Abstract
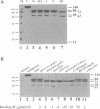
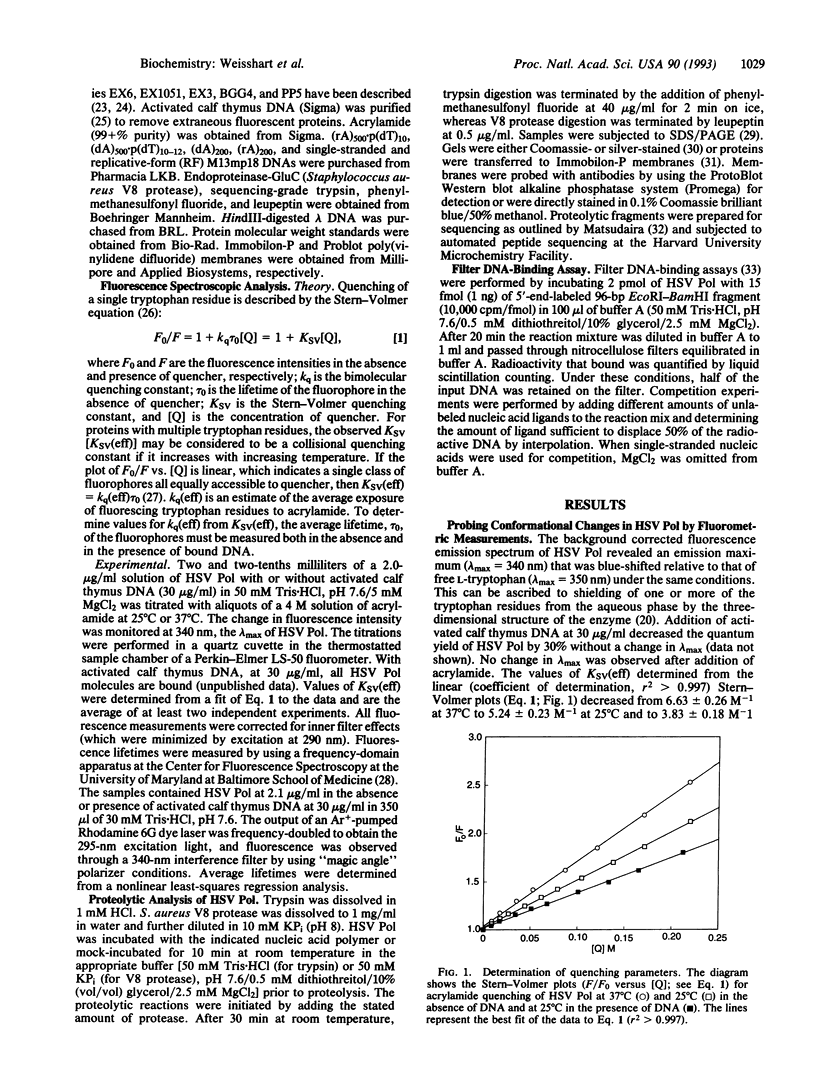
Herpesvirus DNA polymerases are prototypes for alpha-like DNA polymerases and important targets for antiherpesvirus drugs. We have investigated changes in the catalytic subunit of herpes simplex virus DNA polymerase following DNA binding by using the techniques of endogeneous fluorescence quenching and limited proteolysis. The fluorescence studies revealed a reduction in the rate of quenching by acrylamide in the presence of DNA without changes in the wavelength of the emission peak or in the lifetime of the fluorophore, consistent with the possibility of conformational changes. Strikingly, the proteolysis studies revealed that binding to a variety of natural and synthetic DNA and RNA molecules induced the appearance of a new cleavage site for trypsin near residue 1060 of the protein and increased cleavage by trypsin near the center of the protein. The extent of these cleavages correlated with the affinity of the polymerase for these ligands. These data provide strong evidence that binding to nucleic acid polymers induces substantial localized conformational changes in the polymerase. The locations of enhanced tryptic cleavage near sites implicated in substrate recognition and interaction with a processivity factor suggest that the conformational changes are important for catalysis and processivity of this prototype alpha-like DNA polymerase. Inhibition of these changes may provide a mechanism for antiherpesvirus drugs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Catalano C. E., Allen D. J., Benkovic S. J. Interaction of Escherichia coli DNA polymerase I with azidoDNA and fluorescent DNA probes: identification of protein-DNA contacts. Biochemistry. 1990 Apr 17;29(15):3612–3621. doi: 10.1021/bi00467a004. [DOI] [PubMed] [Google Scholar]
- Catalano C. E., Benkovic S. J. Inactivation of DNA polymerase I (Klenow fragment) by adenosine 2',3'-epoxide 5'-triphosphate: evidence for the formation of a tight-binding inhibitor. Biochemistry. 1989 May 16;28(10):4374–4382. doi: 10.1021/bi00436a038. [DOI] [PubMed] [Google Scholar]
- Chen M., Horwitz M. S. Dissection of functional domains of adenovirus DNA polymerase by linker-insertion mutagenesis. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6116–6120. doi: 10.1073/pnas.86.16.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri N. Conformational adaptability in enzymes. Adv Enzymol Relat Areas Mol Biol. 1973;37:397–648. doi: 10.1002/9780470122822.ch7. [DOI] [PubMed] [Google Scholar]
- Coen D. M. The implications of resistance to antiviral agents for herpesvirus drug targets and drug therapy. Antiviral Res. 1991 May;15(4):287–300. doi: 10.1016/0166-3542(91)90010-o. [DOI] [PubMed] [Google Scholar]
- Cowart M., Gibson K. J., Allen D. J., Benkovic S. J. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry. 1989 Mar 7;28(5):1975–1983. doi: 10.1021/bi00431a004. [DOI] [PubMed] [Google Scholar]
- Crute J. J., Lehman I. R. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5'----3' exonuclease with ribonuclease H activity. J Biol Chem. 1989 Nov 15;264(32):19266–19270. [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Bebrin W. R., Weisshart K., Coen D. M. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J Virol. 1993 Jan;67(1):398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Coen D. M. A novel functional domain of an alpha-like DNA polymerase. The binding site on the herpes simplex virus polymerase for the viral UL42 protein. J Biol Chem. 1990 Oct 15;265(29):17393–17396. [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976 Feb 10;15(3):672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Ollis D. L., Steitz T. A., Joyce C. M. A domain of the Klenow fragment of Escherichia coli DNA polymerase I has polymerase but no exonuclease activity. Proteins. 1986 Sep;1(1):66–73. doi: 10.1002/prot.340010111. [DOI] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Bastow K. F., Cheng Y. C., Coen D. M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Hall J. D., Mount D. W., Retondo M. J., Weller S. K., Coen D. M. Sequence and mapping analyses of the herpes simplex virus DNA polymerase gene predict a C-terminal substrate binding domain. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7969–7973. doi: 10.1073/pnas.82.23.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S., Weisshart K., Digard P., deBruynKops A., Knipe D. M., Coen D. M. Polymerization activity of an alpha-like DNA polymerase requires a conserved 3'-5' exonuclease active site. Mol Cell Biol. 1991 Sep;11(9):4786–4795. doi: 10.1128/mcb.11.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990 Dec;64(12):5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffey M. L., Novotny J., Bruccoleri R. E., Carroll R. D., Stevens J. T., Matthews J. T. Structure-function studies of the herpes simplex virus type 1 DNA polymerase. J Virol. 1990 Oct;64(10):5008–5018. doi: 10.1128/jvi.64.10.5008-5018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991 Aug 11;19(15):4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kuchta R. D., Benkovic P., Benkovic S. J. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988 Sep 6;27(18):6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marcy A. I., Olivo P. D., Challberg M. D., Coen D. M. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 1990 Mar 11;18(5):1207–1215. doi: 10.1093/nar/18.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Yager D. R., Coen D. M. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J Virol. 1990 May;64(5):2208–2216. doi: 10.1128/jvi.64.5.2208-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Philips A. V., Coleman M. S., Maskos K., Barkley M. D. Time-resolved fluorescence spectroscopy of human adenosine deaminase: effects of enzyme inhibitors on protein conformation. Biochemistry. 1989 Mar 7;28(5):2040–2050. doi: 10.1021/bi00431a012. [DOI] [PubMed] [Google Scholar]
- Quinn J. P., McGeoch D. J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985 Nov 25;13(22):8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wang Y. S., Woodward S., Hall J. D. Use of suppressor analysis to identify DNA polymerase mutations in herpes simplex virus which affect deoxynucleoside triphosphate substrate specificity. J Virol. 1992 Mar;66(3):1814–1816. doi: 10.1128/jvi.66.3.1814-1816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K., Knopf C. W. The herpes simplex virus type I DNA polymerase. Polypeptide structure and antigenic domains. Eur J Biochem. 1988 Jul 1;174(4):707–716. doi: 10.1111/j.1432-1033.1988.tb14155.x. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Yager D. R., Marcy A. I., Coen D. M. Translational regulation of herpes simplex virus DNA polymerase. J Virol. 1990 May;64(5):2217–2225. doi: 10.1128/jvi.64.5.2217-2225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]