Abstract
Background
Echinostomes are cosmopolitan digenean parasites which infect many different warm-blooded hosts. Their classification is extremely confused; the host spectrum is wide, and morphological similarities often result in misidentification. During our long-term studies on the helminth fauna of rodents and carnivores we have collected 27 collar-spined echinostomes which differ in morphology to an extent that suggests the presence of more than one species. Here, we describe this material, and the extent of host-related variation in this parasite.
Methods
Specimens of Isthmiophora isolated from four host species (badger, American mink, hedgehog, striped field mouse) were subject to morphological and molecular examination; the data were statistically analysed.
Results
Our results show that genetically all the Isthmiophora specimens obtained from all the examined hosts are conspecific and represent I. melis. On the other hand, the individuals isolated from Apodemus agrarius are morphologically distinct and, based on this criterion alone, should be described as a new species.
Conclusions
The morphological traits of Isthmiophora melis are much variable and host-dependent; without molecular analysis they would suggest a necessity to describe a new species or even genus. Such a high level of intraspecific variability may be affected by the host’s longevity.
Keywords: Isthmiophora melis, Rodents, Phenotypic plasticity, Molecular taxonomy
Background
Since molecular techniques became commonly used in taxonomic studies, the list of valid taxa in different groups of organisms has been changing, and in many cases the results of molecular investigations are radically different from those obtained with classical methods. However, while museum collections dating from the pre-molecular period remain the cornerstone of taxonomy, morphology must continue to provide a starting point for molecular studies [1]. Molecular taxonomy has also contributed to revealing the common occurrence of cryptic species in nature, in virtually all major taxa. Although such species are genetically distinct from each other, they are morphologically very similar [2, 3].
On the other hand, free-living organisms and parasites can adjust their life-history strategies and a given genotype may produce a variety of phenotypes under different environmental conditions [4]. Due to their exposure to widely differing environmental conditions (i.e. different host species, host’s immune system), parasites often display a phenotypic plasticity which is expressed as differences in body size or fecundity [4]. In the case of Digenea, most species-diagnostic features are the body proportions or the shape and location of internal organs. Phenotypic variation may be induced by differences in the intensity of infection (“crowding effect”) and in the host’s identity (“host-induced variation”) [5]. These phenotypic effects may lead to species-specific variation resulting in misidentification [6].
Echinostomes are cosmopolitan digenean parasites which mainly infect many different warm-blooded hosts [7]. The taxonomy and classification of the echinostomes is highly confused. The wide host spectrum of echinostomes is a result of phylogenetic, physiological, and ecological adjustments between the parasite and the host in a dynamic evolutionary process, where the main factor influencing the host specificity is the host’s behaviour, particularly the feeding habits of vertebrate hosts [7]. Species misidentifications have arisen because of similarities in morphology and because of the lack of isolates in molecular databases [8]. Most studies have focused on the genus Echinostoma, especially the “revolutum” group, e.g. [7–14], see Kostadinova and Gibson [11] for review. Despite numerous studies, two recent papers [15, 16] showing that even by the use of molecular tools the taxonomy of this group is still not straightforward. However, taxonomic difficulties are also known in other groups of the Echinostomatinae [17, 18]. One of the remaining interesting issues concerns the 27-collar-spined echinostomes of the genus Isthmiophora Luhe, 1909. The type-species, I. melis, is a parasite reported mainly from European, Asian and American carnivores. However, Radev et al. [19], in his review of literature, lists c. 30 species as definitive hosts of this parasite. Host-induced morphological variation within this digenean, which apparently lacks host specificity, should be clearly visible.
During our long-term studies on the helminth fauna of rodents and carnivores we have collected 27 collar-spined echinostomes which differ in morphology to an extent suggesting the presence of more than one species. Molecular studies, on the other hand, suggested that these worms belong to Isthmiophora. Here we describe this material, and the extent of host-related variation in this parasite.
Methods
Parasite sampling
Representatives (N = 148) of Isthmiophora used for the morphological analysis were collected from four host species: striped field mouse (Apodemus agrarius, N = 37), European badger (Meles meles, N = 13), American mink (Neovison vison, N = 64) and European hedgehog (Erinaceus europaeus, N = 34), all captured during parasitological and faunistic studies carried out by the Department of Parasitology in cooperation with the Polish Academy of Sciences. The rodents were captured in Lower Silesia (Dolina Baryczy, Nature Reserve “Stawy Milickie”, 51°31′56″N/17°20′12″E) in 2010 – permission 46/2008 issued by the Second Local Commission for Animal Experiments, worms form the mink (N. Poland; Marzęcino, 54°13′1.54″N 19°13′20.43″E) captured in 2010 were obtained from the Polish Academy of Sciences, trematodes from the hedgehog and badger (2010) were obtained from the Czech Republic (Zahlinice, 49°17′06″N/17°28′41″E). After washing in tap water, the worms were fixed in 70 % ethanol. Some of the collected trematodes were stained in iron-aceto-carmine [20], dehydrated in a graded ethanol series, cleared in clove oil, mounted in Canada balsam and identified according to Kostadinova and Gibson [17]. The voucher specimens of trematodes obtained from each hostare deposited in the polish helminthological collection of Natural History Museum of Wroclaw University (MNHW).
Statistical analysis
All the examined specimens of I. melis were subject to detailed morphological and morphometric analysis, including the following measurements: body length (L), maximum body width (W), body area (BA), maximum body width as a proportion of body length (BW), forebody length (FB), forebody as a proportion of body length (FO), hindbody length (HB), hindbody as a proportion of body length (H), post-testicular region length (PTR), post-testicular region length as a proportion of body length (T), oral sucker area (OSA), ventral sucker area (VSA), anterior testis area (ATA), posterior testis area (PTA), ovary area (OA), gonad area/body area (GA/BA), ventral sucker to ovary distance as a proportion of body length (U), egg length (EL), egg width (EW). The body and gonad areas were calculated using the following equations: body area = π*(body length/2)*(body widith/2); gonads area = π*r2. All the measurements were expressed in micrometers and proportions as percentage. Prior to the analysis the data were log-transformed (log10). The mean (M), minimum/maximum values and coefficients of variation (CV %; defined as the ratio of standard deviation to the mean) were calculated for all the variables. One-way analysis of variance (ANOVA) was carried out to test if the particular morphological features of Isthmiophora differed between the host species. In the next step we performed discriminant analysis. To avoid the size effect of the worms (Isthmiophora spp. isolated from the badger was much bigger than those from the other hosts) only variables expressed as ratios (BW, FO, H, T, U, GA/BA) were included in this analysis. Moreover, according to the literature data, the major diagnostic characters in this taxon are based on ratios (i.e. BW, FO, T and U). All the analyses were conducted using Statistica 10.0 software.
Molecular analysis
Molecular analysis was performed for I. melis collected from four host species studied, from which a set of two worms was used for the analysis (N = 8). DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen), and amplified using PCR specific for 2 nuclear markers (internal transcribed spacers 1 and 2 [ITS1, ITS2] and a fragment of mitochondrial cytochrome oxidase I [CO1] gene) (Table 1). Two additional molecular markers (SSU and LSU of rDNA) were amplified for the specimens from A. agrarius. PCR conditions included initial denaturation in 95 °C for 5 min, followed by 35 cycles: 45 s denaturation (95 °C), 30 s annealing (52 °C for SSU, LSU, ITS 1, ITS 2 and 48 °C for COI), 30 s elongation (72°), and a 5 min step of final elongation (72 °C). PCR products were sequenced using the same primer pairs, and chromatograms inspected visually for ambiguities. In order to elucidate any homologies with the previously deposited sequences in GenBank, we conducted a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD = Web&PAGE_TYPE = BlastHome). Multiple alignment was done using CLUSTAL W in MEGA 5.0 package [21]. The sequences obtained in this study were deposited in GenBank under the following accession numbers: [GenBank: KT359582] and [GenBank: KT359583] for SSU and LSU; [GenBank: KT359584] for ITS complex; [GenBank: KT359580] and [GenBank: KT359581] for COI (Table 1).
Table 1.
The list of host species used for molecular identification of I. melis with the Gen Bank accession numbers of newly obtained sequences
Results
Molecular analysis
The morphological distinctness of I. melis from the striped field mouse did not permit unambiguous identification of the parasite to specific or even generic level. Two markers 18S rDNA (1078 bp) and 28S rDNA (1352 bp) were therefore used for preliminary identification. BLAST analysis showed 99 % similarity with the sequences of I. melis [GenBank: AY222131] and [GenBank: AF151941] and I. hortensis [GenBank: AB189982] for both loci. Amplification of COI from the four host species generated sequences of 222–261 bp. Two haplotypes were observed, one for the sequences from N. vison and another for the sequences from A. agrarius, M. meles and E. europaeus. The overall variation between these haplotypes amounted to 1.4 % (3 nucleotides out of 219). In the case of ITS, amplification and sequencing generated four sequences of 1029–1042 bp. However, a 1014 bp alignment revealed that all the sequences from each host species were identical.
Morphological analysis
The means, ranges and CV % values of I. melis from the four host species are shown in Table 2. The observed values of the analysed parameters demonstrate a high level of both intra- and between-host variation in the morphometric characters of the worms. The CV % values calculated for the variables expressed as ratios (BW, FO, H, T, U, GA/BA) were at the same level and did not show any statistically significant differences between the host species (F = 0.01; df = 3; p = 0.998). However, one-way analysis of variance (ANOVA) showed that host species played an important role in shaping the characteristics of I. melis (F = 20.3; df = 51; p < 0.001). The results of post hoc Tukey test showed that the differences were mostly associated with the trematodes from A. agrarius (Table 3). These specimens were characterised by a relatively smaller body size, higher values of maximum body width, expressed as proportion of body length (BW), a very short post-testicular region and therefore low values of the post-testicular region as a proportion of body length (T) (Fig. 1). The worms from A. agrarius displayed the highest values of relative gonad area/body area (GA/BA) (Fig. 1). In discriminant analysis (Table 4) the model was generated by the use of 6 variables. The chi-square test showed that the first three roots were required to separate I. melis among the four host species. The roots accounted for 91.5 % (Root 1), 96.6 % (Root 2) and 100 % (Root 3) of the overall variation. Root 1 separated the worms from A. agrarius based on the following variables, in order of descending importance: GA/BA, T, FO and U. The analysis also revealed that according to these criteria all the specimens of I. melis from A. agrarius were classified correctly (Table 5). Roots 2 and 3 separated trematodes from M. meles based on GA/BA and U, however this explained only 8.5 % of the variation. These results are also visible in the plot of canonical scores (Fig. 2) where I. melis from the striped field mouse are clearly separated from those isolated from the remaining hosts.
Table 2.
Morphology of Isthmiophora melis from various hosts obtained in this study
| Apodemus agrarius (N = 37) | Erinaceus europaeus (N = 34) | Neovison vison (N = 64) | Meles meles (N = 13) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | Range | CV % | M | Range | CV % | M | Range | CV % | M | Range | CV % | |
| L | 2,625 | 1,075–4,100 | 32 | 4,476 | 3,070–5,675 | 15 | 3,778 | 2,350–5,975 | 19 | 6,821 | 5,950–7,725 | 8 |
| W | 623 | 260–1,050 | 34 | 876 | 590–1,160 | 14 | 762 | 500–1,250 | 25 | 1,389 | 1,250–1,700 | 10 |
| BA | 1,411 | 219–2,967 | 57 | 3,113 | 1,422–4,344 | 24 | 2,356 | 1,021–5,863 | 45 | 7,468 | 5,838–10,309 | 16 |
| BW | 24 | 19–30 | 11 | 20 | 16–26 | 14 | 20 | 15–25 | 12 | 20 | 17–23 | 8 |
| FB | 665 | 315–950 | 26 | 904 | 630–1,030 | 10 | 754 | 530–1,090 | 16 | 1,083 | 960–1,210 | 8 |
| FO | 26 | 20–32 | 11 | 21 | 18–33 | 15 | 20 | 15–27 | 12 | 17 | 15–18 | 7 |
| HB | 1,558 | 550–2,750 | 39 | 2,997 | 1,900–3,925 | 19 | 2,487 | 1,680–4,050 | 21 | 4,613 | 4,225–4,940 | 6 |
| H | 58 | 31–69 | 12 | 67 | 54–83 | 7 | 66 | 47–80 | 8 | 71 | 69–72 | 2 |
| PTR | 692 | 293–1,225 | 33 | 1,640 | 1,010–2,060 | 18 | 1,486 | 1,000–2,300 | 19 | 2,570 | 2,100–3,025 | 11 |
| T | 27 | 20–32 | 11 | 37 | 31–46 | 8 | 39 | 34–46 | 7 | 38 | 35–41 | 5 |
| OSA | 22,566 | 8,247–53,066 | 42 | 35,747 | 22,687–43,352 | 12 | 26,897 | 15,386–57,227 | 33 | 57,060 | 46,163–70,650 | 14 |
| VSA | 127,434 | 24,732–277,910 | 49 | 266,453 | 79,133–468,454 | 29 | 185,377 | 54,091–515,036 | 51 | 508,892 | 424,077–653,635 | 15 |
| ATA | 118,293 | 36,287–250,592 | 53 | 152,747 | 37,994–237,463 | 32 | 93,766 | 35,448–248,379 | 57 | 447,761 | 110,391–671,666 | 35 |
| PTA | 129,210 | 28,339–296,907 | 53 | 167,942 | 37,994–270,948 | 28 | 104,482 | 39,740–259,541 | 60 | 475,179 | 186,560–671,665 | 31 |
| OA | 26,674 | 3,190–53,066 | 55 | 30,758 | 13,523–61,544 | 31 | 21,441 | 7,850–68,315 | 61 | 84,499 | 70,650–93,435 | 9 |
| GA/BA | 158 | 99–267 | 23 | 112 | 63–142 | 15 | 90 | 67–129 | 18 | 148 | 56–192 | 30 |
| U | 4 | 1–8 | 36 | 4 | 1–7 | 40 | 3 | 1–5 | 40 | 4 | 3–9 | 50 |
| EL | 131 | 120–140 | 5 | 129 | 120–140 | 5 | 121 | 115–125 | 4 | 127 | 115–140 | 8 |
| EW | 81 | 70–90 | 8 | 79 | 75–85 | 5 | 89 | 80–95 | 5 | 82 | 75–90 | 7 |
All measurements are expressed in micrometers; M mean, Min minimal value, max maximal value, CV % coefficient of variation; L-body length, W-maximum body width, BA-body area, BW-maximum body width as a proportion of body length, FB-forebody length, FO-forebody as a proportion of body-length, HB-hindbody length, H-hindbody as a proportion of body length, PTR-post-testicular region length, T-post-testicular region length as a proportion of body length, OSA-oral sucker area, VSA-ventral sucker area, ATA-anterior testis area, PTA-posterior testis area, OA-ovary area, GA/BA-gonad area/body area, U-ventral sucker to ovary distance as a proportion of body length, EL-egg length, EW-egg width
Table 3.
Results of post hoc Tukey test of one-way analysis of variance (ANOVA)
| Aa/Ee | Aa/Nv | Aa/Mm | Ee/Nv | Ee/Mm | Nv/Mm | |
|---|---|---|---|---|---|---|
| L | + | + | + | + | + | + |
| W | + | n/s | + | + | + | + |
| BA | + | + | + | + | + | + |
| BW | + | + | + | n/s | n/s | n/s |
| FB | + | n/s | + | + | + | + |
| FO | + | + | + | n/s | + | + |
| HB | + | + | + | + | + | + |
| H | + | + | + | n/s | n/s | n/s |
| PTR | + | + | + | n/s | + | + |
| T | + | + | + | + | n/s | n/s |
| OSA | + | n/s | + | + | + | + |
| VSA | + | n/s | + | + | + | + |
| ATA | n/s | n/s | + | + | + | + |
| PTA | n/s | n/s | + | + | + | + |
| OA | n/s | n/s | + | + | + | + |
| U | n/s | + | n/s | + | n/s | n/s |
| GA/BA | + | + | n/s | + | + | + |
The data are presented pairwise for particular host species (Aa – A. agrarius, Ee – E. europaeus, Nv – N. vison, Mm – M. meles) and indicate statistical significance (+) or its lack (n/s)
Fig. 1.
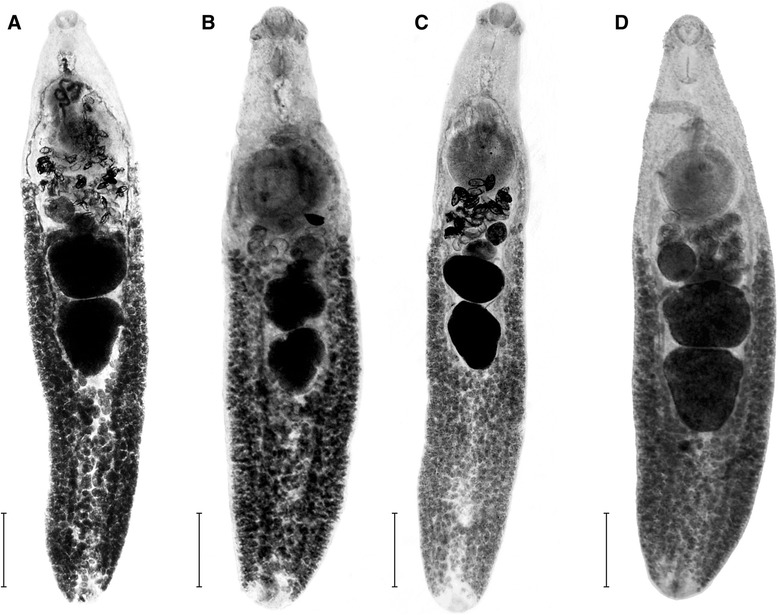
Morphology and body proportions of Isthmiophora melis. a – M. meles, scale bar – 1 mm; b – N. vison, scale bar – 0.5 mm; c – E. europaeus, scale bar – 0.6 mm; d – A. agrarius, scale bar – 0.3 mm
Table 4.
Summary of DFA; the table presents the full list of variables included in the analysis
| Roots removed | Eigenvalue | Canonical R | Wilks’ lambda | Chi-square | df | p-value |
|---|---|---|---|---|---|---|
| 0 | 5.207 | 0.916 | 0.104 | 259.857 | 18 | < 0.001 |
| 1 | 0.298 | 0.479 | 0.648 | 49.909 | 10 | < 0.001 |
| 2 | 0.189 | 0.399 | 0.841 | 19.931 | 4 | < 0.001 |
| Wilks’ lambda | Partial lambda | p-value | Root 1 | Root 2 | Root 3 | |
| BW | 0.108 | 0.966 | 0.274 | −0.001 | 0.079 | 0.487 |
| FO | 0.132 | 0.794 | < 0.001* | −0.501 | −0.307 | −0.698 |
| H | 0.111 | 0.944 | 0.093 | 0.282 | 0.201 | 0.079 |
| T | 0.123 | 0.854 | < 0.001* | 0.483 | 0.275 | −0.460 |
| U | 0.127 | 0.824 | < 0.001* | −0.334 | 0.230 | −0.955 |
| GA/BA | 0.137 | 0.759 | < 0.001* | −0.541 | 0.718 | −0.063 |
| Eigenvalue | 5.207 | 0.298 | 0.190 | |||
| Cumulative proportion | 0.915 | 0.966 | 1.000 | |||
Statistically significant variables are marked with asterisk (*). Chi-square tests with successive roots removed are presented in the upper part of the table. Columns Root 1, Root 2 and Root 3 present standardized coefficients for canonical variables
Table 5.
Classification efficiency of Isthmiophora melis from each host species
| % correct class. | M. vison (p = 0.422) | E. europaeus (p = 0.273) | M. meles (p = 0.057) | A. agrarius (p = 0.248) | Root 1 | Root 2 | Root 3 | |
|---|---|---|---|---|---|---|---|---|
| M. vison | 92.2 | 47 | 4 | 0 | 0 | 1.825 | −0.353 | 0.225 |
| E. europaeus | 72.8 | 8 | 24 | 1 | 0 | 0.364 | 0.256 | −0.664 |
| M. meles | 71.4 | 2 | 0 | 5 | 0 | 1.132 | 1.919 | 0.769 |
| A. agrarius | 100 | 0 | 0 | 0 | 30 | −3.768 | −0.129 | 0.168 |
| Total | 87.6 | 57 | 28 | 6 | 30 | |||
Columns Root 1, Root 2 and Root 3 reflecting the means of canonical values
Fig. 2.
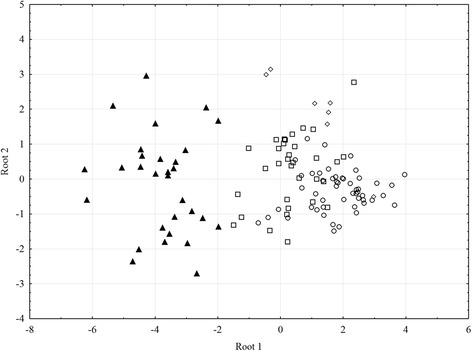
Results of canonical analysis of Isthmiophora melis obtained from four host species. Plot generated based on 6 variables measured in 148 specimens. Symbols denoting host species: circles – N. vison, squares – E. europaeus, diamonds – M. meles, black triangles – A. agrarius
Discussion
The history of the genus Isthmiophora Luhe, 1909, especially in relation to the genus Euparyphium Dietz, 1909, is long and complicated, but both genera were established as valid by Kostadinova and Gibson [17]. The main characteristic features of Isthmiophora are: anterior position of the testes (proportion of length of post-testicular region to body length = 30–50 %), short forebody (FO = 10–20 %), presence of an armed cirrus, small head collar with 27 collar spines, varied size of dorsal spines (oral longer than aboral), short uterus and large eggs [17]. The life cycle of Isthmiophora includes lymnaeid snails, tadpoles and fish as intermediate hosts and carnivores as definitive hosts. Six species (I. melis, I. hortensis (= Echinostoma hortense), I. beaveri, I. citellicola, I. inermis, I. lukjanovi) are currently regarded as valid [17]. I. melis is widespread in Europe, Asia and North America and uses more than 30 species of vertebrates as definitive hosts [19], including humans and rodents: Apodemus agrarius, A. sylvaticus, Rattus norvegicus and Mus musculus [22–24]. In Poland the species has been reported from fox, marten, badger, hedgehog and rodents [22, 24].
The specimens of I. melis from A. agrarius collected in Lower Silesia did not fully correspond to the description of I. melis [17], and two of the key features: forebody as a proportion of body length (FO) and post-testicular field as a proportion of body length (T), were distinct. According to Kostadinova [18], Isthmiophora possessed an intestinal bifurcation just anterior to the ventral sucker, the cirrus was armed and T = 30–50 % while Euparyphium was characterised by the intestinal bifurcation located halfway between the pharynx and the ventral sucker, unarmed cirrus and T = 20–30 %. The worms from A. agrarius had a short post-testicular field as a proportion of body length (T = 26.6 %), an armed cirrus and the intestinal bifurcation situated halfway between the pharynx and the ventral sucker. These features did not permit unambiguous identification of the trematodes as Isthmiophora. Additionally, Radev et al. [19] showed that in experimental infections of hamsters, specimens of I. melis still corresponded to the general description of the species, and the key features did not change significantly. Molecular identification, based on SSU and LSU of rDNA, of I. melis from the striped field mouse definitively confirmed their identity as Isthmiophora, while the less conservative markers (ITS1/ITS2 of rDNA and COI of mtDNA) pointed to a specific identity as I. melis. Additional specimens of I. melis, isolated from different hosts (M. meles, N. vison and E. europaeus), shared this molecular identity, with minimal (1.4 %) variation within the COI gene. Based on this molecular analysis, we must conclude that the echinostomatids collected from A. agrarius did represent I. melis. Nolan and Cribb [6] presented an extensive discussion of the role of ITS sequences in digenean taxonomy. Internal transcribed spacers in this group in general showed a small intraspecific variation, which was however sufficient to explore the validity of species boundaries in the group [6]. Morgan and Blair [12] also investigated the taxonomic position of eight 37-collar-spined echinostomatid species using ITS sequences and found that these spacer regions provided sufficient variation to distinguish 5 of the 8 nominal species examined, and the level of interspecific variation ranged between 1.1 % and 19.2 %. The remaining three species had identical ITS sequences and were indistinguishable. The same authors [13] also re-examined the same material using mitochondrial markers (CO1 and ND1), which allowed for unambiguous identification of the analysed material. Based on the reliability of the combination of nuclear and mitochondrial markers in these studies [6, 12, 13], we are confident that our specimens from A. agrarius do indeed represent I. melis.
The observed morphological variation must therefore be host-induced phenotypic variation, and its scale affects the diagnostic features at both generic and specific level. There is an extensive literature on the influence of population density on the echinostome morphology (e.g. the “crowding effect” of Fried and Freeborne [25, 26]), but we suspect an effect of host longevity as well. In general, the lifespan of carnivorous species is considerably longer than that of small rodents. The lifespan of A. agrarius in the wild approximates a few months (5–8) only, while, for example, the lifespan of M. meles is up to 15 years. The growth of body size and internal organs of trematodes is correlated at the initial phase. At a later period, when the gonads are fully developed, the body continues to grow with a simultaneous slower growth rate of gonads. For example, in experimental studies on the development of E. revolutum, Franco et al. [27] observed that gonads were fully developed 20–25 days post infection while full body size was only attained 55 days post infection. In our studies the highest values for the coefficient of relative gonad area to body area was observed in the trematodes from A. agrarius, indicating that the growth of the body had ceased; the values of this coefficient in the striped field mouse were almost identical as those observed in the badger – the type host for I. melis.
Genetic markers constitute a powerful tool in the studies on intraspecific variation in many taxa, including helminths, but the morphology still plays a crucial part in species descriptions [28]. However it is evident that morphology alone may not provide adequate taxonomic resolution and may lead to misidentifications. The phenotypic plasticity of helminths has been reported frequently in the literature, e.g. [29–32]. For example, Boyce et al. [30] explain the differences in the morphology of Notocotylus malhamensis Boyce et al. 2012 as a result of the presence of young adults in one of the hosts, i.e. the specimens of N. malhamensis in Microtus agrestis have not fully developed. The second possible reason of host-induced morphological differences in N. malhamensis is crowding effect. The wide host range of I. melis combined with the very different sizes of the hosts (e.g. badger vs. field striped mouse) makes the phenotypic plasticity even more spectacular. Our studies suggest that species identification is very subjective and, when descriptions of new species or even higher taxa are based on few specimens, misidentification is very likely. Thus the combination of morphology with molecular analysis and studies on life histories is most desirable when identifying parasites.
Conclusions
The morphological traits of Isthmiophora melis are highly variable and host-dependent, and without molecular analysis they might lead to a description of a new species or even genus. Such a high level of intraspecific variation may be affected by the host’s longevity.
Acknowledgements
Project supported by the Wrocław Centre of Biotechnology, programme The Leading National Research Centre (KNOW) for 2014–2018. The study was partially supported also by the National Science Centre, Poland, project no. N303 580939.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Molecular analyses carried out by GZ, JH and ZL; statistical analysis by GZ. Morphological work and collection of material by JH, MA and GZ. MS drafted by GZ with help from JH. All authors have read and approved the final manuscript.
Contributor Information
Joanna Hildebrand, Email: joanna.hildebrand@uwr.edu.pl.
Maja Adamczyk, Email: maja.adamczyk@uni.wroc.pl.
Zdzisław Laskowski, Email: laskowz@twarda.pan.pl.
Grzegorz Zaleśny, Email: grzegorz.zalesny@up.wroc.pl.
References
- 1.Perez Ponce De Leon G. Host-induced morphological variability in adult Posthodiplostomum mimimum (Digenea: Neodiplostomidae) J Parasitol. 1995;81:118–120. doi: 10.2307/3284022. [DOI] [PubMed] [Google Scholar]
- 2.Miura O, Kuris AM, Torchin ME, Hechinger RF, Dunham EJ, Chiba DS. Molecular-genetic analyses reveal cryptic species of trematodes in the intertidal gastropod, Batillaria cumingi (Crosse) Int J Parasitol. 2005;35:793–801. doi: 10.1016/j.ijpara.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Georgieva A, Selbach C, Faltynkova A, Soldanova A, Sures B, Skrinisson K, et al. New Cryptospecies of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) reveled by molecular and morphological data. Parasites Vectors. 2013;6:64. [DOI] [PMC free article] [PubMed]
- 4.Poulin R. Evolutionary Ecology of Parasites. UK: Princeton University Press; 2007. [Google Scholar]
- 5.Stunkard HW. Intraspecific variation in parasitic flatworms. Syst Zool. 1957;6:7–18. doi: 10.2307/2411703. [DOI] [Google Scholar]
- 6.Nolan MJ, Cribb TH. The use and implications of ribosomal DNA sequencing for the discrimination of digenean species. Adv Parasitol. 2005;60:102–63. [DOI] [PubMed]
- 7.Toledo R, Esteban J-G, Fried B. Chapter 3. Recent advances in the biology of echinostomes. Adv Parasitol. 2009;69:147–204. doi: 10.1016/S0065-308X(09)69003-5. [DOI] [PubMed] [Google Scholar]
- 8.Kostadinova A, Herniou EA, Barrett J, Littlewood DTJ. Phylogenetic relationships of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) and related genera re-assessed via DNA and morphological analyses. Syst Parasitol. 2003;54:159–176. doi: 10.1023/A:1022681123340. [DOI] [PubMed] [Google Scholar]
- 9.Munfioz-Antoli C, Carpena I, Espert A, Esteban JG, Toledo R. The effect of host species on the development of Echinostoma friedi (Trematoda: Echinostomatidae) adult worms. Rev IbeÏrica Parasitol. 2004;64:81–7. [Google Scholar]
- 10.Kostadinova A, Gibson DI, Biserkov V, Ivanova R. A quantitative approach to the evaluation of the morphological variability of two echinostomes, Echinostoma miyagawai Ishii, 1932 and E. revolutum (Frolich, 1802), from Europe. Syst Parasitol. 2000;45:1–15. doi: 10.1023/A:1006232612469. [DOI] [PubMed] [Google Scholar]
- 11.Kostadinova A, Gibson DI. The systematics of the echinostomes. In: Fried B, Graczyk TK, editors. Echinostomes as Experimental Models for Biological Research. The Netherlands: Kluwer Academic Publishers; 2000. pp. 31–57. [Google Scholar]
- 12.Morgan JA, Blair D. Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar-spine group. Parasitology. 1995;111:609–15. doi: 10.1017/S003118200007709X. [DOI] [PubMed] [Google Scholar]
- 13.Morgan JA, Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda) Parasitology. 1998;116:289–297. doi: 10.1017/S0031182097002217. [DOI] [PubMed] [Google Scholar]
- 14.Toledo R, Espert A, Carpena I, Munfioz-Antoli C, Fried B, Esteban JG. The comparative development of Echinostoma caproni (Trematoda: Echinostomatidae) adults in experimentally infected hamsters and rats. Parasitol Res. 2004;93:439–444. doi: 10.1007/s00436-004-1161-1. [DOI] [PubMed] [Google Scholar]
- 15.Faltýnková A, Georgieva S, Soldánová M, Kostadinova A. A re-assessment of species diversity within the ‘revolutum’ group of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) in Europe. Syst Parasitol. 2015;90:1–25. doi: 10.1007/s11230-014-9530-3. [DOI] [PubMed] [Google Scholar]
- 16.Georgieva S, Faltýnková A, Brown R, Blasco-Costa I, Soldánová M, Sitko J, et al. Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasites Vectors. 2014;7:520. [DOI] [PMC free article] [PubMed]
- 17.Kostadinova A, Gibson DI. Isthmiophora Lühe, 1909 and Euparyphium Dietz, 1909 (Digenea: Echinostomatidae) re-defined, with comments on their nominal species. Syst Parasitol. 2002;52:205–17. doi: 10.1023/A:1015789703396. [DOI] [PubMed] [Google Scholar]
- 18.Kostadinova A. Family Echinostomatidae Looss, 1899. In: Jones A, Bray RA, Gibson DI, editors. Keys to Trematoda, vol. 2. London: CAB International and the Natural History Museum; 2005.
- 19.Radev V, Kanev I, Khrusanov D, Fried B. Reexamination of the life cycle of Isthmiophora melis (Trematoda: Echinostomatidae) on material from southeast Europe. Parazitologiia. 2009;43:445–53. [PubMed] [Google Scholar]
- 20.Georgiev BB, Biserkov VY, Genov T. In toto staining method for cestodes with iron acetocarmine. Helminthologia. 1986;23:279–281. [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrand J, Popiołek M, Okulewicz A, Zaleśny G. Helmintofauna myszy z rodzaju Apodemus z okolic Wrocławia. Wiad Parazytol. 2004;50:623–8. [PubMed] [Google Scholar]
- 23.Genov T. Helminths of insectivores mammals and rodents in Bulgaria. Sofia: Publishing House of the Bulgarian Academy of Sciences; 1984.
- 24.PojmanÏska T, Niewadomska K, Okulewicz A. Pasożytnicze helminty Polski. Gatunki, żywiciele, białe plamy. Warszawa: Polskie Towarzystwo Parazytologiczne; 2007. [Google Scholar]
- 25.Fried B, Freeborne NE. Effects of Echinostoma revolutum (Trematoda) adults on various dimensions of the chicken intestine, and observations on worm crowding. Proc Helminth Soc Washington. 1984;51:297–300. [Google Scholar]
- 26.Stillson LL, Platt TR. The crowding effect and morphometric variability in Echinostoma caproni (Digenea: Echinostomatidae) from ICR mice. J Parasitol. 2007;93:242–6. doi: 10.1645/GE-1015R.1. [DOI] [PubMed] [Google Scholar]
- 27.Franco J, Huffman JE, Fried B. Infectivity, growth and development of Echinostoma revolutum (Digenea: Echinostomatidae) in the golden hamster Mesocricetus auratus. J Parasitol. 1986;72:142–7. doi: 10.2307/3281807. [DOI] [PubMed] [Google Scholar]
- 28.Nadler S, Perez-Ponce De Leon G. Integrating molecular and morphological approaches for characterizing parasite cryptic species: implications for parasitology. Parasitology. 2011;138:1688–1709. doi: 10.1017/S003118201000168X. [DOI] [PubMed] [Google Scholar]
- 29.Matejusová I, Koubková B, Gelnar M, Cunningham CO. Paradiplozoon homoion Bychowsky & Nagibina, 1959 versus P. gracile Reichenbach-Klinke, 1961 (Monogenea): Two species or phenotypic plasticity? Syst Parasitol. 2002;53:39–47. doi: 10.1023/A:1019945921143. [DOI] [PubMed] [Google Scholar]
- 30.Boyce K, Hide G, Craig PS, Harris PD, Reynolds C, Pickles A, et al. Identification of a new species of digenean Notocotylus malhamensis n. sp. (Digenea: Notocotylidae) from the bank vole (Myodes glareolus) and the field vole (Microtus agrestis). Parasitology. 2012;139:1630–9. [DOI] [PubMed]
- 31.Miller TL, Cribb TH. Dramatic phenotypic plasticity within species of Siphomutabilus n. g. (Digenea: Cryptogonimidae) from Indo-Pacific caesionines (Perciformes: Lutjanidae) Syst Parasitol. 2013;86:101–112. doi: 10.1007/s11230-013-9436-5. [DOI] [PubMed] [Google Scholar]
- 32.Catalano S, Lejeune M, Van Paridon B, Pagan CA, Wasmuth JD, Tizzani P, et al. Morphological variability and molecular identification of Uncinaria spp. (Nematoda: Ancylostomatidae) from grizzly and black bears: New species or phenotypic plasticity? J Parasitol. 2015;101:182–92. [DOI] [PubMed]
- 33.Littlewood DTJ, Olson PD. Small subunit rDNA and the phylum Platyhelminthes: signal, noise, conflict and compromise. In: Littlewood DTJ, Bray RA, editors. Interrelationships of the Platyhelminthes. London: Taylor & Francis; 2001. pp. 262–278. [Google Scholar]
- 34.Tkach V, Pawlowski J, Mariaux J. Phylogenetic analysis of the suborder Plagiorchiata (Platyhelminthes, Digenea) based on partial lsrDNA sequences. Int J Parasitol. 2000;30:83–93. doi: 10.1016/S0020-7519(99)00163-0. [DOI] [PubMed] [Google Scholar]


