Abstract
The cross-talk between ovarian cancer (OvCa) cells and the metastatic microenvironment is an essential determinant of successful colonization. Micro(mi)RNAs play several critical roles during metastasis; however, the role of microenvironmental cues in the regulation of miRNAs in metastasizing cancer cells has not been studied. Using a 3D culture model that mimics the human omentum, one of the principal sites of OvCa metastasis, we identified and characterized the microenvironment-induced downregulation of a tumor suppressor miRNA, miR-193b, in metastasizing OvCa cells. The direct interaction of the OvCa cells with mesothelial cells, which cover the surface of the omentum, caused a DNA methyltransferase 1 (DNMT1) mediated decrease in the expression of miR-193b. The reduction in miR-193b enabled the metastasizing cancer cells to invade and proliferate into human omental pieces ex vivo and into the omentum of a mouse xenograft model of OvCa metastasis. The functional effects of miR-193b were mediated, in large part, by the concomitant increased expression of its target, urokinase-type plasminogen activator (uPA), a known tumor-associated protease. These findings link paracrine signals from the microenvironment with the regulation of a key miRNA that is essential for the initial steps of OvCa metastatic colonization. Targeting miR-193b could prove effective in the treatment of OvCa metastasis.
Keywords: miR-193b, ovarian cancer, metastasis, mesothelial cells, uPA
Introduction
Ovarian cancer (OvCa) rarely metastasizeshematogenously, and is generally confined to the peritoneal cavity1, 2. The cancer cells exfoliate from the primary tumor and are carried around the peritoneal cavity by the peritoneal fluid. One of the predominant sites of metastasis is the omentum, which is a large adipose-rich fold of the peritoneum that covers the bowels3. The key and rate limiting step for successful metastasis is the colonization of the metastatic organs following attachment of cancer cells4, 5. Most of our current knowledge of OvCa metastasis is derived from comparisons of the factors expressed in metastatic tumors with those in the primary tumor. Therefore, the regulation of the steps involved as the cancer cells spread from the primary tumor to the distant metastatic site is not well understood.
MicroRNAs(miRNA) have well known pleiotropic effects in cancer6 and a specific role in metastasis, which has recently begun to be explored (reviewed in7). In particular, studies have investigated the role of miRNAs in the regulation of processes that aid metastasis: miR-10b, miR-373 and miR-520c have been shown to promote invasion8, 9; miR-374a, miR-200, and miR-22 to induce EMT10, 11; and miR-126 was found to increase angiogenesis12. Recent reports have demonstrated that miRNAs are regulated by the cellular microenvironment13, 14. However, it is not yet clear whether miRNA changes in cancer cells that contribute to the initial colonization of distant metastatic sites arise independently, or if they are dependent on microenvironmental cues.
Given the bidirectional communication between the tumor-microenvironment and cancer cells15, we reasoned that mesothelial cells, the first cell type an OvCa cell encounters when metastasizing to tissues in the peritoneal cavity, could affect miRNA expression in OvCa cells. Using an organotypic 3D culture model that mimics the surface of the human peritoneum and omentum16, we examined how miRNAs regulateOvCametastatic colonization. We report here that the most downregulated miRNA, miR-193b, has a role in promoting omental colonization through the upregulation of its target urokinase (uPA) and that miR-193b is epigenetically repressed in cancer cells through their interactions with the mesothelial cells covering the surface of the omentum.
Results
miR-193b is the most downregulated miRNA in colonizing OvCa cells
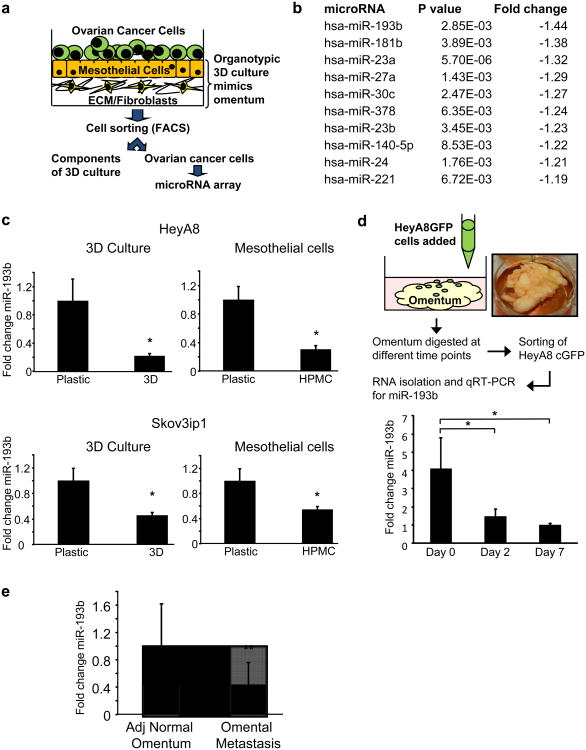
An in vitro organotypic 3D culture system that mimics the surface of the human omentum was used to identify miRNAs that could potentially regulate early metastatic colonization16. The 3D culture system was assembled by seeding a confluent monolayer of human primary mesothelial cells (HPMC) over a layer of collagen I and normal omental fibroblasts (NOFs). HeyA8 OvCa cells expressing GFP were added to the 3D culture and sorted after 2 days by FACS (Fig. 1a). A miRNA array analysis was performed to compare miRNA expression levels of OvCa cells seeded on the 3D culture with those seeded on plastic (Fig. 1a). Since most miRNAs are globallydownregulated in OvCa17, we focused on miRNAs whose expression was further decreased in cancer cells when seeded on the 3D culture. The most downregulated miRNA was miR-193b (Fig. 1b). Since mesothelial cells cover the surface of the entire abdominal cavity, including the omentum, and are the first cell type with which OvCa cancer cells interact as they metastasize18, 19, OvCa cells were seeded ona confluent monolayer of HPMCs and miRNA expression profiling was repeated (Supplementary Fig. 1). Again, miR-193b was one of the 5 most downregulated miRNAs in HeyA8 cells seeded on HPMCs (Supplementary Table 1). These results were confirmed by qRT-PCR for miR-193b in 2 OvCa cell lines seeded on the 3D culture or on HPMCs (Fig. 1c). A similar decrease in the expression of miR-193b was also seen in primary OvCa cells obtained from patient ascites and in RKO1 colon cancer cells when seeded on the 3D culture (Supplementary Fig. 2a and c). To approximate the in vivo situation encountered by OvCa cells more closely, cells were seeded on pieces of full human omentum and cultured ex vivo for up to 7 days (Fig. 1d). At each time point, the cancer cells were isolated by enzymatic digestion followed by FACS to separate the fluorescently labeled OvCa cells. qPCR for miR-193b showed that miR-193b was decreased in HeyA8 cells colonizing the omentum for 2 and 7 days (Fig. 1d), suggesting that the decrease was an early but sustained response to interactions with the microenvironment. We also compared the miR-193b expression levels in omental metastasis and the adjacent normal omentum in 7 high grade serous OvCa patients. miR-193b expression was significantly decreased in the metastatic tumors (Fig. 1e). Since adipocytes are a major constituent of the omentum, their effect on miR-193b expression in OvCa cells was tested by co-culturing Skov3ip1 cells with adipocytes isolated from human omentum. Co-culture with adipocytes had no effect on Skov3ip1 miR-193b expression (Supplementary Fig. 2b). These results suggest that miR-193b downregulation is an early event in omental colonization, and that interactions with mesothelial cells alone are sufficient to downregulate miR-193b expression in cancer cells.
Figure 1. miR-193b is the most downregulated miRNA in metastasizing OvCa cells.
(a) Schematic representation of a human orthotopic 3D culture model mimicking the surface layers of the omentum. GFP-expressing OvCa cells were grown on the 3D culture for 2 days and then separated by FACS. RNA isolated from these OvCa cells was used for miRNA array analysis to determine the deregulated miRNAs during early metastatic colonization. (b) miRNA array. Shown is a list of significantly downregulated miRNAs which were sorted according to the fold-change in expression. (c) qRT-PCR validation of miR-193b expression in HeyA8 and Skov3ip1 OvCa cells seeded on the 3D culture or on mesothelial cells (HPMCs) (mean±SD; 3 independent experiments). (d) HeyA8 cells stably expressing GFP were seeded on pieces of full human omentum and cultured ex vivo for 2 or 7 days. The omentum pieces were digested, fluorescent cancer cells separated by FACS and miR-193b expression was quantified by qRT-PCR (mean±SD; 3 independent experiments). (e) qRT-PCR for miR-193b expression in paired omental metastasis and adjacent (Adj.) normal omentum from 7 high grade serous OvCa patients (mean±SD, 3 replicates, * p<0.01, **p<0.05, Student's t test).
miR-193b suppresses cancer cell growth and motility
Because miR-193b was downregulated during metastatic colonization, we studied the effect of overexpression or inhibition of miR-193b on OvCa cell growth, motility, invasiveness and adhesion. HeyA8 OvCa cells, made to stably overexpress miR-193b by lentiviral infection (Supplementary Fig. 3a), exhibited both reduced colony formation (Fig. 2a) and transwell migration (Fig. 2b). Similar results were obtained when Skov3ip1 and ES2 OvCa cells were transiently transfected with pre-miR-193b(Supplementary Fig. 4a-d). We also found that stable overexpression of miR-193b inhibited the ability of HeyA8 cells to attach to the 3D culture or to mesothelial cells (Fig. 2c) and impaired invasion through the 3D culture (Fig. 2d). Conversely, transient transfection of HeyA8 cells with a miR-193b inhibitor (LNA anti-miR-193b, Supplementary Fig. 3b) increased colony formation as well as migration (Fig. 2e and f). Similar results were obtained with Skov3ip1 cells (Supplementary Fig. 4e and f). In order to more realistically mimic the in vivo scenario found in patients, the role of miR-193b in cancer cells colonizing human omentum pieces ex vivo was investigated. HeyA8 cells stably expressing miR-193b and GFP were seeded on pieces of normal omentum in culture dishes and allowed to grow for 7 days (as described in Fig. 1d). The GFP-expressing cancer cells growing on the omentum were imaged with a fluorescent microscope, digested, and cancer cell growth was quantified by measuring fluorescence. Fluorescent imaging and H&E staining showed a decrease in the growth of the HeyA8 cells that stably expressed miR-193b, as compared to control vector. (Fig. 3a). To test the effect of the overexpression of miR-193b in vivo, an orthotopic xenograft mouse model of OvCa metastasis was used. This model recapitulates the pattern of human OvCa metastasis, as evidenced by disseminated nodules on the peritoneum and omental metastasis20, 21. HeyA8 cells, stably expressing either miR-193b or the control vector, were injected intraperitoneally (i.p.) into female nude mice. Stable overexpression of miR-193b (Supplementary Fig. 5) decreased tumor burden by more than 50% in the mice (Fig. 3b). This decrease was paralleled by a significant reduction of proliferation in the miR-193b overexpressing tumors (Ki-67 staining). The miR-193b tumors also had a greater rate of apoptosis, as evidenced by increased staining of cleaved caspase 3 in tumor sections (Fig. 3b). To determine the effect of miR-193b inhibition, HeyA8 cells stably expressing miR-Zip-193b, an inhibitor of miR-193b, or control vector, were injected i.p. in female nude mice (Fig. 3c). Inhibition of miR-193b resulted in a 1.6 fold increase in tumor weight, indicating that the decreased miR-193b activity promoted metastasis in vivo.
Figure 2. miR-193b inhibits motility and growth of ovarian cancer cells.
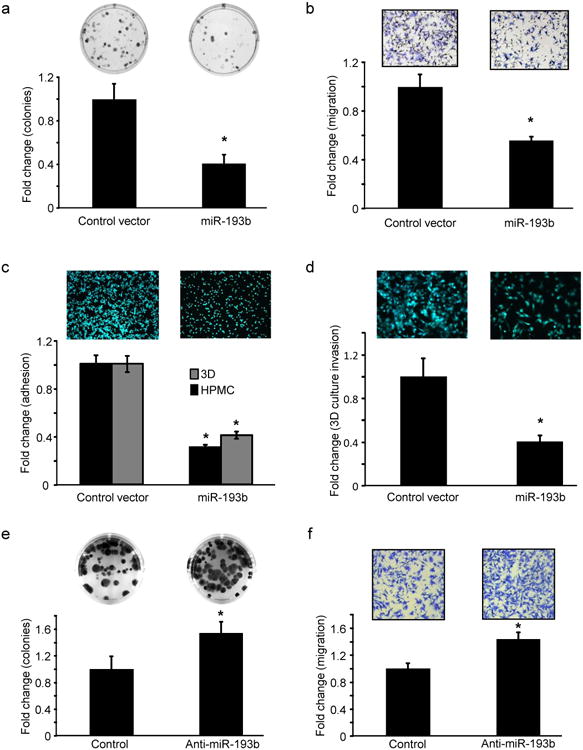
(a-d) miR-193b stable overexpression. (a) Colony formation. HeyA8 cells stably expressing miR-193b or vector controls were seeded in 6 well plates for a colony formation assay. Colonies were fixed after 2 weeks, stained and counted (mean ± SD; 3 independent experiments). (b) Migration. HeyA8 cells stably expressing miR-193b or control vector were seeded on 8 μm pore size inserts and allowed to migrate towards medium containing 10% FB. Migrated cells were fixed, imaged and quantified (mean±SD; 3 independent experiments). (c) Adhesion. HeyA8 GFP cells stably expressing miR-193b or control vector were seeded on the 3D culture model and allowed to adhere for 1 hour and quantified using a fluorescent plate reader (mean±SD; 3 independent experiments). (d) Invasion. HeyA8 GFP cells stably expressing miR-193b were allowed to invade through the 3D culture assembled in the upper chamber of 8 μm pore transwell inserts to assess their ability to invade through the surface of the omentum. The invaded HeyA8GFP cells were imaged and quantified (mean±SD; 3 independent experiments). (e-f) miR-193b inhibition. (e) Colony formation. HeyA8 cells were transfected with anti-miR-193b or negative control oligomers and a colony formation assay performed as described in (a) (mean±SD; 3 independent experiments). (f) Migration. HeyA8 cells were transfected with anti-miR-193b and used in a transwell migration assay (mean±SD; 3 independent experiments). * p<0.01, Students t test.
Figure 3. miR-193b inhibits ovarian cancer cells metastasis.
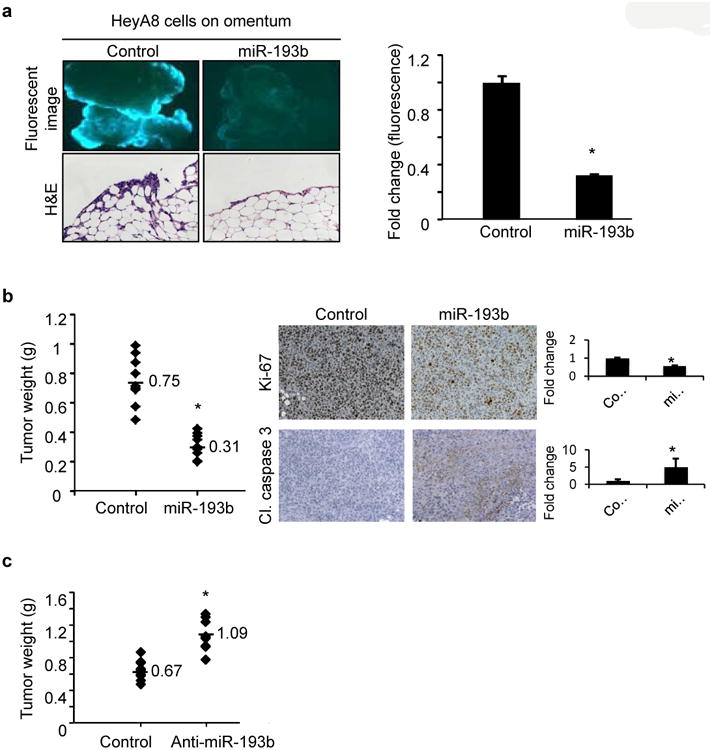
(a) Ex vivo omental assay. HeyA8 cells stably expressing GFP and miR-193b or control vector were seeded on pieces of full, freshly removed, human omentum and cultured ex vivo for 7 days. The growth of GFP-expressing cells overexpressing miR-193b or control vector on the pieces of omentum were imaged with a fluorescent microscope (top) and quantified on a fluorescent plate reader after enzymatic digestion of the tissue (right). Representative images of Hematoxylin and Eosin staining of omental sections (bottom)(mean±SD; 2 independent experiments with 3 replicates) (b) Xenograft mouse model of metastasis. Female athymic nude mice were injected i.p. with 1×106 HeyA8 cells stably expressing miR-193b or the vector control (10 mice per group). Mice were euthanized after 15 days and the tumors surgically resected and weighed. Left: Quantification of the tumor weight of HeyA8 tumors stably expressing miR-193b compared to control vector. The mean tumor weights are indicated and marked by dashes. Right:The tumor sections (n=3) were stained for Ki-67 and cleaved (Cl.) caspase 3 expression to test for proliferation and apoptosis, respectively. (c) HeyA8 cells (1×106) stably expressing miR-193b inhibitor miR-Zip-193b(Anti-miR-193b) or control vector were injected i.p. in female athymic nude mice (n=10/group). Mice were euthanized after 15 days and the tumors surgically resected and weighed. Quantification of the tumor weight of HeyA8 tumors stably expressing miR-Zip-193b(Anti-miR-193b) compared to control vector is plotted. The mean tumor weights are indicated and marked by dashes. * p<0.01, Students t test.
miR-193b is regulated by promoter hypermethylation during colonization
miRNAs are transcribed and subsequently undergo sequential nucleolytic processing in the nucleus and in the cytoplasm22. The miR-193b downregulation in the colonizing cancer cells could be due to decreased transcription or changes in post-transcriptional processing. To understand the effect of the microenvironment on miR-193b regulation, HeyA8 cells were grown on the 3D culture for 48 hours, RNA was extracted and primary miR-193b (pri-miR-193b) levels were analyzed by Northern Blot. Figure 4a shows that pri-miR-193b was transcriptionally downregulated when the OvCa cells were seeded on the 3D culture. We tested the possibility that miR-193b was downregulated in OvCa through promoter hypermethylation induced by the interaction of OvCa cells with the microenvironment. Treatment of HeyA8 cells with the DNA methyltransferase (DNMT) inhibitor, 5-aza-2′deoxycytidine (decitabine), led to re-expression of miR-193b when OvCa cells were seeded either on HPMCs or on the 3D culture (Fig. 4b), suggesting that miR-193b was downregulated by mesothelial cell-induced promoter hypermethylation. Expression of the three DNMT isoforms23 was characterized, using qPCR, in HeyA8 cells seeded on the 3D culture and on mesothelial cells. Only DNMT1 expression showed a statistically significant increase in both the HeyA8 cells grown on the 3D culture and on mesothelial cells (Supplementary Fig. 6). Silencing DNMT1 in HeyA8 cells (Supplementary Fig. 7a) and seeding them on the 3D culture increased their expression of miR-193b (Fig. 4c). Because DNMT1 may affect multiple genes, we sought to determine if silencing DNMT1 specifically reverses the functional effect of miR-193b inhibition in cancer cells. HeyA8 cells were transiently transfected with either aDNMT1 siRNA or anti-miR-193b, with both together, or with an equimolar amount of scrambled control oligomers, and then used in proliferation and migration assays. miR-193b inhibition resulted in increased proliferation (Fig. 4d) and migration (Fig. 4e) while DNMT1 knockdown decreased both proliferation (Fig. 4d) and migration (Fig. 4e). Co-transfection of anti-miR-193b and DNMT1 siRNA neutralized the effects of DNMT1 inhibition on both proliferation and migration (Fig. 4d and e), suggesting that DNMT1 modulates miR-193b transcription through promoter methylation.
Figure 4. miR-193b is downregulated by promoter hypermethylation induced by the interaction of ovarian cancer cells with mesothelial cells.
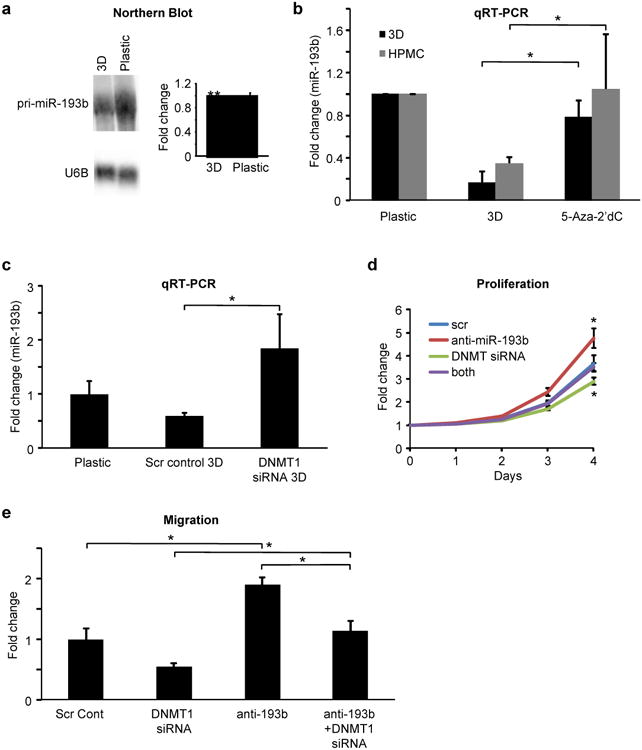
(a) Northern blot analysis of the primary transcript of miR-193b (pri-miR-193b) in HeyA8 cells seeded on the 3D culture or on plastic culture dishes. U6B served as loading control. Quantification of the northern blots normalized to U6B using Image J (mean±SD; 2 independent experiments). (b) qRT-PCR for miR-193b expression in HeyA8 cells pre-treated with 1 μM 5-Aza-2′ deoxycitidine (5-Aza-2′ dC) for 2 days prior to seeding them on the 3D culture model or on mesothelial cells (HPMC). Controls were treated with DMSO. The GFP-expressing cancer cells were sorted by FACS following 2 days of co-culture and RNA was isolated for qRT-PCR(mean±SD; 3 independent experiments). (c) qRT-PCR for miR-193b expression. HeyA8 cells were transfected with DNMT1 siRNA or scrambled control oligomer and 24h later the cells were seeded on the 3D culture model (control, plastic). After 48 hours on 3D culture, HeyA8 cells were sorted by FACS and miR-193b expression in these cells detected by qRT-PCR (mean±SD; 3 independent experiments). (d) Proliferation. HeyA8 cells stably expressing GFP were transfected with either DNMT1 siRNA or anti-miR-193b, transfected with both together, or transfected with equimolar amounts of scrambled control oligomers. The cells were used 24h later in a proliferation assay measuring the change in green fluorescence every 24 hours or (e) in a migration assay through 8 μm pore inserts(mean±SD; 3 independent experiments). * p<0.01, ** p<0.05, Students t test.
miR-193b regulates the expression of urokinase-type plasminogen activator
We next endeavored to determine which target proteins are activated when miR-193b is downregulated during early metastasis. We first considered the urokinase-plasminogen activator (uPA) because 1) we and others had previously identified uPA as a critical factor in the metastatic spread and progression of OvCa24, 25 and 2) uPA is a predicted miR-193b target by miRNA target prediction algorithms (TargetScan and microRNA.org/miRanda). uPA is a secreted serine protease which is upregulated in OvCa25, 26 and plays a role in enhancing the proliferation, migration and adhesion of cancer cells27. When HeyA8 or Skov3ip1 OvCa cells were seeded on the 3D culture, uPA protein expression was induced (Fig. 5a). The overexpression of miR-193b in HeyA8 cells inhibited uPA protein expression while inhibition of miR-193b resulted in an increase in uPA expression (Fig. 5b and Supplementary Fig. 8a), confirming that uPA is regulated by miR-193b in OvCa cells. To investigate the direct binding of miR-193b to its seed sequence in uPA 3′UTR, we used wild type and miR-193b seed match mutated human uPA 3′UTR constructs (Supplementary Fig. 8b). Transfection of the wild type construct along with miR-193b inhibited luciferase activity, where as no inhibition was observed for the mutated uPA 3′UTR, indicating direct targeting of uPA by miR-193b (Fig. 5c). Subsequently, we investigated the possible role of uPA in mediating the functional effects of miR-193b in OvCa metastasis. HeyA8 cells were transfected with uPA siRNA (Supplementary Fig. 7b) and the effect of uPA knockdown was studied. Silencing uPA in HeyA8 cells resulted in decreased colony formation (Fig. 5d), transwell migration (Fig. 5e) and invasion through the 3D culture (Fig. 5f). It did not, however, have a significant effect on adhesion to the 3D culture (Fig. 5g). These results indicate that blocking uPA mimics most of the effects of miR-193b overexpression. We therefore reasoned that silencing uPA should abrogate most of the pro-tumorigenic effects of miR-193b inhibition. To this end, HeyA8 cells were transfected with either anti-miR-193b or uPA siRNA, or with both. Figure 5h shows that co-transfection of uPA siRNA with anti-miR-193b abrogated the increase in migration due to miR-193b inhibition, suggesting that uPA is a mediator of the effects of miR-193b downregulation.
Figure 5. miR-193b inhibits metastatic colonization of ovarian cancer cells through its target, uPA.
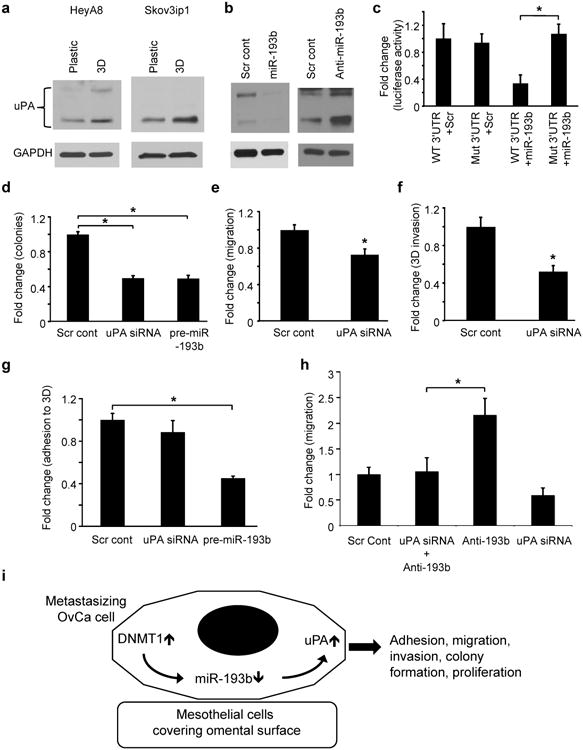
(a) Immunoblot for uPA expression in HeyA8 and Skov3ip1 cells that were seeded on the 3D culture or on plastic dishes. (b) Immunoblot for uPA in (left) HeyA8 cells transiently transfected with pre-miR-193b or scrambled control oligomer or (right) HeyA8 cells stably expressing miR-Zip-193b(Anti-miR-193b) or control vector. Immunoblots are representative of 3 independent experiments. (c) 3′UTR luciferase reporter assay for the binding of miR-193b to the 3′UTR of uPA. Wild type (WT) or miR-193b seed match mutated (Mut) uPA 3′UTR luciferase constructs were co-transfected with pre-miR-193b or scrambled control (Scr) in 293 T cells and the luciferase activity was measured (mean±SD; 3 independent experiments). (d) Colony formation. HeyA8 cells were transiently transfected with siRNA against uPA or with scrambled control oligomer and seeded for colony formation, (e) Migration. HeyA8 cells transfected with uPA siRNA were seeded on 8 μm pore inserts for transwell migration. (f) Invasion. uPA was transiently silenced in GFP-expressing HeyA8 cells and allowed to invade through the 3D culture assembled in the upper chamber of 8 μm pore inserts. The invaded HeyA8 GFP cells were imaged and quantified. (g) Adhesion. GFP-expressing HeyA8 cells transiently transfected with uPA siRNA were allowed to adhere to the 3D culture in 96 well plates for 1 hour and the adherent cells were quantified using a fluorescent plate reader (h) Rescue of miR-193b inhibition effects: HeyA8 cells were transfected either with anti-miR-193b or uPA siRNA, both together, or with equimolar scrambled control oligomers and used in a trans-well migration assay. Error bars represent mean±SD of three independent experiments (* p<0.01, Students t test). (i) Proposed model. Interaction of metastasizing OvCa cells with mesothelial cells covering the omentum results in a DNMT1-mediated downregulation of miR-193b by methylation promoting metastasis through an induction of uPA expression.
Discussion
miRNAs have been reported to play a role in the regulation of metastasis
miR-373 and miR-520c were identified as metastasis promoters by a genetic screen used to detect miRNAs that aid in the migration and invasion of breast cancer cells in vivo9. The downregulation of miR-31 was also found to promote metastasis through the simultaneous induction of several metastasis-promoting targets28. Whether colonization of the distant organ, which is a first and critical step in successful metastasis4, 29, 30, is regulated by miRNA (s) has not been previously explored because of a lack of suitable experimental models. In order to dissect out the early events in the process of colonization during OvCa metastasis, we used an organotypic 3D culture system that closely mimics the surface of the human omentum (reviewed by White et al31). By employing an unbiased miRNA array approach, we identified miR-193b as the most downregulated miRNA in colonizing OvCa cells.
miR-193b has been previously reported to act as a tumor suppressor in prostate cancer, acute myeloid leukemia and hepatocellular carcinoma32-34. Furthermore, a decrease in miR-193b has been found to promote breast cancer tumor progression through an increased expression of urokinase35. We have demonstrated, using a variety of models, that a reduction in miR-193b expression in cancer cells facilitates the early steps of OvCa metastasis, while miR-193b overexpression curtails metastatic functions. We now provide the first report that the mesothelial cell mediated repression of miR-193b is critical for OvCa metastatic colonization and the subsequent early invasion of OvCa cells into the omentum (Fig. 5i). All OvCa cell lines examined, as well as short-term cultures of primary OvCa cells isolated from the ascites of patients with OvCa, and colon cancer cells showed decreased miR-193b expression when seeded on the 3D organotypic culture. Furthermore, miR-193b expression was found to be lower in the omental metastasis than in the adjacent normal omentum of patients with high grade serous OvCa, indicating that the metastatic process results in a sustained decrease in miR-193b expression. This was confirmed with a novel assay in which OvCa cells were seeded on organ cultures of full human omentum ex vivo. In this experiment, a decrease in miR-193b was maintained 7 days after the cancer cells were seeded, demonstrating that it is a sustained effect and not just an acute response. That this downregulation of miR-193b in OvCa cells is dependent on contact with mesothelial cells, is supported by our co-culture experiments of OvCa cells with the 3D culture or mesothelial cells. While the omental microenvironment harbors other cell types, most notably adipocytes and fibrobasts, further experiments demonstrated that neither adipocytes nor fibroblasts significantly affected miR-193b expression in OvCa cells. Although the co-culture of cancer cells with adipocytes was planned with the smallest amount of media allowing the different cell types to touch, the buoyant adipocytes might not completely replicate the more intimate cell-cell interactions in vivo. Therefore, we do not rule out the possible contribution of omental adipocytes to the miR-193b downregulation in the metastasizing cancer cells. Mesothelial cells, however, induced down-regulation of miR-193b in the cancer cells, resulting in a pronounced increase in biologic effects that include important first steps of abdominal metastasis; adhesion, invasion, migration, and colony formation. Conditioned medium from mesothelial cells did not alter miR-193b expression in the cancer cells (data not shown), indicating that juxtacrine interactions are probably involved in the cross-talk between OvCa and mesothelial cells. To the best of our knowledge, this is the first report showing that mesothelial cells directly regulate miRNA expression in cancer cells.
The sustained downregulation of miR-193b in OvCa cells, even after a short interaction with mesothelial cells, suggested a possible epigenetic regulation of miR-193b. Significantly, it has been demonstrated that miR-193b is downregulated in prostate cancer cells via hypermethylation of CpG islands in its promoter32. We found that pre-treatment of HeyA8 cells with the DNMT inhibitor, decitabine, completely abrogated a decrease in miR-193b when the cancer cells were seeded on the 3D culture or on mesothelial cells. Our data further revealed that DNMT1 repressed miR-193b in OvCa cells cultured on mesothelial cells. Silencing DNMT1 in HeyA8 cells could abrogate the effects of miR-193b inhibition on their migration as well as proliferation, indicating a role for DNMT1 in miR-193b regulation. This is consistent with recent data showing that DNMTs are overexpressed in OvCa and regulated by activated EGFR36, 37. Moreover, HeyA8 OvCa cells have a three-fold higher expression of DNMT1 when compared to normal ovarian surface epithelial cells38. Although DNMT1 has a preference for hemi-methylated DNA, it can also act as a de novo methyltransferase, since it has greater specific activity than DNMT3A and DNMT3B23.
The pro-colonizing effect of miR-193b loss is mediated, at least in part, by regulation of the serine protease uPA, consistent with reports that miR-193b directly targets uPA in breast and prostate cancer cells35, 39, 40. Urokinase has been shown to promotelung cancer cell invasion through the recruitment of integrins and ECM proteins into the uPA/uPA receptor complex41. In OvCa, uPA expression in serum or ascites is correlated with an adverse prognosis and inhibition of uPA by antibodies, siRNA or anti-sense oligonucleotides reduced metastasis in several preclinical models42, 43. Given that uPA mediates key tumor functions, the up-regulation of uPA as a consequence of miR-193b down-regulation during early metastasis is consistent with the identification of miR-193b as a tumor suppressor. Taken together, the data are consistent with a model in which mesothelial cell-induced downregulation of miR-193b facilitates metastatic colonization of OvCa cells through the increased expression of its target, uPA (Fig. 5i). These findings support the recent concept that the microenvironment is recruited by tumor cells to support critical functions required for metastasis14, 44.
Materials and methods
Reagents
Trypsin, Dulbecco's Modified Eagle Medium (DMEM), MEM vitamins, MEM nonessential amino acids and Penicillin-Streptomycin were purchased from Media Tech (Manassas, VA). TaqMan miRNA assay for hsa-miR-193b was obtained from Applied Biosystems (Foster City, CA). Pre-miR-193b, and scrambled pre-miR negative control were from Ambion (Austin, TX), miRCURY LNA anti-miR-193b and scrambled anti-miR negative control were from Exiqon (Vedbæk, Denmark). The lentivirus vector expressing copepod (c) GFP (CD511B-1) and pre-miR-193b, miR-Zip-193b microRNA inhibitor vector, the control vectorsand the lentivirus packaging kit (LV500A-1) were purchased from System Biosciences (Mountain View, CA). The DNMT inhibitor 5-Aza-2′-deoxycytidine (decitabine) was purchased from Sigma-Aldrich, St. Louis, MO (Cat# A3656). The siRNA's used were Dharmacon (Lafayette, CO) SMARTpool siRNAs: DNMT1 siRNA M-004605-01 and siGENOME PLAU siRNA M-006000-02
Cell lines
The human OvCa cell lines Skov3ip1 (first described by Janet Price45) and HeyA8 were from Gordon B. Mills (M.D. Anderson Cancer Center, Houston, TX). Human OvCa cell lines HeyA8, Skov3ip1, and ES2 were validated by short tandem repeat (STR) DNA fingerprinting using the AmpFℓSTR Identifier kit (Applied Biosystems) and compared with known American Type Culture Collection fingerprints, the Cell Line Integrated Molecular Authentication database (CLIMA), and the University of Texas MD Anderson Cancer Center fingerprint database.
Isolation and culture of primary cells and assembly of the 3D culture
Primary human mesothelial cells (HPMC) and normal omental fibroblasts (NOF) were isolated as described previously16 from omentum obtained from female patients undergoing surgery for benign conditions at the University of Chicago. The primary cells were grown in DMEM with 10% FBS. HPMCs were used for experiments between passages 1 and 2 while NOFs were used between 2 and 5. The 3D omental culture was assembled in 10 cm culture dishes by first seeding 360,000 NOFs along with 91μg of Collagen Type I (Rat tail, BD Cat#354236) in DMEM. After attachment, 3.6×106 HPMCs were seeded on top to form a confluent monolayer and the 3D culture was used 24 h later for experiments to investigate the initial events of colonization. GFP expressing OvCa cells (1×106) were seeded on the 3D culture and allowed to grow for 2 days. Cells were trypsinized and fluorescent cancer cells were isolated by fluorescence activated cell sorting (FACS) and used for RNA/protein isolation. The human omental adipocytes were isolated and co-cultured with OvCa cells, as described previously46. Briefly, Skov3ip1 cells (1 × 106) were seeded in 60mm dishes, incubated for 18 – 24 h, washed twice with PBS and 2.1 ml of DMEM/F12 media (1:1 ratio, containing 0.1% Fraction V, fatty-acid-free BSA) was added. 700μl (packed cell volume) of adipocytes was overlaid onto the Skov3ip1 cells, and co-cultured for 8 hours. Adipocytes were removed by aspiration and Skov3ip1 cells were washed 3 – 5 times with PBS. RNA was isolated using TRizol Reagent (Life Technologies, Grand Island, NY) and used for miR-193b qPCR. This research was approved by the Institutional Regulatory Board of the University of Chicago. To set up the 3D culture in transwell inserts or 96 well plates, the number of cells and the amount of collagen were proportionately reduced according to the growth surface area.
Omental metastasis and adjacent normal omentum samples
RNA was extracted from paired, fresh frozen samples of omental metastasis and adjacent normal omentum, obtained from 7 high grade serous OvCa patients. Briefly, tissue was homogenized using the TissueRuptor (Qiagen) and total RNA was extracted using the miRNeasy kit (Qiagen), according to the manufacturer's instructions. The miR-193b expression levels in these samples were assayed by qRT-PCR using a TaqMan assay (Applied Biosystems).
miRNA Array
Total RNA was isolated using the miRNeasy mini kit (Qiagen) according to the manufacturer's protocol and submitted to Exiqon for miRNA profiling using the miRCURY LNA array (v.10.0). First, all miRNAs that were not significantly expressed were eliminated. Thereafter, only those miRNAs with p-values ≤0.05 for HeyA8 cells seeded on 3D culture versus plastic were included in the analysis and were ranked according the fold change in expression.
Real-time PCR
Quantitative real-time–PCR (qPCR) for miR-193b was performed using TaqManmiRNA assay kit according to the manufacturer's protocol on the Prism7500 TaqMan qPCR machine (Applied Biosystems) with U6 or RNU48 as endogenous controls. Similarly, qPCR for uPA, DNMT1, DNMT3A and DNMT3B were carried outusing TaqMan gene expression assay (Applied Biosystems) using GAPDH as an endogenous control.
Northern blotting
Total RNA (30μg) was separated by 1% agarose formaldehyde gel electrophoresis and transferred overnight to a nylon membrane (Hybond-N, GE Healthcare) by the capillary method. RNA was cross-linked to the membrane with UV and hybridized to 0.1nM double-DIG labeled antisense miRCURY LNA probes against miR-193b (Exiqon). The membrane was probed with anti-DIG AP antibody (Roche, Cat#11093274910) and detected using chemiluminescence (Roche CDP-Star, Cat# 12041677001) which was imaged using a SyngeneG:Box imaging system.
Ex vivo culture of human omentum
Omental tissue, obtained from female patients undergoing surgery for benign conditions, was cut into pieces weighing 300 mg and cultured in DMEM containing 10% FBS in 6 well dishes. GFP-expressing HeyA8 cells were added to the omentum pieces and allowed to colonize for up to 7 days. Medium was changed every day. At the end point the cancer cells colonizing the omentum were imaged with an Axio-observer A1 fluorescent microscope using 1.25× objective (Carl Zeiss). Thereafter, the omentum pieces were digested with Liberase (Roche) and the cancer cell growth was quantified by measuring their fluorescence in a plate reader (Synergy HT, BioTek), orcancer cells were isolated by FACS for miR-193b qPCR.
Mouse orthotopic xenograft model of OvCa metastasis
The xenograft model was described previously20. Briefly, 1×106 HeyA8 cells either stably expressing miR-193b, miR-Zip-193b miR-193b inhibitor, or the respective control vectorswere injected intraperitoneally (i.p.) into 6 week old, female, athymic nude mice (10 mice/group). Mice were euthanized 15 days after infection and the tumors were surgically resected and weighed.
Transient transfection
OvCa cells were transfected14 with 30 nM pre-miR-193b, scrambled control oligo (Ambion), 30 nM miRCURY LNA anti-miR-193b, or scrambled control LNA oligo (Exiqon) using siPORTNeoFX (Ambion). The cells were used for experiments 48 h after transfection or as indicated.
Migration
Transwell Migration assays were conducted using 8 μm pore size inserts (BD, Falcon). GFP-expressing OvCa cells transfected with pre- or anti-miR-193b were added to the upper chamber in 200 μl DMEM and allowed to migrate for 3 h at 37°C. DMEM with 10% FBS was used as a chemoattractant in the lower chamber. Cells were fixed in 4% paraformaldehyde and imaged (5 fields/insert) using an Axio-observer A1 fluorescent microscope (Carl Zeiss).
Colony formation
OvCa cells were seeded in 6 well plates (HeyA8, 500 cells/well;Skov3ip1, 2000 cells/well) and allowed to form colonies. Once visible colonies were formed (HeyA8, 2 weeks; Skov3ip1, 3 weeks), they were fixed with 4% paraformaldehyde and stained with 0.005% crystal violet and were imaged using a SyngeneG:Box imaging system and the number of colonies/well were counted.
Invasion through 3D culture
Cellular invasion through the surface of the omentum was assayed in vitro by first assembling the 3D culture on 8 μm pore size Fluoroblocktranswell inserts in 24 well plates. GFP-expressing OvCa cells were then seeded on the 3D culture in the transwell insert in 200 μl serum-free DMEM. DMEM with 10% FBS served as a chemoattractant in the lower chamber. Cells were allowed to invade for 16 h and then were fixed with 4% paraformaldehyde. The fluorescent cancer cells that had invaded were imaged (5 fields/insert) using an Axio-observer A1 fluorescent microscope (Carl Zeiss).
Adhesion to 3D culture
Cellular adhesion to the surface of the omentum was assayed in vitro by first assembling the 3D culture in dark 96 well plates with clear bottoms. OvCa cells expressing GFP were seeded on the 3D culture and allowed to adhere for 1 h. Total fluorescence in each well was measured using a Synergy HT plate reader (BioTek) (excitation, 490 nm; emission, 520 nm) and then the wells were washed 3 times with PBS before the fluorescence of the adherent cells was measured again. Adherent cells were quantified by normalizing their fluorescence to the total fluorescence.
Immunohistochemistry
Immunohistochemical studies were performed by using 5μm thick formalin-fixed deparaffinized sections as previously described20, 21. Xenograft tumor tissue was probed with anti-Ki-67 (1:300)(Thermo Scientific, SP6), and anti-cleaved caspase 3 (1:100) (Cell Signaling, Asp175) antibodies.
Immunoblotting
Immunoblotting was done as previously described 20. Briefly, proteins were separated by 4-20% gradient SDS-PAGE and transferred to nitrocellulose, probed with anti-uPA antibody (American Diagnostica, HD-UK1) and detected using a HRP-linked anti-mouse IgG secondary antibody (Cell Signaling, Cat#7076).
Luciferase Assay
3′UTR luciferase reporter assay was performed as described previously14. Briefly, 293T cells were co-transfected with a firefly luciferase construct (5 ng) containing the wild type PLAU 3′-UTR (SwitchGear Genomics, Menlo Park, CA) or the PLAU 3′-UTR mutated at miR-193b seed sequence, along with 10 nM pre-miR-193b or scrambled oligo, using Lipofectamine 2000 (Invitrogen). Mutations at the seed sequence of hsa-miR-193b in the 3′-UTR of PLAU were made using Quick Change Lightning mutagenesis kit (Stratagene) according to the manufacturer's protocol. Luciferase activity was measured using a LightSwitch Luciferase Assay Kit (SwitchGear Genomics) and read on a BioTek Synergy H1 plate reader.
Statistics
Data analysis was done by unpaired, two-tailed Student's t-test assuming equal variance of the test and the control populations.
Supplementary Material
Acknowledgments
We want to thank Dr. A.F. Haney for collecting omental biopsies, Dr. Abir Mukherjee for helpful discussions, and G. Isenberg for carefully editing the manuscript (all at the University of Chicago, Department of Obstetrics and Gynecology). We are indebted to all the patients, resident and attending physicians in the Department of Obstetrics and Gynecology at the University of Chicago for their participation in tissue collection for these experiments. This research was supported by a Marsha Rivkin Pilot Award (AKM) and by National Cancer Institute R01 CA111882 and RO1 CA169604 grants (EL), and an Ovarian Cancer Research Fund Program Project Development Grant (EL, MEP).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177:1053–1064. doi: 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 5.Kenny HA, Chiang CY, White EA, Schryver EM, Habis M, Romero IL, et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J Clin Invest. 2014;124:4614–4628. doi: 10.1172/JCI74778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 9.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 10.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, et al. MicroRNA-Antagonism Regulates Breast Cancer Stemness and Metastasis via TET-Family-Dependent Chromatin Remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lwin T, Zhao X, Cheng F, Zhang X, Huang A, Shah B, et al. A microenvironment-mediated c-Myc/miR-548m/HDAC6 amplification loop in non-Hodgkin B cell lymphomas. J Clin Invest. 2013;123:4612–4626. doi: 10.1172/JCI64210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–1108. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Kenny HA, Krausz T, Yamada SD, Lengyel E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int J Cancer. 2007;121:1463–1472. doi: 10.1002/ijc.22874. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niedbala MJ, Crickard K, Bernacki RJ. Interactions of human ovarian tumor cells with human mesothelial cells grown on extracellular matrix. An in vitro model system for studying tumor cell adhesion and invasion. Exp Cell Res. 1985;160:499–513. doi: 10.1016/0014-4827(85)90197-1. [DOI] [PubMed] [Google Scholar]
- 19.Iwanicki MP, Davidowitz RA, Ng MR, Besser A, Muranen T, Merritt M, et al. Ovarian cancer spheroids use myosin-generated force to clear the mesothelium. Cancer Discov. 2011;1:144–157. doi: 10.1158/2159-8274.CD-11-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra AK, Sawada K, Tiwari P, Mui K, Gwin K, Lengyel E. Ligand-independent activation of c-Met by fibronectin and alpha (5) beta (1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30:1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, et al. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 2008;68:2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 23.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 24.Schmalfeldt B, Prechtel D, Harting K, Spathe K, Rutke S, Konik E, et al. Increased expression of matrix metalloproteinases (MMP)-2, MMP-9, and the urokinase-type plasminogen activator is associated with progression from benign to advanced ovarian cancer. Clin Cancer Res. 2001;7:2396–2404. [PubMed] [Google Scholar]
- 25.Wang L, Madigan MC, Chen H, Liu F, Patterson KI, Beretov J, et al. Expression of urokinase plasminogen activator and its receptor in advanced epithelial ovarian cancer patients. Gynecol Oncol. 2009;114:265–272. doi: 10.1016/j.ygyno.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Kenny HA, Swindell EP, Mitra AK, Hankins PL, Ahn RW, et al. Urokinase plasminogen activator system-targeted delivery of nanobins as a novel ovarian cancer therapy. Mol Cancer Ther. 2013;12:2628–2639. doi: 10.1158/1535-7163.MCT-13-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 28.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White EA, Kenny HA, Lengyel E. Three-dimensional modeling of ovarian cancer. Adv Drug Deliv Rev. 2014 doi: 10.1016/j.addr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauhala HE, Jalava SE, Isotalo J, Bracken H, Lehmusvaara S, Tammela TL, et al. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127:1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 33.Gao XN, Lin J, Gao L, Li YH, Wang LL, Yu L. MicroRNA-193b regulates c-Kit proto-oncogene and represses cell proliferation in acute myeloid leukemia. Leuk Res. 2011;35:1226–1232. doi: 10.1016/j.leukres.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 35.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 36.Lee JY, Jeong W, Lim W, Lim CH, Bae SM, Kim J, et al. Hypermethylation and post-transcriptional regulation of DNA methyltransferases in the ovarian carcinomas of the laying hen. PLoS One. 2013;8:e61658. doi: 10.1371/journal.pone.0061658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samudio-Ruiz SL, Hudson LG. Increased DNA methyltransferase activity and DNA methylation following Epidermal Growth Factor stimulation in ovarian cancer cells. Epigenetics. 2012;7:216–224. doi: 10.4161/epi.7.3.19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 39.Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S. Impaired MicroRNA Processing Facilitates Breast Cancer Cell Invasion by Upregulating Urokinase-Type Plasminogen Activator Expression. Genes Cancer. 2011;2:140–150. doi: 10.1177/1947601911408888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie C, Jiang XH, Zhang JT, Sun TT, Dong JD, Sanders AJ, et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32:2282–2291. 2291 e2281–2287. doi: 10.1038/onc.2012.251. [DOI] [PubMed] [Google Scholar]
- 41.Tang CH, Hill ML, Brumwell AN, Chapman HA, Wei Y. Signaling through urokinase and urokinase receptor in lung cancer cells requires interactions with beta1 integrins. J Cell Sci. 2008;121:3747–3756. doi: 10.1242/jcs.029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konecny G, Untch M, Pihan A, Kimmig R, Gropp M, Stieber P, et al. Association of urokinase-type plasminogen activator and its inhibitor with disease progression and prognosis in ovarian cancer. Clin Cancer Res. 2001;7:1743–1749. [PubMed] [Google Scholar]
- 43.Zhang W, Ling D, Tan J, Zhang J, Li L. Expression of urokinase plasminogen activator and plasminogen activator inhibitor type-1 in ovarian cancer and its clinical significance. Oncol Rep. 2013;29:637–645. doi: 10.3892/or.2012.2148. [DOI] [PubMed] [Google Scholar]
- 44.Nieman KM, Romero IL, Van Houten B, Lengyel E. Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim Biophys Acta. 2013;1831:1533–1541. doi: 10.1016/j.bbalip.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brader KR, Wolf JK, Hung MC, Yu D, Crispens MA, van Golen KL, et al. Adenovirus E1A expression enhances the sensitivity of an ovarian cancer cell line to multiple cytotoxic agents through an apoptotic mechanism. Clin Cancer Res. 1997;3:2017–2024. [PubMed] [Google Scholar]
- 46.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.