Abstract
By generating and studying mosaic organisms we are learning how intricate tissues form as cells proliferate and diversify through organism development. FLP/FRT-mediated site-specific mitotic recombination permits the generation of mosaic flies with efficiency and control. With heat-inducible or tissue-specific FLP transgenes at our disposal, we can engineer mosaics carrying clones of homozygous cells that come from specific pools of heterozygous precursors. This permits detailed cell lineage analysis followed by mosaic analysis of gene functions in the underlying developmental processes. Expression of transgenes (e.g. reporters) only in the homozygous cells enables mosaic analysis in the complex nervous system. Tracing neuronal lineages by using mosaics revolutionized mechanistic studies of neuronal diversification and differentiation, exemplifying the power of genetic mosaics in developmental biology.
Introduction
Generating mosaics (i.e. organisms with cells of different genotypes) allows manipulation of gene functions in subsets of cells within otherwise unperturbed organisms 1. In diploid organisms, genetic mosaics can be generated by mitotic recombination that produces homozygous daughter cells from heterozygous precursors which have undergone homologous recombination during mitoses 2.
Such mosaic organisms carry patches of cells homozygous for either of the recombinant chromosome arms in heterozygous tissues. Their patterns of mosaicism reflect where and when the events of mitotic recombination have occurred during the organism development. The mosaic pattern, if properly labeled, can faithfully reveal descent. Generating mosaics by mitotic recombination (as opposed to various cell-cycle-independent mosaic techniques) is therefore uniquely suitable for cell lineage analysis to delineate retrospectively the cell proliferation processes from which specific cells of interest have arisen 3.
Cell lineage analysis is fundamental for understanding tissue complexity and development. Retrospective cell lineage analysis is carried out by characterizing the mature cells derived from an earlier precursor whose physical identity is often unknown. It generally involves sparse stochastic permanent labeling of progenitors and thus their descendents are also labeled. In Drosophila, the limited intermingling and migration of cells mean that cells of the same lineage origin typically form a spatially coherent clone in adult tissues. This ‘clonal’ organization greatly simplifies cell lineage analysis. Technically, sampling clones of various sizes induced at different developmental stages should efficiently unveil the overall lineage map. Full-size clones consist of all the descendents of a founding progenitor, while subclones of decreasing sizes carry subsets of related cells with closer pedigree relationships. One can then build the developmental hierarchy of the recovered clones based on clone size, location, timing of induction, and offspring composition to deduce the mode(s) of cell proliferation and learn the role(s) of cell lineage in the derivation of tissues with characteristic cells in specific arrangements.
One can speculate two extreme scenarios as to the relationship between cell lineage and cell fate determination. At one extreme, the precursors may undergo variable rounds of cell divisions and collectively deposit excessive terminal cells that remain naïve and not committed to any particular cell fate, including the possibility of undergoing apoptosis, until exit of cell cycles. In this scenario, random lineages would exist and one could hardly recover similar clones even after clone induction in well-synchronized organisms. At the other extreme, both precursor numbers and cell cycles are tightly controlled such that an intricate tissue always develops from the same invariant lineages. In this case, stage-specific stereotyped clones that are characteristic of each lineage can be obtained repeatedly. One can in theory reconstruct the underlying lineages through the analysis of clones respectively derived from the founding progenitors as well as all the intermediate precursors.
The presence of invariant lineages indicates the derivation of specific terminal cells from specific precursors and implicates the involvement of lineage-intrinsic mechanisms in cell fate determination. This would encourage the identification of the conserved precursors for the investigation of possible cell fate commitments due to progressive restriction in their developmental potentials. One can further scrutinize individual offspring in a completely mapped stereotyped lineage for their detailed structural and functional roles in adult tissues as well as the mechanisms underlying their acquisition of different mature structures and functions. Given that most genes possess (possibly diverse) functions in diverse cells, it is mandatory in most cases to manipulate genes only in the cells of interest to uncover cell-autonomous functions for the genes. Generating mosaics by mitotic recombination, to create clones of homozygous daughter cells from heterozygous precursors, can provide information about lineage and gene function at the same time and thus is a powerful tool in Drosophila melanogaster. It enables foundational cell lineage analyses followed by studies of the genetic requirements of lineage development.
Genetic mosaics prior to site-specific mitotic recombination
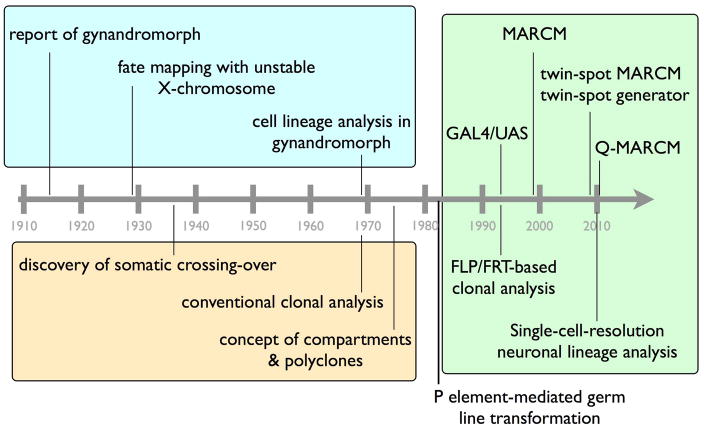
Genetic lineage analyses in Drosophila were initially performed in the gynandromorphs that carry mixtures of female (XX) and male (XO) tissues due to somatic loss of an X chromosome 4,5,6,7 (see Fig. 1 for timeline of genetic chimeras). Using unstable chromosomes or mutations that promote chromosomal loss 2, gynandromorphs can be produced at sufficient frequencies for constructing developmental fate maps to outline the distribution of progenitors for late structures in early embryos 6. Landmark studies in gynandromphs led to a more detailed understanding of the embryonic origins of the imaginal discs and their relative locations in the early embryo. The major limitations of generating mosaics via chromosome loss include (1) no spatiotemporal control over the occurrence of chromosome loss, and (2) the detrimental effects of losing an entire autosome, which restricts the hemizygous mosaic studies only to X-linked genes.
Figure 1. Timeline of Drosophila genetic chimeras and cell lineage analysis.
Major advances in Drosophila cell lineage analysis through gynandromorphy (within blue box), irradiation-induced mitotic recombination (within orange box), or FLP/FRT-mediated site-specific recombination (within green box) are shown chronologically. All the modern genetic mosaic tools were established via germ line transformation, which was made possible by Rubin and Spradling in 1982.
Compared to chromosome loss, generating mosaics by mitotic recombination 8 is genetically much subtler and can be controlled through induction of double-strand DNA breaks historically mediated by ionizing radiation 9,10. Double-strand DNA breaks can be repaired via homologous recombination, leading to sister-chromatid exchanges during mitosis and subsequent loss of heterozygosity following X segregation of the recombinant chromosomes. Instead of losing one entire homologous chromosome, clones derived after mitotic recombination remain diploid but become homozygous for all the genes residing distal to the site of recombination and thus differ from the surrounding heterozygous non-recombinant cells. Clones homozygous for a recessive mutation cell-autonomously affecting cells’ gross appearance (e.g. color or morphology) can be reliably detected 10,11. Moreover, clones can be induced at different developmental stages by stage-specific irradiation 10,2. Systematical analysis of such temporally induced clones has demonstrated the presence of anterior and posterior compartments in the larval imaginal discs 12,13. Each compartment exists as a polyclone composed of all the descendants of a small group of founder cells 14. The apposed anterior-posterior boundary then patterns the development of the adult appendages 15.
The identifiable clones can be mutant for genes of interest and homozygous for the recessive marker mutation when both mutations coexist on the same recombinant chromosome arm. This allows knockout of essential genes in mosaic patches for loss-of-function mutant analysis in otherwise nonviable organisms. A cell-autonomous growth retardation mutation on the other homologous chromosome arm would reduce the number of non-marked cells and increase the number of cells from the marked clones (because the marked clones lost the dominant slow-growing mutation and possess a growth advantage over their slow-dividing neighbors). Such clone competition underlies the Minute technique 16 used to increase clone sizes for studying compartment boundary or employed to promote clone survival for recovering sick mutant clones. Similar genetic tricks underlie the development of germline mosaics, which involves the use of dominant female sterile mutations (placed on the otherwise wild-type chromosome arm) to block the production of any but the homozygous mutant oocytes 17,18. One can therefore eliminate essential genes selectively from female germlines to study gene function in oogenesis or examine the roles of maternal contributions in early organism development.
Despite all these genetic innovations, the poor efficiency of clone production and the potential toxicity of ionizing radiation severely limited the applicability of mosaic studies with mitotic recombination. In addition, the marking of clones based on changes in gross appearance often prevented visualization of internal or immature clones. Besides, having no control over the site(s) of mitotic recombination complicates data interpretation because sometimes the marked clones can lack the mutation in the gene of interest or unmarked background clones with unknown genotypes can exist.
Genetic mosaics by site-specific mitotic recombination
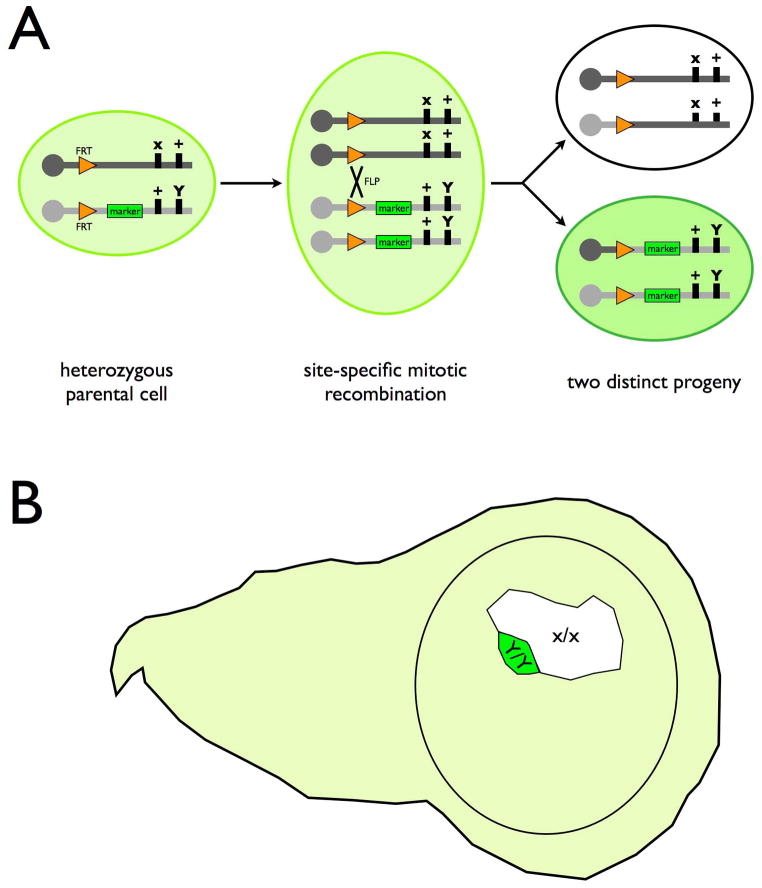
The development of P element-mediated germ line transformation 19 followed by the introduction of the FLP recombinase and its targets, FRTs, into the Drosophila genome 20 fostered the establishment of a much more efficient and controllable genetic mosaic system (Fig. 1). The FLP/FRT-based genetic mosaic system depends on FLP-mediated mitotic recombination between FRTs residing in trans on homologous chromosomes 21,22,23. Mitotic recombination across FRTs allows the production of homozygous clones in heterozygous organisms (Fig. 2). The resulting mosaic organisms thus consist of cells with three distinct genotypes. The non-recombinant cells remain heterozygous and carry both paternal and maternal copies of genes located distal to FRT. By contrast, the homozygous sister clones derived from a common heterozygous precursor would exclusively carry two copies of the paternal alleles and two copies of the maternal alleles, respectively. Placing copies of FRTs near the centromere on a given chromosome arm for all major chromosomes enables genetic mosaic studies for over 95% of the Drosophila genome 23. Reporter transgenes can be placed distal to the FRT site on only one homologous chromosome arm for marking the mutant clones as well as their wild-type sister clones in dosage-dependent manners (two copies versus zero copy of the reporter transgene) 23 (Fig. 2). Markers may carry exogenous epitopes or chromophores and various subcellular targeting motifs for better characterization of external as well as internal clones at different stages of organism development.
Figure 2. Genetic mosaics by site-specific mitotic recombination.
(A) A heterozygous parental cell can give rise to two distinct daughter cells homozygous for different alleles of genes and transgenes residing distal to the site of mitotic recombination (orange triangles: FRTs; recombination between FRTs requires FLP). The resulting homozygous daughter cells and their offspring can be distinguished from the surrounding heterozygous cells based on the copy number of the marker transgene (green). One can further enrich clones homozygous for the recessive x mutation by incorporating a dominant slow-growing Y mutation onto the otherwise wild-type homologous chromosome arm.
(B) A schematic heterozygous wing disc (light green) carries a large x/x mutant clone (unmarked) paired with a tiny Y/Y sister clone (dark green). The mutant clone shows growth advantages due to the absence of the dominant slow-growth Y mutation.
Mitotic recombination across FRTs exclusively depends on the FLP, which could now be expressed in discrete spatiotemporal patterns. This granted an unprecedented level of versatility in the control of clone induction. Under the control of a heat shock promoter, FLP can be acutely induced within a defined developmental window to monitor stage-specific tissue proliferation patterns. One can also restrict FLP expression to certain progenitor pools using various enhancers to generate tissue-specific mosaics 24,25. Incorporation of a dominant slow-growing or recessive cell-lethal mutation into the otherwise wild-type homologous chromosome arm further allows one to enrich mutant clones and even obtain mosaics with tissues that are exclusively homozygous for the mutant FRT chromosome arm 26,24,25 (e.g. Fig. 2). The efficiency of clone production, the control of clone induction, and the reliable clone detection with FLP/FRT have promoted reverse as well as forward genetic mosaic screens, greatly facilitating gene discovery in later developmental processes of interest 27.
Mosaic Analysis with a Repressible Cell Marker
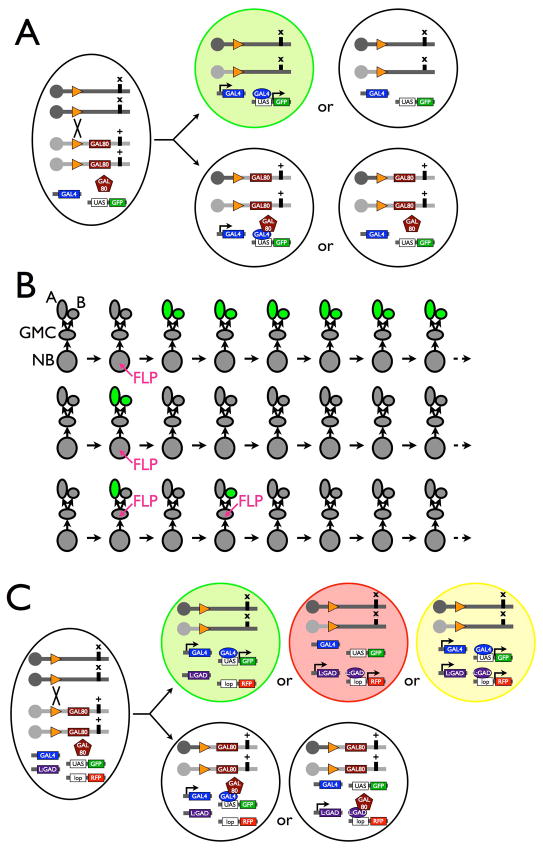
Despite all the advances, mosaic analysis in the complex central nervous system (CNS) remained limited mainly because of the challenges in detecting mutant clones in dense convoluted tissues. This motivated the development of MARCM (Mosaic Analysis with a Repressible Cell Marker) in 1999 28. MARCM controls reporter expression through the GAL4/UAS binary transgene induction system 29. In MARCM, loss of heterozygosity following mitotic recombination results in loss of the sole GAL80 (GAL4 repressor) transgene from one of two homozygous daughter cells and all the offspring (if any) of the GAL80-minus cell. The subsequent unique labeling of the GAL80-minus clones depends on the activation of UAS-reporter by the employed GAL4 driver (Fig. 3A). The brain consists of diverse classes and types of neurons plus glia, such that most tissue-specific GAL4 drivers show rather restricted patterns of activities in the CNS. Use of such GAL4 drivers could limit the visualization of MARCM clones to a specific subset of neurons and greatly simplify the clone patterns. The selective detection of clones among GAL4-positive neurons is often necessary for detailed analysis of neurite trajectories. However, inability to observe all GAL80-minus clones or every single cell in a GAL80-minus clone may bias data interpretation. For instance, an undetected mutant clone might non-autonomously affect the mutant phenotypes of an observed MARCM clone. To exclude such indirect effects on the marked clone requires comparative analysis of the same clone in multiple mosaic organisms that supposedly carry different (unseen) background clones.
Figure 3. MARCM and discrete sizes of neural clones.
(A) MARCM allows the derivation of homozygous mutant cells devoid of the GAL80 transgene (plum) following site-specific mitotic recombination. Depending on the GAL4 (blue) expression pattern, subsets of GAL80-negative mutant cells can be uniquely labeled by UAS-reporter (green).
(B) A neuronal lineage forms as one neuroblast (NB) repeatedly bud off ganglion mother cells (GMC) that each divide once to produce two post-mitotic neurons which often acquire distinct binary cell fates (A versus B) due to differential Notch signaling. Given this general pattern of neurogenesis, MARCM allows labeling of either a multi-cellular NB clone or a two-cell GMC clone following mitotic recombination in a dividing NB and marking of either A or B neuron as a single-neuron clone if mitotic recombination occurs in a dividing GMC. Note only post-mitotic neurons are marked in the panels given the transient nature of precursors.
(C) Dual-expression-control MARCM allows differential labeling of distinct populations of GAL80-minus cells via use of two independent GAL80-repressible binary transgene induction systems (e.g. GAL4/UAS & LexA::GAD/lexAop). Green: GAL4+ & LexA::GAD−; red: GAL4− & LexA::GAD+; yellow: GAL4+ & LexA::GAD+.
The pattern of MARCM clones in the fly CNS primarily depends on the patterns of neurogenesis (e.g. Fig. 3B). In contrast to most non-neural tissues that grow exponentially via repetitive symmetric cell divisions, neurons are sequentially added by common progenitors, called neuroblasts (NBs), during embryogenesis and through the protracted larval/pupal development. A typical NB divides asymmetrically to renew itself and bud off a ganglion mother cell (GMC) that divides once to produce two post-mitotic neurons. Repeating the asymmetric cell division, a single NB can yield a series of sister-neuron pairs comprising a neuronal lineage, in the adult CNS. Given this pattern of neurogenesis, clones derived from a single neuronal lineage could be either a large NB clone or a two-cell GMC clone if mitotic recombination occurred in the NB (Fig. 3B). Independent single-cell clones are the result if mitotic recombination occurred in GMCs (Fig. 3B). One can learn how distinct neuronal lineages constitute a neural network by characterizing multi-cellular NB clones and delineating individual neurite trajectories by analyses of single-cell clones.
The patterns of MARCM clones depend on the induction scheme of the FLP recombinase. MARCM was built upon the pre-assembled FLP/FRT-mediated genetic mosaic system. In most cases the use of hs-FLP is favored, as opposed to tissue-specific FLP, in the induction of neural clones for multiple reasons.
First, the apparently uniform expression of FLP following heat shock permits clone induction in all active neuronal lineages. Second, varying the intensity of heat shock can adjust the frequency of clones. Sparse clones are critical for assessing the lineage relationships among multiple labeled neurons. Third, the ability to induce clones at specific developmental stages by transient heat shock allows one to follow neurogenesis with great temporal resolution through the protracted CNS development. Only the precursors present during the transient expression of FLP can be subject to mitotic recombination. One can therefore retrospectively determine, through the analysis of MARCM clones induced at different developmental times, when a particular neuron type and its immediate precursor(s) were born in a long neuronal lineage.
GAL4 drivers ultimately determine which GAL80-minus cells will be labeled with MARCM. Clone induction with hs-FLP has the potential of hitting all proliferative lineages. Except around larval hatching and after puparium formation, most CNS NBs are actively dividing and often surrounded by multiple recently born GMCs 30,31. Even a brief induction of hs-FLP can elicit many GAL80-minus clones; visualizing all of them makes tracing of individual clones extremely challenging. Given the complexity of the CNS and the obvious self-sufficiency in lineage development, it is desirable to focus on one lineage at one time for detailed phenotypic analysis. This could be achieved through use of GAL4 drivers that each selectively label whole lineages. Unfortunately, such drivers rarely exist (probably reflecting the heterogeneity of NB offspring) and it is hard to determine if any potential lineage driver does faithfully label all the progeny made by a given NB. Nonetheless, an ideal MARCM driver should mask as many background clones as possible without sacrificing the coverage of the target lineage(s). The identification and characterization of such drivers have paved the way for fruitful MARCM studies in the complex CNS.
MARCM with GAL4-OK107 and GAL4-GH146 allowed lineage mapping among the mushroom body (MB) intrinsic neurons and three groups of antennal lobe (AL) projection neurons (PNs), respectively 32,33,34. First, stochastic labeling of NB clones induced at the beginning of larval neurogenesis confirms the presence of four ‘equivalent’ MB NB clones and ascribes the discrete groups of analogous AL PNs to separate lineages. Second, analysis of single-cell clones reveals the diversity of neurons within a given neuronal lineage. Intriguingly, diverse sibling neurons arise in an invariant sequence, as evidenced by the orderly appearance of distinct identifiable neurons (e.g. MB neurons that project into specific MB lobes or AL PNs that target different AL glomeruli) among the sequentially derived single-cell/two-cell clones of the same lineage. Such stereotyped clonal development implicates involvement of lineage-intrinsic mechanisms in the specification of diverse neurons. It encourages systematic identification of all neuron types in the complex CNS through comprehensive single-neuron lineage mapping. However, resolving a highly heterogeneous neuronal lineage that potentially makes two unique neurons from each GMC could be extremely challenging. In fact, a complete single-neuron lineage mapping was not possible in those complex AL PN lineages until the development of twin-spot MARCM (see below).
MARCM further fostered genetic mosaic studies to uncover genes that regulate neural development in cell-autonomous manners 35,36. Such mechanistic studies require groundwork that allows us to catalog identifiable neurons based on single-neuron morphology and cell lineage information. We can then reproducibly target the same lineage or particular neurons for loss-of-function mosaic studies. Single-cell clones provide Golgi-quality single-neuron morphology for detailed morphometric analysis. Abnormal morphology in a single-neuron clone that is homozygous for a loss-of-function mutation in the gene of interest would reveal the gene’s cell-autonomous function(s) in neuronal morphogenesis. Knocking out genes from neural precursors followed by analysis of mutant NB or GMC clones could further reveal genes’ lineage- or cell-autonomous roles in neurogenesis or neuron fate specification 37. In addition, loss of heterozygosity in GMCs prior to the neuron-producing mitoses is often necessary for unraveling the null morphogenetic phenotypes since perdurance of gene functions from GMCs to post-mitotic neurons could significantly alleviate defects in initial neuronal differentiation. Moreover, additional UAS transgenes, encoding engineered proteins or interfering RNAs, can be uniquely expressed in the GAL4-positive, GAL80-minus cells for rescuing mutant phenotypes or dominantly manipulating endogenous gene functions 38. Furthermore, intragenic MARCM with FRTs inside distinct (engineered) alleles of a given gene can allow reconstitution of a functional mutant allele and at the same time disruption of the pre-existing wild-type allele in MARCM clones for protein structure-function analysis at the endogenous expression level and in single-cell mosaics 39.
Twin-Spot positive labeling
Mitotic recombination can yield two sister cells homozygous for distinct homologous chromosome arms. The original version of MARCM permits visualization of only one of the two homozygous sister cells and the descendant(s) of only that sister cell (Fig. 3). An invisible sister clone precludes a comprehensive cell lineage study. For instance, the only way to validate a GMC clone, which may contain one or two neurons (e.g. Fig. 5B–D vs. Fig. 5E) and be easily confused with two independent single-neuron clones, is to observe its pairing with a NB clone. Sorting out the sister relationships among distinct single-neuron clones also requires visualization, and ideally differential labeling, of both post-mitotic neurons made by a given GMC (e.g. Fig. 5E). Marking the mutant clone and its wild-type sister clone helps substantiate the identity of the precursor that underwent the mitotic recombination (e.g. Fig. 6). Learning exactly where and when the mitotic recombination has occurred is critical for determining the prospective cell fate of the mutant clone, to detect fate transformations among endogenous cell types.
Figure 5. High-resolution neuronal lineage analysis by twin-spot MARCM.
(A) The adult Drosophila brain (gray) carries neurons with different characteristic morphologies that arise in clones from specific NBs. Exemplified in this illustration are representative neurons present in two antennal lobe (AL) NB clones (dashed circumferences). The anterodorsal AL (adAL) lineage yields distinct uniglomerular projection neurons (PNs), which each target one of the 50 or so AL glomeruli and then project through the MB calyx into the LH (e.g. the right-hemisphere blue neuron). Besides making uniglomerular PNs that target a different set of AL glomeruli (e.g. the left-hemisphere blue neuron), the lateral AL (lAL) lineage generates diverse AL local interneurons (LNs, e.g. the orange neuron) plus various non-AL PNs (e.g. the green neuron) that connect other neuropils. To map individual neurons and further elucidate the origins of neuron diversity requires tracking single neurons serially made by a common NB.
(B–D) Systematic analysis of GMC clones (red) paired with NB clones (green) of different sizes allows mapping of the serially derived neurons in the adAL lineage. Only one of the sister neurons derived from each adAL GMC survives into the adult stage, as the NB clones consistently associate with a single-neuron GMC clone. Notably, the uniglomerular adAL neurons that innervate different AL glomeruli are born sequentially in a defined order from the adAL NB. For instance, the VM3-, 1-, and DL2v-targeting neurons are born at the early (B), middle (C), and late (D) stages, respectively. Scale bar: 10 μm.
(E) The lAL NB clone (cell bodies marked with black asterisks) exhibits complex morphologies. Differential labeling of twin neurons derived from common GMCs consistently reveals one PN (magenta) paired with one LN (green) in the lAL lineage. Distinct PN/LN pairs are selectively recovered following clone induction (blue asterisks) in different two-hour windows after larval hatching (ALH), suggesting neuron fate specification based on the GMC birth order and the A/B binary fate decision. Scale bar: 10 μm.
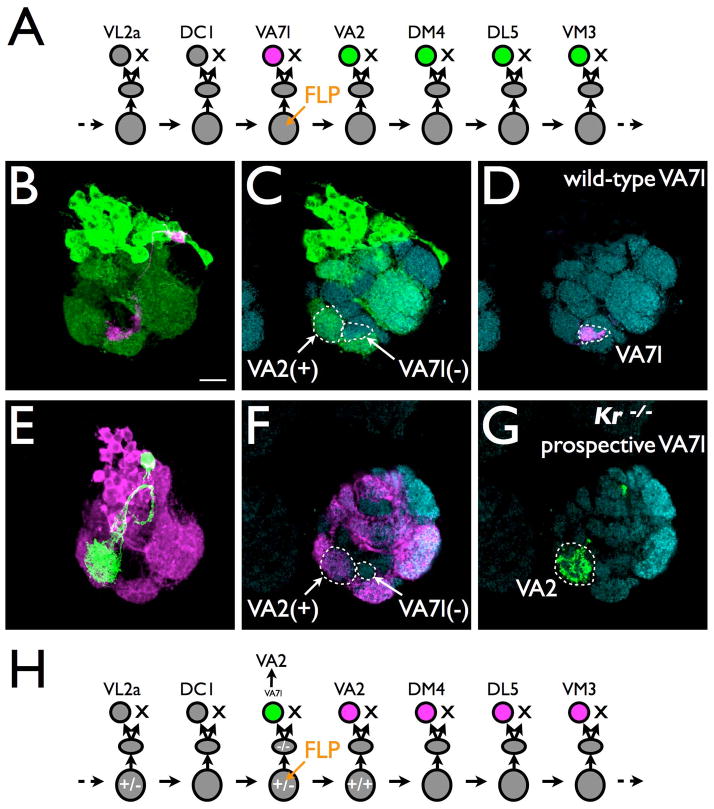
Figure 6. Loss-of-function mosaic analysis of neuronal temporal identity.
(A–D) In the wild-type adAL lineage, the VA7l and VA2 uniglomerular PNs are born sequentially (A), as twin-spot MARCM labeling (B) shows pairing of the smallest VA2-containing NB clone (C) with the VA7l-targeting PN (D).
(E–H) By contrast, the Kr mutant single-neuron GMC clone (green) paired with the equivalent VA2-containing wild-type NB clone (magenta) redundantly innervates the VA2 glomerulus (E–G). This chronologically abnormal phenotype argues for the acquisition of the next VA2 temporal cell fate by the prospective VA7l PN (H). Scale bar: 10 μm.
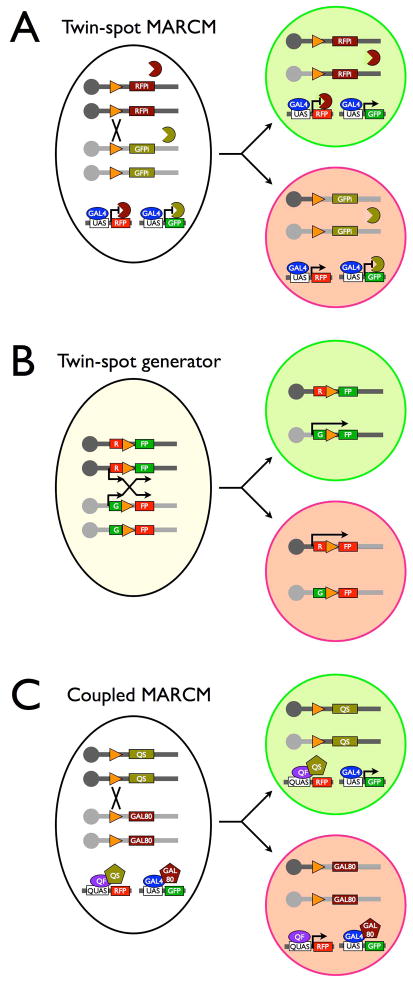
Three transgenic systems allow differential positive labeling of twin spots (paired sister clones) (Fig. 4). First, twin-spot MARCM 40 employs transgenic microRNAs (miRNAs) to silence reporter genes except in the homozygous sister clones that have lost distinct miRNA transgenes and thus express different reporters. The main concerns about twin-spot MARCM include: (1) the efficiency and perdurance of RNA interference, and (2) the challenge in the co-expression of multiple transgenes in the clones of interest. Second, Q-MARCM 41, which involves a different repressible transgene induction system, can be combined with conventional MARCM to label the paired sister clones by QUAS-reporter1 and UAS-reporter2, respectively. To ensure reliable twin-spot labeling with coupled MARCMs requires use of drivers that faithfully label the same lineage(s) in both systems. Third, the twin-spot generator 42 links site-specific recombination with reconstitution of two intact reporter genes that reside in trans and can be segregated by mitotic recombination into different daughter cells for twin-spot labeling. Since site-specific recombination could occur in non-mitotic cells and lead to co-expression of both reporters in heterozygous cells, genetic mosaic studies with twin-spot generator requires additional efforts to identify true twin spots, a non-trivial task in the complex CNS.
Figure 4. Twin-spot unique labeling techniques.
(A) Twin-spot MARCM involves two repressible marker genes (UAS-RFP vs. UAS-GFP) that are respectively silenced by a corresponding transgenic miRNA (UAS-RFPi vs. UAS-GFPi) in heterozygous cells. miRNAs (little Pacman eaters) repress the expression of reporters in heterozygous cells. Opposing de-repression of either marker gene can occur in the sister cells derived from a heterozygous precursor, when the miRNA transgenes residing distal to FRTs (orange triangles) are differentially lost.
(B) Twin-spot generator allows reconstitution of distinct reporter genes (GFP vs. RFP) following site-specific recombination, which can be segregated into distinct daughter cells for differential labeling. Note the heterozygous mother cell is shown in light yellow to indicate the fact that recombination without segregation could result in co-expression of both reporters in heterozygous cells.
(C) Coupled MARCM involves two independent pairs of transcriptional activators and repressors. GAL80 (GAL4 repressor) and QS (QF repressor) reside on the apposing homologous chromosome arms distal to FRTs. Mitotic recombination leads to respective loss of GAL80 and QS from the two distinct homozygous daughter cells that can be differentially labeled by GAL4-dependent UAS-GFP versus QF-dependent QUAS-RFP.
Sister-clone labeling with twin-spot MARCM in the Drosophila central brain shows the power of twin-spot positive labeling in comprehensive cell lineage analysis. It allows one to simultaneously detect the outcomes of both daughter cells derived from a common precursor and to unambiguously determine the sister relationships among distinct MARCM clones. Identifying the sister clone paired with a NB clone is essential for validating the asymmetric cell division of the NB 40, which yields a GMC in the conventional type I NB lineages and an intermediate neural progenitor (INP) in the eight type II NB lineages. One GMC may produce one or two adult neurons or none due to premature neuron loss. By contrast, an INP can deposit a short series of GMCs through a limited run of self-renewing asymmetric cell divisions and thus produce about 10 neurons following each type II NB asymmetric cell division 43,44,45. Based on the cell numbers of the accompanying NB clones, one can further arrange the serially derived GMC clones chronologically to determine the birth order for individual neurons in a group of clonally related GAL4-positive neurons (Fig. 5B–D). Next the sister neurons made by a given GMC can be unambiguously identified and independently traced through characterization of paired single-neuron clones (Fig. 5E). To ensure full coverage, one should employ drivers that label all the offspring in the lineage(s) of interest. The degree of coverage by a given driver can be initially assessed by comparing the extent of labeling between the GAL4 driver and a GAL80-repressible ubiquitous LexA driver in dual-expression-control MARCM (Fig. 3C) 46,47.
Two protracted AL neuronal lineages have been mapped with unprecedented single-neuron resolution by twin-spot MARCM (Fig. 5). The rather restrictive GAL4-acj6 driver permits full coverage of the anterodorsal AL (adAL) lineage. Despite the full coverage, adAL NB clones of various sizes consistently paired with a single-cell GMC clone, arguing that one of the two post-mitotic neurons made by each of the serially derived about 100 GMCs has died prematurely. Single neurons paired with larger NB clones were born prior to single neurons paired with smaller NB clones (Fig. 5B–D). Systematic analysis of such paired NB/GMC clones has revealed the production of 40 AL PN types in an invariant sequence from the adAL NB 48. Monitoring the reduction in the NB clone size as the lineage proceeds further demonstrates the different fixed cell counts of distinct AL PN types. Such stereotyped single-neuron development indicates that the NB keeps track of every cell division and confers specific temporal identity on each newborn neuron to ensure the production of an invariant set of neuron types with specific cell counts from the lineage.
Single-neuron lineage mapping of a more complicated AL lineage made by the lateral AL (lAL) NB unveils comparable yet more intriguing phenomena. Distinct from the adAL lineage, the lAL lineage consists of two distinct hemilineages as the sister neurons derived from each GMC, if survived into the adult stage, respectively develop into one projection neuron (PN) and one local interneuron (LN) which can be identified as paired single-neuron clones by twin-spot MARCM using a pan-neuronal driver (Fig. 5E). Characterizing numerous pairs of differentially labeled single-neuron clones sampled through larval development has revealed the presence of 48 distinct PN/LN pairs that arise sequentially from the lAL NB 49. Notably, PNs of the lAL lineage may connect the AL, the auditory/gravity-sensing center, or the gustatory station with various neuropils through distinct axonal tracks. By contrast, their paired LNs uniformly reside within the AL. The multi-neuropil PNs exhibit higher cellular diversity than the pure-AL LNs, such that a series of distinct PNs may arise sequentially with undistinguishable LNs from common GMCs. The opposite with serial production of distinct LNs and unchanged PNs could also occur and has been well noted in one occasion. All these phenomena indicate that the lAL PN and LN sister hemilineages are independently patterned to yield diverse PNs and LNs in distinct sequences. Taken together, diverse neurons in the Drosophila brain are specified based on cell lineages according to (1) lineage origin, (2) hemilineage identity, and (3) GMC birth order.
Genetic mosaic studies in the complex CNS are greatly enhanced by twin-spot MARCM, which permits unique labeling of not only mutant clones but also their paired wild-type sister clones. Visualizing the wild-type sister clone reveals where and when the mitotic recombination has occurred and therefore provides information about the prospective cell fate of the mutant clone of interest. This way, one can unambiguously detect temporal cell fate transformation involving adjacent neuron types in a rapidly changing neuronal lineage. For instance, the demonstration of the transformation of the 11th temporal fate of the adAL lineage to the next temporal fate upon loss of Krupple (Kr) requires the identification of the exact single-neuron clone made by the Kr mutant GMC that normally yields the 11th type of adAL PN 50 (Fig. 6). Validating the temporal requirement of Chinmo in the LN hemilineage of the lAL lineage also depends on the detection of a chinmo mutant LN with wrong temporal identity as judged from the temporal fate of its paired wild-type PN 49. Chinmo is required for proper temporal fate specification of the paired PN as well. But loss-of-Chinmo has transformed the PN to a PN derived from a much later-born GMC as compared to the transformation into the immediate next temporal fate on the LN side 49. These observations illustrate how sister neurons made by the same intermediate precursors may acquire distinct temporal cell fates through differential responses to common temporal fating factors (e.g. Chinmo). In sum, further sophisticated MARCM technologies have enabled the retrospective tracking of individual precursors and their developmental fates by clonal analysis. Moreover, all the clones obtained by MARCM can to some degree be genetically customized such that one can immediately study the molecular mechanisms governing any MARCM-enlightened developmental phenomena of interest.
Conclusions and Future Perspectives
Cell lineage analysis in Drosophila has been primarily carried out through clonal labeling that allows one to determine retrospectively the progeny derived from a common precursor. Clones generated through mitotic recombination are particularly informative in the characterization of dynamic and heterogeneous pools of precursors for multiple reasons. First, any clone regardless of the clone size, including a single-cell clone, can in theory be ascribed to some precursor that divided during the time of clone induction. Second, it allows independent tracking of sister cells, if properly marked, derived from the precursor that underwent mitotic recombination, providing the necessary sublineage information for documenting sister fate differences. Third, one can manipulate gene functions independently in the sister cells rendered homozygous for distinct homologous chromosome arms that carry different mutations and/or transgenes, for genetic mosaic studies of organism development based on cell lineages.
Over the course of ~20 years, both strategies of clone induction and methods of clone labeling have evolved extensively to meet experimental challenges. The introduction of FLP/FRT-mediated site-specific mitotic recombination has drastically improved the efficiency and the temporal or spatial control of clone induction. Mitotic recombination at defined genomic loci further guarantees the genotypes of the derived homozygous daughter cells, ensuring the linkage of all genetic elements distal to the FRT sites for reliable cell-autonomous marking of clones. The detection of homozygous sister clones within heterozygous organisms first involved marking clones with no expression or two-fold higher expression of a ubiquitous reporter than the background. MARCM was then established to allow unique positive labeling of only one of the paired sister clones, providing the necessary clarity for clonal analysis in the complex nervous system. Twin-spot MARCM ultimately granted the ability to label sister clones in distinct colors in an otherwise unstained background, enabling comprehensive lineage analysis of the CNS.
The contemporary FLP/FRT-based genetic mosaic system and numerous follow-up improvements were made possible due to the awesome power of fly genetics, the development of P element-mediated transgenesis, and the versatility in transgene expression control with the GAL4/UAS binary induction system. As newer sophisticated genetic/genomic/transgenic toolkits become available 51,52,53,54, we should establish even more powerful genetic mosaic and lineage tracing systems. Clearly there is room for optimization in the production as well as visualization of homozygous clones in heterozygous organisms. For example, we need combined and improved spatial and temporal control of FLP activity to target specific precursors so that the resolution of our lineage tracing and mosaic studies come ever closer to provide ‘real-time’ data about cell division and genetic requirements. In addition, we would love to determine and unambiguously trace independent clones that coexist and intermingle. Concurrent analysis of overlapping clones is essential for reconstruction of composite tissue development, to unveil possible developmental plasticity and intricate clone-clone interactions.
Acknowledgments
I thank Chris Zugates for critical reading of the manuscript and anonymous reviewers for inputs to improve both text and figures. Our cell lineage analysis of Drosophila central brain is primarily funded by Howard Hughes Medical Institute and National Institute of Health.
References
- 1.Blair SS. Genetic mosaic techniques for studying Drosophila development. Development. 2003;130:5065–5072. doi: 10.1242/dev.00774. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila, A laboratory handbook. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 3.Buckingham ME, Meilhac SM. Tracing cells for tracking cell lineage and clonal behavior. Dev Cell. 2011;21:394–409. doi: 10.1016/j.devcel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Morgan TH. Mosaics and gynandromorphs in Drosophila. Proc Soc Exp Biol Med. 1914;11:171–172. [Google Scholar]
- 5.Sturtevant AH. The claret mutant type of Drosophila simulans, a study of chromosome elimination and cell lineage. Z wiss Zool. 1929;135:323–356. [Google Scholar]
- 6.Garcia-Bellido A, Merriam JR. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool. 1969;170:61–75. doi: 10.1002/jez.1401700106. [DOI] [PubMed] [Google Scholar]
- 7.Hotta Y, Benzer S. Mapping of behaviour in Drosophila mosaics. Nature. 1972;240:527–535. doi: 10.1038/240527a0. [DOI] [PubMed] [Google Scholar]
- 8.Stern C. Somatic Crossing over and Segregation in Drosophila Melanogaster. Genetics. 1936;21:625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker HJ. Roentgen mosaic spots & defective mutations at the eye of Drosophila & the evolutive physiology of the eye. Z Indukt Abstamm Vererbungsl. 1957;88:333–373. [PubMed] [Google Scholar]
- 10.Bryant PJ, Schneiderman HA. Cell lineage, growth, and determination in the imaginal leg discs of Drosophila melanogaster. Dev Biol. 1969;20:263–290. doi: 10.1016/0012-1606(69)90015-3. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence PA, Johnston P. Observations on cell lineage of internal organs of Drosophila. J Embryol Exp Morphol. 1986;91:251–266. [PubMed] [Google Scholar]
- 12.Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalisation of the wing disk of Drosophila. Nat New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalization in the dorsal mesothoracic disc of Drosophila. Dev Biol. 1976;48:132–147. doi: 10.1016/0012-1606(76)90052-x. [DOI] [PubMed] [Google Scholar]
- 14.Crick FH, Lawrence PA. Compartments and polyclones in insect development. Science. 1975;189:340–347. doi: 10.1126/science.806966. [DOI] [PubMed] [Google Scholar]
- 15.Crozatier M, Glise B, Vincent A. Patterns in evolution: veins of the Drosophila wing. Trends Genet. 2004;20:498–505. doi: 10.1016/j.tig.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 17.Perrimon N, Gans M. Clonal analysis of the tissue specificity of recessive female-sterile mutations of Drosophila melanogaster using a dominant female-sterile mutation Fs(1)K1237. Dev Biol. 1983;100:365–373. doi: 10.1016/0012-1606(83)90231-2. [DOI] [PubMed] [Google Scholar]
- 18.Perrimon N. Clonal Analysis of Dominant Female-Sterile, Germline-Dependent Mutations in DROSOPHILA MELANOGASTER. Genetics. 1984;108:927–939. doi: 10.1093/genetics/108.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 20.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 21.Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 22.Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 24.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 26.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Rubin GM. The effort to make mosaic analysis a household tool. Development. 2012;139:4501–4503. doi: 10.1242/dev.085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 29.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 30.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 31.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 32.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 33.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 34.Zhu S, Chiang AS, Lee T. Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development. 2003;130:2603–2610. doi: 10.1242/dev.00466. [DOI] [PubMed] [Google Scholar]
- 35.Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 38.Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffin R, Sustar A, Bonvin M, Binari R, del Valle Rodriguez A, Hohl AM, Bateman JR, Villalta C, Heffern E, Grunwald D, et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods. 2009;6:600–602. doi: 10.1038/nmeth.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 47.Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- 48.Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 2012;10:e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nern A, Pfeiffer BD, Svoboda K, Rubin GM. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc Natl Acad Sci U S A. 2011;108:14198–14203. doi: 10.1073/pnas.1111704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venken KJ, Bellen HJ. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and PhiC31 integrase. Methods Mol Biol. 2012;859:203–228. doi: 10.1007/978-1-61779-603-6_12. [DOI] [PubMed] [Google Scholar]