, such as H2O2, nitric oxide, or hydrogen sulfide. Considerable efforts have been deployed in redox research in biology and medicine. The following brief comments are meant to exemplify the current state and opportunities in this rapidly developing field. Since enzymes rather than low-molecular-weight micronutrients play the major role in redox biology (102), aspects of genetic polymorphisms of crucial redox enzymes and of transport systems are important (e.g., responders vs. nonresponders). While genetic polymorphisms are not treated here in detail, just one example might illustrate the point: the set point of the concentration of GSH, a low-molecular-weight component, has been described as a heritable trait (117).
An example relating nutrition to redox biology is given by the micronutrient, selenium, which is present as selenocysteine in selenoproteins, as mentioned above, and also as selenomethionine in place of methionine in some proteins. There is a diversity of selenium functions in health and disease (12), including epigenetic effects (107) and a role in adjuvant therapy of viral and bacterial infections (5, 108).
Aging
Irrespective of the question of cause and effect, an association of a more oxidized set point in the redox balance with increasing age has been amply documented. The fraction of cellular proteins present with oxidized side chains increases with age and is more prominent in diseases of premature aging (79). In blood plasma, the small-molecule thiols, cysteine and GSH, are increasingly present as disulfides as individuals are more aged (Fig. 5B) (9). Thus, protein oxidation in aging and the machinery of removal of oxidized proteins has attracted attention (41). Autophagy and its potential regulation by dietary micronutrients have been examined in terms of healthy aging (83). Apart from loss of proteostasis, the hallmarks of aging include genomic instability, telomere attrition, epigenetic alterations, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication (63). Most, if not all, of these processes comprise a redox component (52). The NAD system is increasingly recognized as an attractive target for intervention in age-associated disorders (72). NAD precursors slow down aging and extend the life span in yeast and protect severed axons from degeneration in disease models.
Aspects of intermediary metabolism
Reactive oxygen species, H2O2 in particular, have been identified as signaling molecules in responses to extracellular stimuli such as insulin. Indeed, the insulin-dependent production of H2O2 is indispensable for propagation of intracellular signaling triggered by insulin (112). However, conversely, high levels of intracellular reactive oxygen species inhibit insulin activity and allow activation of stress responses, making for what has been called the peroxide dilemma (112). Interference with insulin signaling can also result from overexpression of hydroperoxide-reducing capacity, for example, by selenoproteins, and there has been an association between excessive selenium intake and incidence of diabetes type 2 (111), pointing to an interplay between selenium and carbohydrate metabolism (109). Likewise, strategies to improve the redox state in terms of obesity-associated oxidative stress have been examined (97). Diabetes type 2 has even been called a redox disease (120).
Cardiovascular arteriosclerosis
Oxidative damage from elevated production of reactive oxygen species contributes to ischemia–reperfusion injury in myocardial infarction and stroke. The enormous literature on this topic is not to be covered here [see Tocchetti et al. (114)]. One recent aspect might deserve special attention in the present context, linking protein redox modification to energy production: a specific cysteine switch on mitochondrial complex I has been identified by selective S-nitrosation of Cys39 on the ND3 unit of complex I (19).
Wound healing
Upon occurrence of a wound, the healing process begins as soon as damage arises. This process starts with conserved transcription-independent damage signals, such as H2O2 (Fig. 4C), calcium ions, and ATP, which initiate cell shape changes, functional actomyosin structures, and recruitment of immune cells (20). There are important redox signals in wound healing, and the recent identification of miRNA in wound inflammation and angiogenesis has led to the concept of OxymiRs, in particular miR-210 and miR-21, in their roles in responses to tissue oxygenation, skin development, and repair, as well as wound healing (91).
Immunology
Host defense is intimately linked to bacterial and virucidal oxidants generated by specialized cells, which are recruited and activated at the appropriate sites. Among the biological defense mechanisms, the production of superoxide by leukocytes was an early hallmark (3). Human fibroblasts release reactive oxygen species in response to interleukin-1 and tumor necrosis factor-α (67). The multiple aspects of redox processes in immunology are beyond the scope of the present article [see Nathan and Cunningham-Bussel (75)]. In inflammation, H2O2 is recognized as the messenger, guide, and assassin (124).
Proliferation of cancer
The relationship of redox processes with cell proliferation and apoptosis has been studied extensively and will not be presented here in detail. A recent overview on oxidative stress and cancer illustrates the range of the topic (106). The use of redox cycling drugs (53) has been a mainstay in cancer therapy.
Circadian rhythm
Chronobiological research has revealed tight coupling of redox reactions to circadian rhythmicity (36). As mentioned above, peroxiredoxins are conserved markers of circadian rhythms (Fig. 5A) (24), and there is diurnal variation in GSH and cysteine redox states (9). Physiological functions such as neuronal excitability are regulated by rhythmic changes in the redox state, and there are numerous implications of rhythmicity to human health (121). This burgeoning field of research has been circumscribed as the circadian topology of metabolism (4), with clocks impacting on fitness in multicellular organisms, ranging in medical aspects from nutrition to chronopharmacology and chronotherapeutics (81). Novel chronoassays as the diurnal exhalome may facilitate practical applications (66).
Concluding Remarks: The Redox Code
Research in redox biology has experienced progress to new levels by emerging methods, which identify redox states and allow monitoring in time and space. Central reactions of energetics and metabolism are controlled through near-equilibrium NAD reactions. The structure and function of cells, including genomics and epigenomics, are controlled through nonequilibrium reactions of the cysteine proteome. The spatiotemporal organization of complex systems is coordinated through activation/deactivation cycles of H2O2 production. Redox changes are a common feature of aging, and disruption is a common factor in disease. Hence, application of the new redox tools to define targets and target interventions affecting the NAD and redox proteome systems opens a new horizon for therapeutics impacting a broad range of human disease. The concept of the redox code integrates spatiotemporal set points in cellular organization.
Abbreviations Used
- BMAL
transcriptional activator, core component of circadian clock
- Cys
cysteine
- CySS
cystine
- EROS
endoplasmic reticulum oxidase system
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
glutathione disulfide
- HIF-1α
hypoxia-inducible factor-1α
- NAD
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- NF-κB
nuclear factor-κB
- Nox
NADPH oxidase
- Nrf2
nuclear factor-E2-related factor 2
- PDI
protein disulfide isomerase
- PGC-1α
peroxisome proliferator-activated receptor γ coactivator 1-α
- PTP1B
protein-tyrosine phosphatase 1B
- Ref-1
redox factor-1
- TR1
thioredoxin reductase-1
- TR2
thioredoxin reductase-2
- Trx
thioredoxin
Acknowledgments
The authors are grateful to Dr. Young-Mi Go and Dr. Wilhelm Stahl for critical comments during article development. H.S. is a Fellow of the National Foundation for Cancer Research (NFCR), Bethesda, MD, and expresses gratitude for the longstanding research support as well as support by Deutsche Forschungsgemeinschaft, Bonn, Germany. D.P.J. is supported, in part, by NIH grants, ES023485, ES009047, AG038746, HL113451, and ES019776.
References
- 1.Abate C, Patel L, Rauscher FJ, 3rd, and Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science 249: 1157–1161, 1990 [DOI] [PubMed] [Google Scholar]
- 2.Adimora NJ, Jones DP, and Kemp ML. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid Redox Signal 13: 731–743, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babior BM, Kipnes RS, and Curnutte JT. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J Clin Invest 52: 741–744, 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass J. Circadian topology of metabolism. Nature 491: 348–356, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Beck MA, Nelson HK, Shi Q, Van Dael P, Shiffrin EJ, Blum S, Barclay D, and Levander OA. Selenium deficiency increases the pathology of an influenza infection. FASEB J 15: 1481–1483, 2001 [PubMed] [Google Scholar]
- 6.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, and Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant defense. J Biol Chem 279: 47939–47951, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Beinert H, Holm RH, and Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277: 653–659, 1997 [DOI] [PubMed] [Google Scholar]
- 8.Berndt C, Lillig CH, and Flohé L. Redox regulation by glutathione needs enzymes. Front Pharmacol 5: 168, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco RA, Ziegler TR, Carlson BA, Cheng PY, Park Y, Cotsonis GA, Accardi CJ, and Jones DP. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr 86: 1016–1023, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Bleier L, Wittig I, Heide H, Steger M, Brandt U, and Dröse S. Generator-specific targets of mitochondrial reactive oxygen species. Free Radic Biol Med 78: 1–10, 2015 [DOI] [PubMed] [Google Scholar]
- 11.Brigelius-Flohé R. and Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal 15: 2335–2381, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigelius-Flohé R, and Sies H. (eds.). Diversity of Selenium Functions in Health and Disease. Boca Raton: CRC Press, 2015 [Google Scholar]
- 13.Buchanan BB. and Balmer Y. Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Bücher T, Brauser B, Conze A, Klein F, Langguth O, and Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem 27: 301–317, 1972 [DOI] [PubMed] [Google Scholar]
- 15.Bücher T. and Klingenberg M. Wege des Wasserstoffs in der lebendigen Organisation. Angew Chem 70: 552–570, 1958. [Pathways of hydrogen in the living organisation] [Google Scholar]
- 16.Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch Biochem Biophys 300: 535–543, 1993 [DOI] [PubMed] [Google Scholar]
- 17.Buettner GR, Wagner BA, and Rodgers VGJ. Quantitative redox biology: an approach to understanding the role of reactive species in defining the cellular redox environment. Cell Biochem Biophys 67: 477–483, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chance B, Sies H, and Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev 59: 527–605, 1979 [DOI] [PubMed] [Google Scholar]
- 19.Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cocheme HM, Reinhold J, Lilley KS, Partridge L, Fearnley IM, Robinson AJ, Hartley RC, Smith RA, Krieg T, Brookes PS, and Murphy MP. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med 19: 753–759, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordeiro JV. and Jacinto A. The role of transcription-independent damage signals in the initiation of epithelial wound healing. Nat Rev Mol Cell Biol 14: 249–262, 2013 [DOI] [PubMed] [Google Scholar]
- 21.D'Autréaux B. and Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8: 813–824, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Di Stefano M. and Conforti L. Diversification of NAD biological role: the importance of location. FEBS J 280: 4711–4728, 2013 [DOI] [PubMed] [Google Scholar]
- 23.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, and Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99: 11908–11913, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, and Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finck BN. and Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flohé L. The fairytale of the GSSG/GSH redox potential. Biochim Biophys Acta 1830: 3139–3142, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Gambini J, Gomez-Cabrera MC, Borras C, Valles SL, Lopez-Grueso R, Martinez-Bello VE, Herranz D, Pallardo FV, Tresguerres JA, Serrano M, and Vina J. Free [NADH]/[NAD(+)] regulates sirtuin expression. Arch Biochem Biophys 512: 24–29, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Ruiz C, Colell A, Morales A, Kaplowitz N, and Fernandez-Checa JC. Role of oxidative stress generated from the mitochondrial electron transport chain and mitochondrial glutathione status in loss of mitochondrial function and activation of transcription factor nuclear factor-kappa B: studies with isolated mitochondria and rat hepatocytes. Mol Pharmacol 48: 825–834, 1995 [PubMed] [Google Scholar]
- 29.Ghesquiere B, Jonckheere V, Colaert N, Van Durme J, Timmerman E, Goethals M, Schymkowitz J, Rousseau F, Vandekerckhove J, and Gevaert K. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics 10: M110.006866, 2011. DOI: 10.1074/mcp.M110.006866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghisla S. and Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur J Biochem 181: 1–17, 1989 [DOI] [PubMed] [Google Scholar]
- 31.Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol 63: 69–172, 1990 [DOI] [PubMed] [Google Scholar]
- 32.Go YM. and Jones DP. The redox proteome. J Biol Chem 288: 26512–26520, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Go YM, Roede JR, Walker DI, Duong DM, Seyfried NT, Orr M, Liang Y, Pennell KD, and Jones DP. Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol Cell Proteomics: 12: 3285–3296, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goncalves RLS, Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, and Brand MD. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J Biol Chem 290: 209–227, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griendling KK, Sorescu D, and Ushio-Fukai M. NAD(P)H oxidase: role in cardiovascular biology and disease. Circ Res 86: 494–501, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Hardeland R, Coto-Montes A, and Poeggeler B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int 20: 921–962, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Hensel M, Klingauf J, and Piehler J. Imaging the invisible: resolving cellular microcompartments by superresolution microscopy techniques. Biol Chem 394: 1097–1113, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Herrmann JM. and Dick TP. Redox biology on the rise. Biol Chem 393: 999–1004, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Higdon A, Diers AR, Oh JY, Landar A, and Darley-Usmar VM. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. Biochem J 442: 453–464, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoek JB. and Rydstöm J. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem J 254: 1–10, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohn A, Konig J, and Grune T. Protein oxidation in aging and the removal of oxidized proteins. J Proteomics 92: 132–159, 2013 [DOI] [PubMed] [Google Scholar]
- 42.Hohorst HJ, Kreutz FH, and Buecher T. [On the metabolite content and the metabolite concentration in the liver of the rat]. Biochem Z 332: 18–46, 1959 [PubMed] [Google Scholar]
- 43.Holzer H, Lynen F, and Schultz G. [Determination of diphosphopyridine nucleotide/reduced diphosphopyridine nucleotide quotient in living yeast cells by analysis of constant alcohol and acetaldehyde concentrations]. Biochem Z 328: 252–263, 1956 [PubMed] [Google Scholar]
- 44.Hondal RJ, Marino SM, and Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal 18: 1675–1689, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, and Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497: 451–457, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hung YP, Albeck JG, Tantama M, and Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 14: 545–554, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacob C, Giles GI, Giles NM, and Sies H. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew Chem 42: 4742–4758, 2003 [DOI] [PubMed] [Google Scholar]
- 48.Jang SY, Kang HT, and Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J Biol Chem 287: 19304–19314, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones DP, Mody VC, Jr., Carlson JL, Lynn MJ, and Sternberg P., Jr. Redox analysis of human plasma allows separation of pro-oxidant events of aging from decline in antioxidant defenses. Free Radic Biol Med 33: 1290–1300, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295: C849–C868, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones DP. Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Intern Med 268: 432–448, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DP. The redox theory of aging. Redox Biol 5: 71–79, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kappus H. and Sies H. Toxic drug effects associated with oxygen metabolism: redox cycling and lipid peroxidation. Experientia 37: 1233–1241, 1981 [DOI] [PubMed] [Google Scholar]
- 54.Kawahara T, Quinn MT, and Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol 7: 109, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kemp M, Go YM, and Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med 44: 921–937, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kensler TW. and Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis 31: 90–99, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klomsiri C, Karplus PA, and Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal 14: 1065–1077, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich AK, and Jakob U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell 47: 767–776, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4: 181–189, 2004 [DOI] [PubMed] [Google Scholar]
- 60.Le Moan N, Clement G, Le Maout S, Tacnet F, and Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem 281: 10420–10430, 2006 [DOI] [PubMed] [Google Scholar]
- 61.Levonen AL, Hill BG, Kansanen E, Zhang J, and Darley-Usmar VM. Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radic Biol Med 71: 196–207, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lillig CH, Berndt C, and Holmgren A. Glutaredoxin systems. Biochim Biophys Acta 1780: 1304–1317, 2008 [DOI] [PubMed] [Google Scholar]
- 63.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G. The hallmarks of aging. Cell 153: 1194–1217, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, and Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol 15: 222–228, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J. and Holmgren A. The thioredoxin superfamily in oxidative protein folding. Antioxid Redox Signal 21: 457–470, 2014 [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Lozano Sinues P, Zenobi R, and Kohler M. Analysis of the exhalome: a diagnostic tool of the future. Chest 144: 746–749, 2013 [DOI] [PubMed] [Google Scholar]
- 67.Meier B, Radeke HH, Selle S, Younes M, Sies H, Resch K, and Habermehl GG. Human fibroblasts release reactive oxygen species in response to interleukin-1 or tumour necrosis factor-alpha. Biochem J 263: 539–545, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michan S. and Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 404: 1–13, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, and Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal 10: 1941–1988, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miseta A. and Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol 17: 1232–1239, 2000 [DOI] [PubMed] [Google Scholar]
- 71.Morgan B, Ezerina D, Amoako TN, Riemer J, Seedorf M, and Dick TP. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat Chem Biol 9: 119–125, 2013 [DOI] [PubMed] [Google Scholar]
- 72.Mouchiroud L, Houtkooper RH, and Auwerx J. NAD(+) metabolism: a therapeutic target for age-related metabolic disease. Crit Rev Biochem Mol Biol 48: 397–408, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, and Auwerx J. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154: 430–441, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakahata Y, Sahar S, Astarita G, Kaluzova M, and Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nathan C. and Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat Rev Immunol 13: 349–361, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nauser T, Steinmann D, Grassi G, and Koppenol WH. Why selenocysteine replaces cysteine in thioredoxin reductase: a radical hypothesis. Biochemistry 53: 5017–5022, 2014 [DOI] [PubMed] [Google Scholar]
- 77.Niethammer P, Grabher C, Look AT, and Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459: 996–999, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okamoto K, Tanaka H, Ogawa H, Makino Y, Eguchi H, Hayashi S, Yoshikawa N, Poellinger L, Umesono K, and Makino I. Redox-dependent regulation of nuclear import of the glucocorticoid receptor. J Biol Chem 274: 10363–10371, 1999 [DOI] [PubMed] [Google Scholar]
- 79.Oliver CN, Ahn BW, Moerman EJ, Goldstein S, and Stadtman ER. Age-related changes in oxidized proteins. J Biol Chem 262: 5488–5491, 1987 [PubMed] [Google Scholar]
- 80.Oronsky B, Oronsky N, Scicinski J, Fanger G, Lybeck M, and Reid T. Rewriting the epigenetic code for tumor resensitization: a review. Transl Oncol 7: 626–631, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ortiz-Tudela E, Mteyrek A, Ballesta A, Innominato PF, and Levi F. Cancer chronotherapeutics: experimental, theoretical, and clinical aspects. Handb Exp Pharmacol 261–288, 2013 [DOI] [PubMed] [Google Scholar]
- 82.Oshino N, Chance B, Sies H, and Bucher T. The role of H2O2 generation in perfused rat liver and the reaction of catalase compound I and hydrogen donors. Arch Biochem Biophys 154: 117–131, 1973 [DOI] [PubMed] [Google Scholar]
- 83.Pallauf K. and Rimbach G. Autophagy, polyphenols and healthy ageing. Ageing Res Rev 12: 237–252, 2013 [DOI] [PubMed] [Google Scholar]
- 84.Papp LV, Lu J, Holmgren A, and Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal 9: 775–806, 2007 [DOI] [PubMed] [Google Scholar]
- 85.Prigogine I. Time, structure, and fluctuations. Science 201: 777–785, 1978 [DOI] [PubMed] [Google Scholar]
- 86.Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839, 1998 [DOI] [PubMed] [Google Scholar]
- 87.Quinlan CL, Goncalves RLS, Hey-Mogensen M, Yadava N, Bunik VI, and Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem 289: 8312–8325, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rainwater R, Parks D, Anderson ME, Tegtmeyer P, and Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol 15: 3892–3903, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, and Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–654, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rhee SG, Woo HA, Kil IS, and Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem 287: 4403–4410, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roy S. and Sen CK. miRNA in wound inflammation and angiogenesis. Microcirculation 19: 224–232, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rushmore TH, Morton MR, and Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266: 11632–11639, 1991 [PubMed] [Google Scholar]
- 93.Rytkonen KT. and Storz JF. Evolutionary origins of oxygen sensing in animals. EMBO Rep 12: 3–4, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saccoccia F, Angelucci F, Boumis G, Desiato G, Miele AE, and Bellelli A. Selenocysteine robustness versus cysteine versatility: a hypothesis on the evolution of the moonlighting behaviour of peroxiredoxins. Biochem Soc Trans 42: 1768–1772, 2014 [DOI] [PubMed] [Google Scholar]
- 95.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, Jiao X, Li A, Zhang X, Lu Y, Wang C, Byers S, Nicholson R, Link T, Shemluck M, Yang J, Fricke ST, Novikoff PM, Papanikolaou A, Arnold A, Albanese C, and Pestell R. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol 26: 5449–5469, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sassone-Corsi P. Minireview: NAD+, a circadian metabolite with an epigenetic twist. Endocrinology 153: 1–5, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Savini I, Catani MV, Evangelista D, Gasperi V, and Avigliano L. Obesity-associated oxidative stress: strategies finalized to improve redox state. Int J Mol Sci 14: 10497–10538, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schreck R, Albermann K, and Baeuerle PA. Nuclear factor kappa B: an oxidative stress-responsive transcription factor of eukaryotic cells (a review). Free Radic Res Commun 17: 221–237, 1992 [DOI] [PubMed] [Google Scholar]
- 99.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol 9: 47–71, 2014 [DOI] [PubMed] [Google Scholar]
- 100.Sies H. Nicotinamide nucleotide compartmentation. In: Metabolic Compartmentation, edited by Sies H, London: Academic Press, 1982, pp. 205–231 [Google Scholar]
- 101.Sies H. On metabolic compartmentation: introductory remarks. In: Metabolic Compartmentation, edited by Sies H, London: Academic Press, 1982, pp. 1– 6 [Google Scholar]
- 102.Sies H. Strategies of antioxidant defense. Eur J Biochem 215: 213–219, 1993 [DOI] [PubMed] [Google Scholar]
- 103.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J Biol Chem 289: 8735–8741, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol 4: 180–183, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sies H, Bücher T, Oshino N, and Chance B. Heme occupancy of catalase in hemoglobin-free perfused rat liver and of isolated rat liver catalase. Arch Biochem Biophys 154: 106–116, 1973 [DOI] [PubMed] [Google Scholar]
- 106.Sosa V, Moline T, Somoza R, Paciucci R, Kondoh H, and ME LL. Oxidative stress and cancer: an overview. Ageing Res Rev 12: 376–390, 2013 [DOI] [PubMed] [Google Scholar]
- 107.Speckmann B. and Grune T. Epigenetic effects of selenium and their implications for health. Epigenetics 10: 179–190, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steinbrenner H, Al-Quraishy S, Dkhil MA, Wunderlich F, and Sies H. Dietary selenium in adjuvant therapy of viral and bacterial infections. Adv Nutr 6: 73–82, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Steinbrenner H, Speckmann B, Pinto A, and Sies H. High selenium intake and increased diabetes risk: experimental evidence for interplay between selenium and carbohydrate metabolism. J Clin Biochem Nutr 48: 40–45, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Storz G, Tartaglia LA, and Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248: 189–194, 1990 [DOI] [PubMed] [Google Scholar]
- 111.Stranges S, Marshall JR, Natarajan R, Donahue RP, Trevisan M, Combs GF, Cappuccio FP, Ceriello A, and Reid ME. Effects of long-term selenium supplementation on the incidence of type 2 diabetes: a randomized trial. Ann Intern Med 147: 217–223, 2007 [DOI] [PubMed] [Google Scholar]
- 112.Szypowska AA. and Burgering BM. The peroxide dilemma: opposing and mediating insulin action. Antioxid Redox Signal 15: 219–232, 2011 [DOI] [PubMed] [Google Scholar]
- 113.Tanaka T, Hosoi F, Yamaguchi-Iwai Y, Nakamura H, Masutani H, Ueda S, Nishiyama A, Takeda S, Wada H, Spyrou G, and Yodoi J. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J 21: 1695–1703, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tocchetti CG, Stanley BA, Murray CI, Sivakumaran V, Donzelli S, Mancardi D, Pagliaro P, Gao WD, van Eyk J, Kass DA, Wink DA, and Paolocci N. Playing with cardiac “redox switches”: the “HNO way” to modulate cardiac function. Antioxid Redox Signal 14: 1687–1698, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Toledano MB. and Leonard WJ. Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc Natl Acad Sci U S A 88: 4328–4332, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tu BP. and Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van't Erve TJ, Wagner BA, Ryckman KK, Raife TJ, and Buettner GR. The concentration of glutathione in human erythrocytes is a heritable trait. Free Radic Biol Med 65: 742–749, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ventura-Clapier R, Garnier A, and Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res 79: 208–217, 2008 [DOI] [PubMed] [Google Scholar]
- 119.Wang GL. and Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270: 1230–1237, 1995 [DOI] [PubMed] [Google Scholar]
- 120.Watson JD. Type 2 diabetes as a redox disease. Lancet 383: 841–843, 2014 [DOI] [PubMed] [Google Scholar]
- 121.Wilking M, Ndiaye M, Mukhtar H, and Ahmad N. Circadian rhythm connections to oxidative stress: implications for human health. Antioxid Redox Signal 19: 192–208, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Williamson DH, Lund P, and Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J 103: 514–527, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Winterbourn CC. and Hampton MB. Signaling via s peroxiredoxin sensor. Nat Chem Biol 11: 5–6, 2015 [DOI] [PubMed] [Google Scholar]
- 124.Wittmann C, Chockley P, Singh SK, Pase L, Lieschke GJ, and Grabher C. Hydrogen peroxide in inflammation: messenger, guide, and assassin. Adv Hematol 2012: 541471, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wong HL. and Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol 21: 796–804, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xanthoudakis S, Miao G, Wang F, Pan YC, and Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J 11: 3323–3335, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yin F, Boveris A, and Cadenas E. Mitochondrial energy metabolism and redox signalling in brain aging and neurodegeneration. Antioxid Redox Signal 20: 353–371, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]