Abstract
Background
Bladder cancer is the most common malignancy of the urinary system, yet our molecular understanding of this disease is incomplete, hampering therapeutic advances.
Methods
Here we used a genome-wide functional short-hairpin RNA (shRNA) screen to identify suppressors of in vivo bladder tumor xenograft growth (n = 50) using bladder cancer UMUC3 cells. Next-generation sequencing was used to identify the most frequently occurring shRNAs in tumors. Genes so identified were studied in 561 patients with bladder cancer for their association with stratification of clinical outcome by Kaplan-Meier analysis. The best prognostic marker was studied to determine its mechanism in tumor suppression using anchorage-dependent and -independent growth, xenograft (n = 20), and metabolomic assays. Statistical significance was determined using two-sided Student t test and repeated-measures statistical analysis.
Results
We identified the glycogen debranching enzyme AGL as a prognostic indicator of patient survival (P = .04) and as a novel regulator of bladder cancer anchorage-dependent (P < .001), anchorage-independent (mean ± standard deviation, 180 ± 23.1 colonies vs 20±9.5 in control, P < .001), and xenograft growth (P < .001). Rescue experiments using catalytically dead AGL variants revealed that this effect is independent of AGL enzymatic functions. We demonstrated that reduced AGL enhances tumor growth by increasing glycine synthesis through increased expression of serine hydroxymethyltransferase 2.
Conclusions
Using an in vivo RNA interference screen, we discovered that AGL, a glycogen debranching enzyme, has a biologically and statistically significant role in suppressing human cancer growth.
RNA interference (RNAi) is a cellular mechanism that negatively regulates gene expression, and has been utilized in loss-of-function studies (1). There are two primary methods of performing RNAi in animal cells, small interfering RNAs (siRNAs) and plasmid-encoded short-hairpin RNAs (shRNAs) (2); however, only the latter can be used to study complex in vivo xenograft phenotypes where long-term target depletion is required. Here, we used a genome-wide lentiviral shRNA library coupled with next-generation sequencing (NGS) to identify new genes that are important in tumor growth, using human xenograft models of bladder cancer, a common malignancy affecting the urinary system with an estimated 72570 new cases and 15210 deaths in 2013 in the United States (3).
Our genome-wide screen identified the glycogen debranching enzyme amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) as a regulator of in vitro and in vivo human cancer cell growth and prognostic marker in patients. Germline mutation of AGL causes glycogen storage disease III (GSD III, Cori disease) via abnormal glycogen breakdown. However, our studies demonstrate that AGL reduces tumor growth independent of its enzymatic activity, and loss of AGL in cancer cells promotes tumor growth in vitro and in vivo through increased glycine synthesis via induction of the glycine synthesizing enzyme serine hydroxymethyltransferase 2 (SHMT2). In summary, our discovery approach appears to be valuable in uncovering new functional pathways driving tumor growth, complementing current approaches in human tumors, and paving the way for novel therapeutic strategies and biomarkers for selecting patients for personalized approaches.
Methods
Cell Lines
Bladder cancer cell lines UMUC3 and T24 were from ATCC. T24T is a metastatic derivative of T24 (4). Cell lines were validated by DNA fingerprinting March 30, 2012 (UMUC3) and February 28, 2012 (T24, T24T). Unless otherwise stated, cells were cultured as follows: UMUC3: modified Eagle’s medium + 1mM sodium pyruvate, 10% fetal bovine serum (FBS). T24 and T24T: Dulbecco’s modified Eagle’s medium/F12 + 5% FBS. All reagents for media components were obtained from Invitrogen.
Pooled shRNA Library Screen
UMUC3 cells were transduced with MISSION® LentiPlex Human Pooled shRNA Library, targeting more than 15000 human genes (Sigma-Aldrich), at a multiplicity of infection of 1. Transductions were incubated for 18 hours with 8 µg/mL polybrene. Thirty hours after transduction, cells were selected with 2 µg/mL puromycin for 96 hours prior to use in experiments. Genomic DNA isolations of the library infected cells and of the nontarget control cells were prepared for downstream analysis.
Infected UMUC3 cells were inoculated in mice. Mice that developed tumors were killed and tumors harvested; DNA was extracted using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich). shRNA sequences present in each cell line were determined by NGS. Next-generation sequencing was verified using polymerase chain reaction (PCR) amplification of the shRNA-encoding and -flanking sequences from genomic DNA using the primers LAP1, TACAAAATACGTGACGTAGAAA; LAP2, TTTGTTTTTGTAATTCTTTA. PCR products were cloned using TA cloning (Invitrogen). The insert from 10 plasmids per tumor were sequenced using standard sequencing.
Next-Generation Sequencing
Library samples were diluted to 10nM based on Pico Green quantitation and given average base pair fragment size. Diluted libraries were run amplified with quantitative PCR (qPCR) p5 and p7 primers to determine final flow cell loading concentration, then normalized to a PhiX control library. Lane 4 in each flow cell was designated as a PhiX control lane. Clustering/amplification performed using TruSeq SR Cluster Kit v2-cBot-GA (Illumina) was used for sequencing on Genome Analyzer IIx (Illumina). For base-calling and de-multiplexing, *.bcl files were used as input, then converted into the compressed FASTQ format. Demultiplexing was performed during the BCL to FASTQ conversion step. Data were mapped to the human hg19 reference genome using the ELANDv2e alignment tool from the Casava 1.8 software (Illumina).
In Vitro and In Vivo Growth
Anchorage-independent growth was assessed by plating cells in 0.4% agar in triplicate. Colonies were stained with Nitro-BT (Sigma) and counted using Image J. Cell proliferation and viability was assessed by plating 103 cells per well in 96-well plates in triplicate for proliferation studies (5). To determine the effects of 2-deoxyglucose (Sigma), PX866 (Oncothyreon, Seattle, WA), AZD6244 (AstraZeneca, Macclesfield, Cheshire, UK) on cell viability, cells were plated as described, and treated with varying concentrations of drugs. Cell viability was determined by CyQUANT Assay (Invitrogen).
All animals used in this study were treated according to University of Colorado Denver and Institutional Animal Care and Use Committee (IACUC) guidelines. Animal protocols were reviewed and accepted in IACUC protocol number B-93410(12)1E. Four-week-old NCr nu/nu male mice (NCI-Frederick, Frederick, MD) were injected with UMUC3 (2.5×104) or T24T (1.0×105) cells stably expressing AGL shRNA or nontarget shRNA control in the right and left flanks of each mouse for subcutaneous tumor growth. Tumors were measured and tumor volumes calculated as described previously (6). In brief, the length (L) and width (W) of each tumor were measured using calipers, and tumor volume was calculated using the equation (L × W2)/2. Similar experiments were carried out with UMUC3 stably expressing human AGL (vectorEX-E2057-M11, purchased from GeneCopoeia, Rockville, MD). Xenografts were collected and analyzed by immunohistochemical (IHC) and mass spectroscopic analysis for lactate (see Supplementary Methods, available online).
Immunohistochemistry of Human Tissues and Microarray Analysis
Human tissue microarrays (TMAs) were prepared from normal urothelium, metastatic (nodal), and nonmetastatic urothelial carcinoma. Each tissue specimen was represented by four 1.0-mm cores on the TMAs to account for intratumoral heterogeneity. Slides were processed as previously described (7). Details are shown in the Supplementary Methods (available online). The IHC study was based on Cleveland Clinic Institutional Review Board number 06-641 with 112 samples.
Microarray analyses were performed on previously published and publicly available datasets. The raw data of the Stransky et al. (8) dataset (n = 103) were downloaded from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/, accession number E-TABM-147) and normalized using the Robust Multichip Average algorithm (9). The processed data of the Kim et al. (10) dataset (n = 255) were downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/,accession GSE13507). The Sanchez-Carbayo et al. (11) dataset (n = 129) was downloaded from the article’s supplementary data and duplicate arrays for individual patients’ tumors (or normal urothelium) were averaged and log (base 2) transformed. The Blaveri et al. (12) dataset (n = 74) was downloaded from http://waldman.ucsf.edu/Blaveri.html. Patient clinicopathologic characteristics are shown in Supplementary Table 2, and detailed methods can be found in the Supplementary Methods (both available online). All microarray analyses were carried out using the statistical programming language R (http://www.R-project.org).
Statistical Analysis
Nuclear magnetic resonance (NMR), gas chromatography-mass spectrometry (GC-MS), Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS), anchorage-independent growth, quantitative glucose and lactate experiments, real-time PCR quantification, and IHC assessment of angiogenesis, proliferation, and apoptosis in xenografts were analyzed by two-tailed Student t test with unequal variances. Error bars denote standard deviation (SD). Proportions of AGL IHC scores in primary versus metastatic tumors were compared using two-tailed Fisher exact test. Cox proportional hazard models and Log-rank tests were used to compare survival in patients with low IHC scores (0 and 1) and patients with high scores (2 and 3). The assumption of proportionality was verified by examining the slope of a generalized linear regression of the scaled Schoenfeld residuals on log scale of survival time.
Tumor volume and in vitro anchorage-dependent growth experiments were analyzed using a repeated-measures analysis, with unique mean values for each group treatment and each time point, unequal variances between groups and at each time point, and an autocorrelation structure of order 1 (AR(1)). For each experiment, we examined AR(1) models with and without a continuous time covariate and chose the correlation structure that had the minimum deviance. The AR(1) model without a continuous time covariate was the best fit for all experiments. For each experiment, this full model was compared to a nested null model with no treatment effects. P values were calculated based on the deviance between the full and null models based on a one-sided χ2 test statistic. These analyses were carried out using the library nlme (13) in R. The function gls was used to fit each model and the autocorrelation structure was specified by the function corCAR1. ANOVA was used to calculate the deviances and P values. The last observation was carried forward for mice terminated due to tumor size approaching the IACUC limit or for in vitro anchorage-dependent growth experiments where cells in one group reach confluence.
Unless specified otherwise, all P values are two-sided. A P value of less than .05 was considered statistically significant.
Materials and methods, including GC-MS, FT-ICR-MS, and NMR analysis of 13C6-glucose metabolism, NGS, reverse transcription PCR, western blotting, NMR, media glucose analysis, and MS analysis of lactate are described in detail in the Supplementary Methods (available online).
Results
Genome-wide In Vivo shRNA Screen Identification of Inhibitors of Tumor Growth
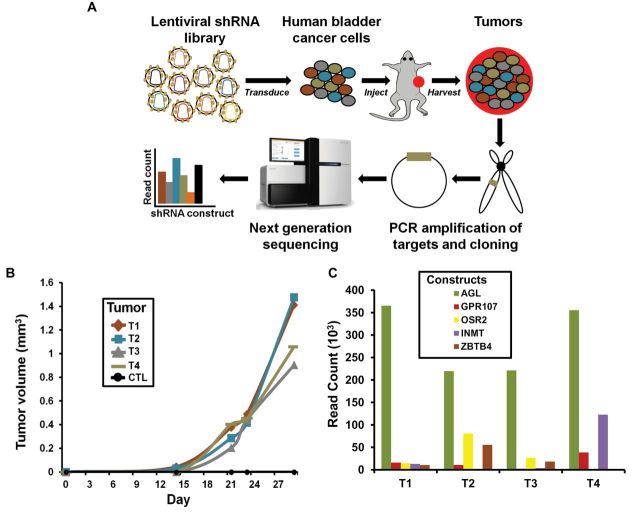
To find previously unidentified genes for which decreased expression leads to enhanced tumor growth, we developed a novel in vivo functional screen. We transduced UMUC3 human bladder cancer cells with the MISSION LentiPlex Human Pooled shRNA Library (Figure 1A). Using serially diluted xenografts (14), we determined that injection of 50000 control shRNA-transfected UMUC3 cancer cells was nontumorigenic in vivo. Therefore, we inoculated mice with 25000 or 12000 UMUC3 cells containing the library. From a total of 50 injection sites, only four tumors grew greater than 900mm3 (Figure 1B), each in different mice. Genomic DNA from these tumors was harvested and evaluated for the amount (read counts) of different shRNA constructs by NGS. The top five shRNA constructs (most read counts) across the four tumors were AGL, indolethylamine N-methyltransferase (INMT), odd-skipped related 2 (OSR2), zinc finger and BTB domain containing 4 (ZBTB4), and G protein-coupled receptor 107 (GPR107) (Figure 1C; Supplementary Table 1, available online).
Figure 1.
Discovery of amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) as a putative tumor growth suppressor. A) Diagram of the pooled short-hairpin (shRNA) library experiment. UMUC3 cancer cells were transduced with either Sigma-Aldrich pooled shRNA library or corresponding control (CTL) shRNA. Limiting dilution experiments were conducted to determine cell number at which UMUC3 cells transduced with control shRNA would not give tumors; number was determined to be 50000 cells. Transduced UMUC3 cells were injected at 25 000 and 12 000 cells per site into 25 immunocompromised mice (two injection sites per mouse). B) Four tumors that grew more than 900mm3 30 days after initial injection were harvested. The y-axis is ×103. C) DNA from harvested tumors was sequenced to determine shRNA constructs. AGL was the most represented shRNA construct in four harvested tumors. The y-axis is ×103.
AGL Expression and Patient Prognosis and Metastasis
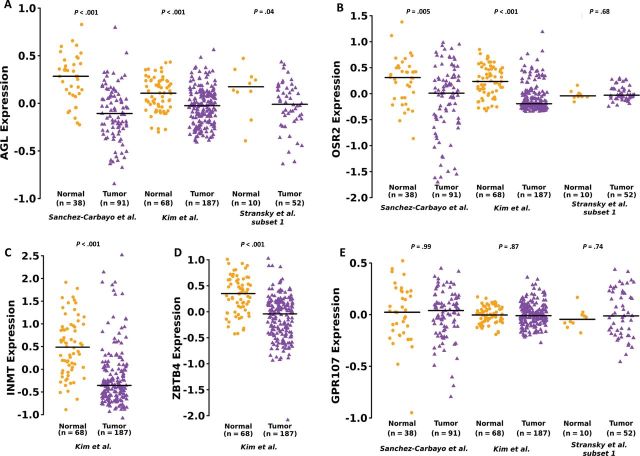
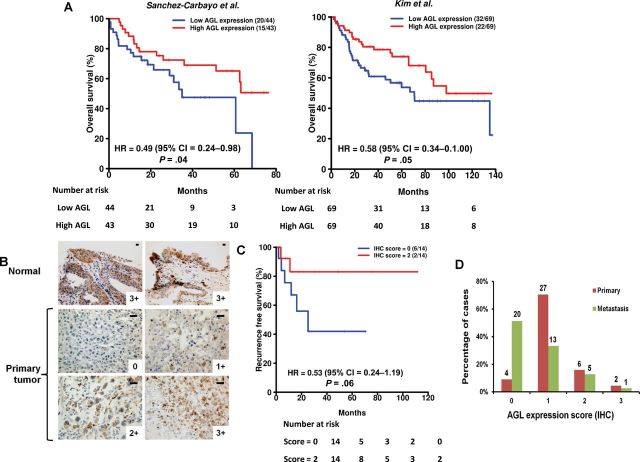
To focus mechanistic work on the most clinically relevant candidates, we investigated mRNA expression of these genes in human bladder cancer. Analysis across multiple patient datasets suggested that low AGL expression is robustly associated with tumor formation and clinically aggressive disease (Figure 2; Supplementary Tables 2 and 3, available online). Because AGL was also the top shRNA sequence read for each tumor (Supplementary Table 1, available online), we selected AGL for further investigation. We investigated if AGL mRNA or protein expression predicts patient survival. For mRNA analysis, we used three microarray datasets (Supplementary Table 2, available online) with survival information. Decreased overall survival was observed in patients with low tumor AGL expression in two patient datasets (Figure 3A; hazard ratio [HR] = 0.49, 95% CI = 0.24 to 0.98, P = .04; and HR = 0.58, 95% CI = 0.34 to 1.00, P = .05), although the exact nature of the relationship between continuous AGL expression and survival is not clear based on plots of smoothed martingale residuals against AGL expression (Supplementary Figure 1, available online). AGL was no longer predictive of survival when examined in a multivariate model that included age, sex, stage, and grade. IHC was performed on normal human urothelium and tumor samples (Figure 3B) with staining intensity scored on a scale of 0–3. All eight normal human urothelial samples had a score of 3. Consistent with the mRNA expression data, AGL protein expression stratified recurrence-free survival (Figure3C; HR = 0.53, 95% CI = 0.24 to 1.19, P = .06). When primary tumors and nodal metastases found at cystectomy were examined, 4 of 39 and 27 of 39 primary urothelial tumor samples were scored 0 or 1, respectively (Figure 3D), whereas 20 of 39 and 13 of 39 lymph node metastasis samples were so scored (P = .001, P = .03, respectively, compared to primary tumors).
Figure 2.
Expression of genes identified by RNA interference screen in bladder tumors. A) Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) mRNA expression in human urothelial tumors (total n = 330) compared to human normal bladder tissue (total n = 116) in three independent datasets. B) OSR2 mRNA expression in tumor samples (total n = 330) compared to normal urothelium (total n = 116) in three independent microarray datasets. C, D) INMT and ZBTB4 mRNA expression in tumors (total n = 187) compared to normal (total n = 68) in one dataset each. E) GPR107 mRNA expression between tumor (n = 330) and normal urothelium (total n = 116) in all datasets. Horizontal lines represent median of gene expression. Differences in distributions were tested by Wilcoxon rank-sum tests (P values shown in the figure). The statistical tests conducted were two-sided. Datasets are described in Supplementary Table 2 (available online).
Figure 3.
Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) expression in human cancer. A) Patients in two independent datasets were divided into two groups using median AGL expression. The two patient groups were compared using Cox proportional hazards models. Hazard ratios (HRs) with 95% confidence intervals (CIs) and log-rank P values are shown. Numbers in parentheses are the number of deaths followed by the number of patients in each group. B) Immunohistochemical (IHC) analysis of AGL expression in normal urothelium and bladder cancer. Scales represent a size of 20 µm. C) Recurrence-free survival rates in patients with low AGL IHC scores (0) and patients with high AGL IHC scores (2). HRs and two-sided log-rank P values are shown. D) AGL staining intensity in primary tumors compared to bladder cancer metastases to investigate AGL protein expression in relation to cancer progresses. All statistical tests were two-sided.
AGL Expression and Abnormally Branched Glycogen in Bladder Cancer Cells
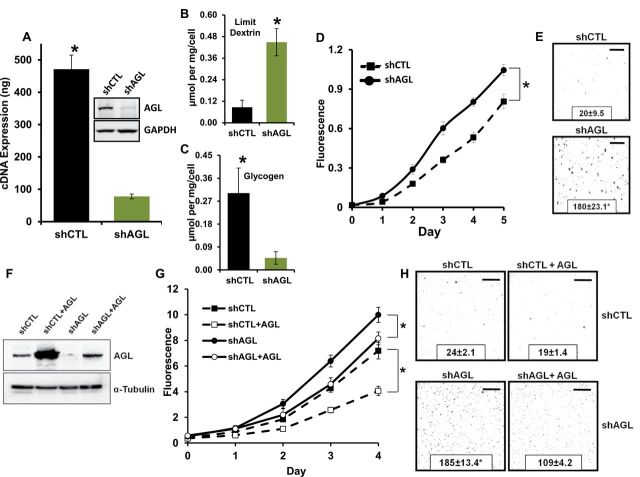
In GSD III, loss of AGL function results in cellular accumulation of limit dextrin (abnormally branched glycogen) in liver (15,16). To evaluate this in cancer cells, we transduced UMUC3 and T24T cancer cells with a different AGL shRNA (TRCN0000035082, Sigma-Aldrich) than the one used in the screening library (TRCN0000035079, Sigma-Aldrich). Expression of AGL was reduced at both the mRNA and protein levels in these cells (Figure 4A; Supplementary Figure 2A, available online, P < .001). Quantitative NMR analysis revealed that limit dextrin was increased in both UMUC3 and T24T shAGL cells (Figure 4B; Supplementary Figure 2B, available online, P < .001) and associated with an expected reciprocal decrease in normal glycogen (Figure 4C; Supplementary Figure 2C, available online, P < .001), confirming that depletion of AGL has biochemical sequelae similar to that of GSD III.
Figure 4.
Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) loss and tumor growth in vitro. A) AGL gene knockdown was validated by western blot analysis and real-time polymerase chain reaction in UMUC3 cell lines. Results are shown as mean ± SD (n = 3). B, C) Nuclear magnetic resonance was used to assess intracellular limit dextrin and glycogen levels. B) Limit dextrin and (C) normal glycogen in the UMUC3 shCTL (cells transduced with control plasmid) and shAGL (cells with stable knockdown of AGL) cell lines. Error bars represent SD. D, E) UMUC3 shCTL and shAGL cell growth in anchorage-dependent (D) and -independent (E) growth (n = 6) for proliferation assays (y-axis is ×103) and (n = 3) for colony formation assays. F) An shRNA-insensitive AGL construct was transiently overexpressed in shCTL and shAGL UMUC3 cells and overexpression was validated by western blot analysis 72 hours after transfection. G and H) Transient overexpression of AGL in shCTL and shAGL UMUC3 cells followed by proliferation assay (G) (y-axis is ×103) and colony formation assay (H) (n = 6) for proliferation assays and (n = 3) for colony formation assays. Details of assays described in the Methods. For proliferation assay, error bars represent SD; for colony formation assay, mean ± SD is represented. *P < .001 by two-sided Student t test (A–C, E, H) or repeated-measures analysis (D, G) based on a one-sided χ2 test statistic. All scale bars represent a size of 0.4cm.
AGL Depletion and In Vitro Bladder Cancer Cell Growth and Glycogenolysis
To generalize the screen findings on tumor growth, we examined anchorage-dependent and -independent growth of UMUC3 and T24T cells transduced with shRNA against AGL (shAGL) and control shRNA (shCTL) (Figure 4A; Supplementary Figure 2A, available online). AGL reduction enhanced growth in anchorage-dependent growth assays (Figure 4D, P < .001; Supplementary Figure 2D, available online, P < .001) and anchorage-independent growth assay (Figure 4E, mean ± SD, 180 ± 23.1 vs 20±9.5, P < .001; Supplementary Figure 2E, 218±22.9 vs 144±5.7, P < .001) compared to shCTL cell lines. Classical mitogenic pathways were not differentially regulated with AGL depletion (Supplementary Figure 3, available online). Additionally, an shRNA-insensitive AGL construct overexpressed in UMUC3 shCTL and shAGL (Figure 4F) reversed anchorage-dependent growth for both treatments (Figure 4G, P < .001) and anchorage-independent growth in shAGL cells (Figure 4H, mean ± SD, 185 ± 13.4 vs 109±4.2, P < .001), confirming the phenotypic specificity of AGL depletion.
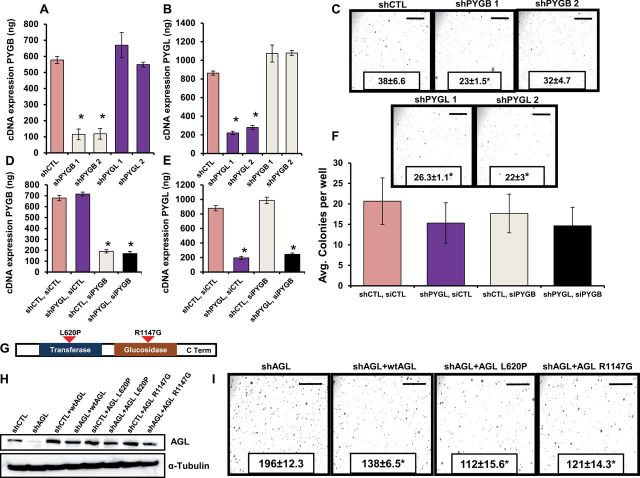
To determine if decreased glycogen breakdown due to AGL loss was driving increased bladder cancer cell growth, we evaluated the effect of glycogenolysis inhibition. Along with AGL, glycogen phosphorylase (PYG) isoforms (brain, liver, and muscle) contribute to glycogenolysis (17,18). The mRNA expression analysis of PYG isoforms in bladder cancer cell lines showed that brain and liver isoforms are expressed in these cells (data not shown). Unlike AGL, depletion of PYG isoforms (brain and liver) either alone (Figure 5, A and B, P < .001) or together (Figure 5, D and E, P < .001) did not increase anchorage-independent growth (Figure 5C; mean ± SD, shCTL = 38±6.6; shPYGB1 = 23±1.5, P = .01; shPYGB2 = 32±4.7; shPYGL1 = 26±1.1, P = .02; shPYGL2 = 22±3, P = .01, Figure 5F). These results suggest that AGL effects on bladder cell growth are not via reduced glycogenolysis.
Figure 5.
Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL)’s enzymatic function, inhibition of glycogenolysis, and tumor progression. Reverse transcription polymerase chain reaction (RT-PCR) analysis for glycogen phosphorylase brain (PYGB) (A) and liver (PYGL) (B) in UMUC3 cells transduced with short-hairpin (sh) RNAs. The terms shPYGB and shPYGL 1/2 indicate two different shRNAs used against glycogen phosphorylase brain and liver isoforms, respectively; shPYGB 1 and 2 are the shRNAs TRCN0000153339 and TRCN0000157479, respectively, described in the Supplementary Methods (available online); shPYGL 1 and 2 are the shRNAs TRCN0000119082 and TRCN0000119083, respectively, described in the Supplementary Methods. *P < .001 by two-sided Student t test. C) Anchorage-independent growth of UMUC3 cells transduced with shRNAs against glycogen phosphorylase brain (shPYGB1 and shPYGB2) and liver (shPYGL1 and shPYGL2) isoforms (n = 3). D, E) RT-PCR analysis for glycogen phosphorylase brain (D) and liver (E) isoform 48 hours after transient knockdown of PYGB in UMUC3 cells having stable PYGL knocked down. The small interfering RNA SMART pool from Dharmacon was used for PYGB knockdown (Supplementary Methods, available online); PYGL stable knockdown achieved with shRNA TRCN0000119082 was used for the experiment. *P < .001 by two-sided Student t test. F) Anchorage-independent growth of UMUC3 cells with dual knockdown of glycogen phosphorylase brain and liver isoform (n = 3). G) Cartoon of AGL showing the enzymatic domains. H) UMUC3 shCTL (cells transduced with control plasmid) and shAGL (cells with stable knockdown of AGL) cells were transfected with wild-type AGL, AGL transferase-null (L620P), or glycosidase-null (R1147G) variant constructs. AGL wild-type and variant overexpression was detected by western blot analysis. I) UMUC3 shAGL cells transfected with wild-type AGL and AGL enzymatic null variants were tested in an anchorage-independent growth assay (n = 3).*P < .001 by two-sided Student t test. Mean ± SD is presented for all the data in the figure. All scale bars represent a size of 0.4cm.
Because many metabolic genes impact tumor growth through their enzymatic activity (19,20), we sought to determine if the AGL effect on tumor growth is functionally dependent on either of its two enzymatic activities, glucotransferase and glucosidase. In UMUC3 shCTL and shAGL cells, we overexpressed shAGL-insensitive wild-type AGL or AGL variants lacking either of these enzymatic functions (21,22) (Figure 5G). Overexpression of wild-type AGL or either enzymatically inactive variant in UMUC3 shAGL cells (Figure 5H) reversed the increase in anchorage-independent growth observed with loss of AGL (Figure 5I, mean ± SD, shAGL = 196±12.3; shAGL + wtAGL = 138±6.5; shAGL + AGL-L620P = 112±15.6; shAGL + AGL-R1147G = 121±14.3; P < .001), again demonstrating that increased tumor growth observed with loss of AGL is unrelated to its role in glycogenolysis.
AGL Depletion and Tumor Growth In Vivo
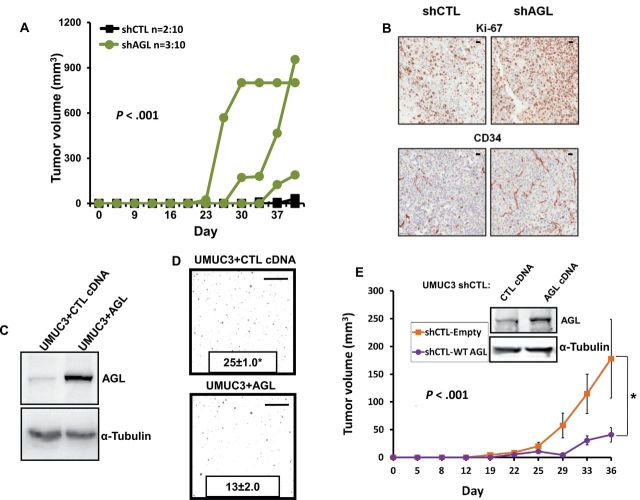
We evaluated the impact of AGL depletion on subcutaneous tumor growth by injecting 25000 UMUC3 or 100000 T24T shCTL or shAGL cells into immunodeficient mice. We observed enhanced tumor growth in mice injected with shAGL cells compared to mice injected with shCTL cells (Figure 6A; Supplementary Figure 2F, available online, P < .001). Immunohistochemical analysis of shAGL UMUC3 and T24T tumors showed statistically significant increased proliferation (Ki67: mean 127 vs 92, P = .042) and angiogenesis (CD34: mean 338 vs 169, P = .031) but no difference in apoptosis (caspase-3, 2.4 vs 2.1, P = .91) with respect to shCTL tumors (Figure 6B). We then evaluated the effect of AGL overexpression in vivo. Wild-type AGL or vector control was transiently transfected into UMUC3 and T24T parental cells (Figure 6C; Supplementary Figure 4A, available online) and stably transfected into UMUC3 shCTL cells. Cells overexpressing AGL had reduced anchorage-independent growth (Figure 6D, mean ± SD, UMUC3 + CTL = 25±1.0, UMUC3 + AGL = 13±2.0, P < .001; Supplementary Figure 4B [available online], T24T + CTL = 133±12.2, T24T + AGL = 60±6.7, P < .001) and subcutaneous tumor formation (Figure 6E, P < .001) compared to control cells. These experiments suggest that AGL expression levels inversely associate with bladder tumor growth.
Figure 6.
Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) expression and tumor growth in vivo. A) Nu/nu mice were injected subcutaneously on both flanks with UMUC3 expressing AGL short-hairpin RNA (shRNA) or the nontarget shRNA. Tumor volume was measured over time. P values represent the statistical difference by autocorrelation structure of order 1 method (13) of the total tumor volumes between the shAGL and shCTL tumors including tumors that did not grow from the total of 10 injection sites. B) Subcutaneous tumors formed from shAGL UMUC3 cells have statistically significant increases in proliferation (Ki-67, P = .04) and angiogenesis (CD-34, P = .03) markers compared to tumors with the control shRNA (shCTL) cells as determined by immunohistochemistry. Scales represent a size of 20 µm. C) UMUC3 cells were transiently transfected with an AGL-expressing construct. Cell lysates were collected 72 hours after transfection, and overexpression of AGL was determined by western blot analysis. D) UMUC3 cells transiently overexpressing AGL had reduced anchorage-independent growth in soft agar (n = 3, *P < .001). Mean ± SD is represented in the figure. Scale bars represent a size of 0.4cm. E) AGL was stably overexpressed in UMUC3 cells already transfected with shCTL and overexpression was determined by western blot analysis. Five mice each were injected on both flanks with 250000 cells per site of shCTL UMUC3 cells stably overexpressing AGL and control plasmid. Mice injected with shCTL UMUC3 cells overexpressing AGL had reduced tumor growth compared to mice injected with shCTL UMUC3 cells harboring empty vector. Error bars represent SD. *P < .001 by two-sided Student t test (D) or by repeated-measures analysis using an autocorrelation structure of order 1 (A, E).
AGL Expression and Aerobic Glycolysis and Dependency on Exogenous Glucose
Initial metabolite analyses revealed glucose levels in the cellular media of shAGL cells were lower compared to shCTL cells (Supplementary Figure 5A, P = .03; Supplementary Figure 5C, P = .02), whereas lactate in the media was greater in shAGL versus shCTL (Supplementary Figure 5, B and D, P = .04). These results suggested that reduction in AGL increases aerobic glycolysis (Warburg effect) (23). Importantly, in vivo, UMUC3 shAGL tumor xenografts (Figure 6A) had statistically significant higher lactate than shCTL tumors (P = .04). Similar observations were made with T24 cells (Supplementary Figure 5, E–G, available online; P = .04 for all). Because loss of AGL increases aerobic glycolysis, we investigated whether cells with reduced AGL levels were more dependent on exogenous glucose. Indeed, glucose starvation led to greater reduction of growth in shAGL compared to shCTL cells (Supplementary Figure 6A, available online; P < .001). We next treated shCTL and shAGL cells with 2-deoxyglucose (Sigma-Aldrich), a known glycolytic inhibitor (24), and found that proliferation of shAGL cells was decreased more than shCTL cells (Supplementary Figure 6B, available online; P = .03 and P = .04 for UMUC3 and T24T, respectively). These data suggest that reduced AGL in cancer cells leads to a greater dependence on exogenous glucose.
AGL Loss and De Novo Glycine Synthesis From Glucose
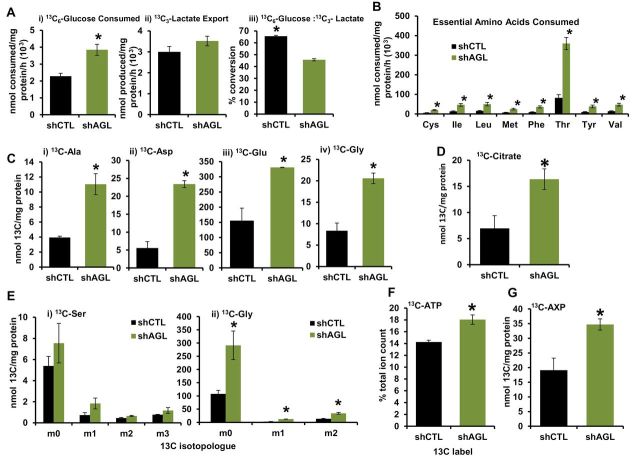
To gain insight into utilization of exogenous glucose upon AGL depletion, we performed metabolomic profiling of shCTL- and shAGL-UMUC3 cells. Cells were treated with 13C6-glucose, and then NMR, GC-MS, and FT-ICR-MS (25–32) analyses were used to determine the fate of glucose. Analysis of the media showed increased 13C6-glucose consumption and 13C3-lactate export in cells with AGL depletion (33,34) (Figure 7A i-ii, P = .04 and P > .05), implying greater glycolytic activity in shAGL cells. However, the fraction of glucose consumed and converted to lactate was lower in shAGL cells compared to shCTL cells (Figure 7A iii, P = .003), suggesting that the increased uptake of glucose is for processes beyond normal conversion to lactate (33,34). Similarly, shAGL cells consume more essential amino acids from the media (Figure 7B, P < .05), suggesting increased anabolic processes. We used heteronuclear single quantum coherence (HSQC) NMR analysis to assess the 13C-labeling of direct transamination and anaplerotic products of glycolysis and the tricarboxylic acid cycle (TCA) cycle from 13C6-glucose, including alanine, aspartate, and glutamate. Increased formation of these free amino acids from glucose occurred upon AGL depletion (Figure 7C i-iii, P = .02, .009, .02). Incorporation of 13C6-glucose into these amino acids is consistent with an increased contribution of glucose from glycolytic and TCA cycle flux, as represented by 13C-citrate levels (Figure 7D, P = .04), followed by transamination of the intermediates oxaloacetate, pyruvate, and α−ketoglutarate (34,35). Protein labeling paralleled these results, with approximately 25% greater accumulation of labeled amino acids (P = .031) in total cellular protein in AGL-depleted cells.
Figure 7.
Amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL) loss and metabolic reprogramming. UMUC3 shCTL (cells transduced with control plasmid) and shAGL (cells with stable knockdown of AGL) cells were treated with 13C6–glucose–containing media followed by collection of cellular extracts and media for analysis by NMR, gas chromatography-mass spectrometry (GC-MS), and Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS). A) 13C6–glucose consumption (P = .04) (i), 13C3–lactate export (P > .05) (ii), and 13C6–glucose to 13C3–lactate conversion (P = .003) (iii) in the media 24 hours after 13C6–glucose treatment by UMUC3 shCTL and shAGL cells as analyzed by 1H NMR. B) Uptake of essential amino acids from cell culture media, as analyzed by GC-MS. Cys, Ile, Leu, Met, Phe, Thr, Tyr, and Val indicate cysteine, isoleucine, leucine, methionine, phenylalanine, threonine, tryptophan, and valine, respectively. *P = .02, .04, .04, .04, .04, .01, .04, and .03, respectively. C, i–iv) 13C-amino acid accumulation from glucose in UMUC3 shCTL and shAGL cells by 1D 1H HSQC NMR analysis. Ala, Asp, Glu, and Gly stand for alanine, aspartate, glutamate, and glycine, respectively. *P = .02, .009, .02, and .01, respectively. D) Krebs cycle intermediate 13C-citrate buildup in UMUC3 shCTL and shAGL cells. *P = .04. E) GC-MS analysis for buildup of (i) serine (Ser) and (ii) glycine (*P = .03, <.001, and = .03, respectively) (Gly) from 13C6-glucose and nonglucose sources in the above-mentioned cells. m0 indicates all 12C isotopologue derived from nonglucose sources, whereas m1-3 indicates 1-3 13C carbon-containing isotopologues derived from labeled glucose. F) Increased incorporation of 13C carbon from 13C6–glucose into the adenine base of ATP in UMUC3 cells without AGL, as analyzed by FT-ICR-MS (*P = .04). G) Increased 13C carbon incorporation from glucose into the ribose moiety of adenine nucleotides (AXP) as analyzed by 1D 1H HSQC NMR (*P = .03). P value by two-sided Student t test. Mean ± SD is represented in the figure.
We observed increased incorporation of 13C6-glucose into glycine in shAGL cells (Figure 7C iv, P = .01). Glycine can be synthesized in cells from glucose via serine (36). Increased glycine synthesis from glucose is noteworthy because glycine was provided in the medium (0.4mM), yet its de novo synthesis from glucose via serine was increased. Thus, we examined changes in labeling patterns of serine and glycine between shCTL and shAGL cells. For serine, we found no statistically significant change in unlabeled (m0) or labeled (13C3-serine [m3]) levels in shAGL versus shCTL cells (Figure 7E i). However, labeled (13C2-glycine [13C2-glycine or m2]) and unlabeled glycine was statistically significantly increased in AGL-depleted cells (Figure 7E ii, P = .03, P < .001, P = .03), suggesting that more de novo synthesized serine is diverted to glycine synthesis in AGL-depleted cells.
Glycine is a precursor of the purines (19,36) required for DNA synthesis. Because enhanced synthesis of glycine may drive this process, we analyzed nucleotides by FT-ICR-MS and showed that more 13C6-glucose was incorporated into the purine ring of ATP (Figure 7F, P = .04) and GTP (data not shown) in shAGL cells versus shCTL cells. Consistently, NMR analysis showed that shAGL cells increased the fraction of de novo synthesized ribose moiety of adenine (Figure 7G, P = .03) and uracil nucleotides (data not shown) from 13C-glucose. These data are consistent with an increased flux through the pentose phosphate pathway, increased incorporation of ribose into purine and pyrimidine nucleotides, or both. FT-ICR-MS analysis confirmed increased pyrimidine ring synthesis from glucose in shAGL cells (data not shown). Thus, in shAGL cells, glucose becomes a more important carbon source for nucleotide ring and ribose moiety de novo synthesis, both required to make RNA and DNA in proliferating cells (20). Together, these detailed analyses suggest that cells with lower AGL undergo substantial metabolic reprogramming, and an important manifestation of this is increased de novo glycine synthesis from serine.
AGL Loss, Tumor Growth, and Expression of Serine Hydroxymethyltransferase 2
The above-mentioned findings and the lack of any statistically significant findings with 13C-glucose incorporation into lipids (phospholipids, de novo synthesized fatty acids, and cholesterol) (Supplementary Figure 7, available online), coupled with the knowledge that glycine synthesis is required for rapid proliferation of cancer cells (19,37,38), led us to hypothesize that AGL depletion drives tumor growth via inducing increased glycine synthesis from glucose and serine. To define the genes responsible for this, we performed gene expression profiling and focused on genes that affect glycine synthesis. We discovered that SHMT2 (19,37), a gene that drives mitochondrial conversion of serine to glycine, is higher in AGL-depleted UMUC3 cells (data not shown).
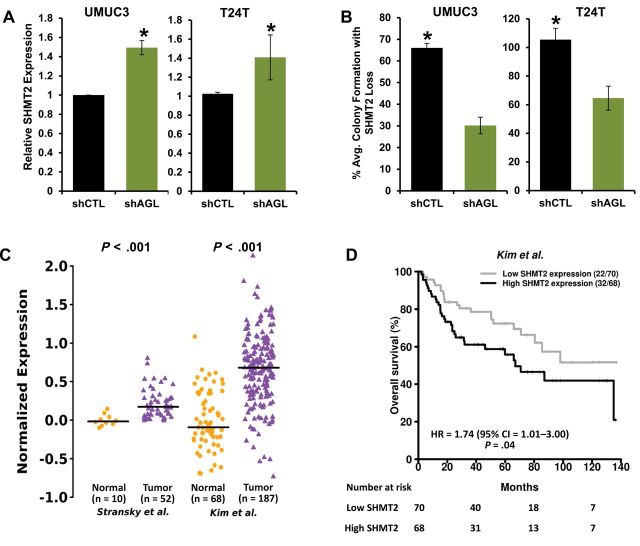
Using qPCR, we observed that SHMT2 was elevated upon depletion of AGL in both UMUC3 and T24T cells (Figure 8A, P = .02 and .01). To determine whether SHMT2-driven glycine synthesis preferentially drives proliferation of cancer cells with low AGL, we developed stable cell lines depleted of both AGL and SHMT2 (Supplementary Figure 8, A–D, available online). We observed a decrease in anchorage-independent growth of UMUC3 and T24T cells deficient in both AGL and SHMT2, compared to cells with AGL depletion only (Figure 8B, P < .001). To evaluate the clinical relevance of the relationship between AGL and SHMT2, we analyzed AGL and SHMT2 mRNA expression in bladder cancer patients (patient and tumor demographics provided in Supplementary Table 2, available online). Expression of AGL and SHMT2 were negatively correlated (Supplementary Figure 9, A–C, available online; r = –0.24 [P = .08], r = –0.37 [P = .02], and r = –0.39 [P < .001]), consistent with an observed increase in SHMT2 levels with AGL loss. SHMT2 also showed higher expression in tumors versus normal bladder tissues (Figure 8C, P < .001), high-grade versus low-grade tumors (Stransky et al. (8), P = .004) and muscle-invasive versus non-muscle-invasive tumors (Blaveri et al. (12), P = .02) in two independent patient datasets. Additionally, high expression of SHMT2 mRNA associated with poor overall survival in one cohort (Figure 8D, HR = 1.74, 95% CI = 1.01 to 3.00, P = .04), although the exact relationship between SHMT2 expression and survival is not clear (Supplementary Figure 10, available online). Here we conclusively illustrate that cells with AGL depletion are dependent on SHMT2 for rapid tumor growth.
Figure 8.
Serine hydroxymethyltransferase 2 (SHMT2)– and amylo-α-1, 6-glucosidase, 4-α-glucanotransferase (AGL)–mediated tumor growth. A) Quantitative reverse transcription polymerase chain reaction for SHMT2 mRNA expression with AGL loss in UMUC3 and T24T cells (*P = .02 and .01, respectively, by two-sided Student t test). Mean ± SD is represented. B) SHMT2 loss and anchorage-independent growth of UMUC3 and T24T cells with AGL loss compared to cells transduced with control short-hairpin RNA (*P < .001 by two-sided Student t test). Mean ± SD is represented. C) SHMT2 mRNA expression in human urothelial tumors (total n = 239) compared to human normal bladder tissue (total n = 78) in two independent datasets. Horizontal lines represent median of SHMT2 expression. Differences in distributions were tested by Wilcoxon rank-sum test. D) SHMT2 expression and survival rates. Patients in the independent dataset were divided into two groups using median SHMT2 expression. The two patient groups were compared using Cox proportional hazards models. Hazard ratios (HRs) with 95% confidence intervals (CIs) and log-rank P values are shown. Numbers in parentheses are number of deaths followed by number of patients in each group. All statistical tests were two-sided.
Discussion
Here, we use a genome-wide lentiviral shRNA library for in vivo identification of functionally important growth regulators of bladder cancer. This approach identified several candidate genes, including AGL, as novel players in tumor growth. Interestingly, germline loss of AGL leads to GSD III (38), a rare condition with an incidence of 1:100000 in North America (16). Recent studies suggest that the only increased rate of cancer seen in patients with GSD III is in hepatocellular carcinoma (15). However, this may be due to the chronic stresses placed on the liver over a prolonged period of time and not due to defects in cellular signaling in liver cells. We investigated whether patients with GSD III have increased rates of bladder cancer. Analysis of two well-maintained patient databases from the Faroe Islands (which has a high rate of individuals with GSD III) and the Netherlands revealed that patients in the databases do not have a higher rate of bladder cancer (personal communications from Drs David Weinstein, Christiaan P. Sentner, and G. Peter A. Smit). However, it is important to note that bladder cancer is a disease predominantly diagnosed at an advanced age, and the aforementioned databases currently have only a small number of individuals older than 60 years. Thus, it will be important to monitor these GSD III patients as they age.
AGL has glucanotransferase and glucosidase catalytic activities (39) and, along with glycogen phosphorylase, is responsible for complete breakdown of glycogen to glucose-6-phosphate (39). We showed that the enzymatic activities of AGL are not required for its growth-suppressive role in bladder cancer. Such findings are consistent with other metabolic enzymes, such as hexokinase, lactate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, glucose-6-phosphate isomerase, pyruvate kinase isozyme M2, and aldolase also have nonmetabolic functions (20,40–42). Furthermore, both metabolic and nonmetabolic activities of these proteins play a role in tumor progression (41,43). We validated that inhibition of glycogenolysis in general is not the primary driver of bladder cancer growth. Thus, there exists a role for AGL in tumor suppression that exists outside its glycogenolysis role.
Our metabolomic analysis showed reduced levels of AGL led to increased glycine synthesis. Recent reports show that an increase in mitochondrial glycine synthesis is required for rapid proliferation of cancer cells (19). Because glucose is not the only source of glycine in cells, and AGL-depleted cells had increased accumulation of glycine from nonglucose sources, we suspected that glycine synthesis was reprogrammed in cells with diminished AGL. Indeed, expression of SHMT2, which promotes glycine synthesis, is increased in cells depleted of AGL and is important for their rapid proliferation. The clinical relevance of this finding was supported by an inverse correlation of expression of these two genes in patient tumor samples. Our findings that cells with low AGL levels have increased reliance on glycine represent a metabolic vulnerability for selectively targeting rapid cancer cell proliferation in patients with low AGL.
Clinical data suggest that AGL and SHMT2 expression may be important prognostic indicators for bladder cancer patients. This supports our functional data and indicates that low AGL and high SHMT2 expression contribute to bladder cancer growth in patients. Both are clinically relevant candidates for further study as mediators of a therapeutically targetable pathway. However, several questions still remain regarding AGL loss in bladder cancer. First, there are no recorded mutations of AGL in patients with bladder cancer; therefore, it is important to determine how AGL expression is being reduced in these patients. Furthermore, although depletion of SHMT2 decreased the growth advantage of shAGL cells, rescue of the AGL phenotype was not complete, suggesting that additional downstream effectors may be playing a role. It will be important to uncover these other effectors that are induced upon reduced AGL expression. Finally, although we have shown here that AGL contributes to tumor growth, we do not yet know the effect of AGL loss on carcinogenesis, and whether AGL is “turned off” during formation of the initial tumor or during progression. This information will be important for the development of personalized therapies in bladder cancer patients with reduced AGL expression.
Funding
This work was supported in part by the National Institutes of Health (grants CA142163 to CP, CA143971 to DT, and ES022191 and CA118434 to TF); the Cancer League of Colorado, Inc (grant AWD-132739 to SGui); the National Science Foundation (EPSCoREPS-0447479 to TF); the Kentucky Challenge for Excellence (to ANL); the National Center for Research Resources (grant P20RR018733); and the J.G. Brown Foundation for NMR support. None of the study sponsors had a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
We thank Dr David Brautigan for critical review of the manuscript; Dr Erin Griner for technical assistance; Andrea Merz and Dr Natalie Serkova in the University of Colorado Cancer Center Shared Resources supported by National Institutes of Health grant P30CA046934 for analysis of cellular metabolites and AGL immunohistochemistry on the tumor xenografts; Drs Adam Heuberger and Jessica Prenni at Colorado State University in the Proteomics and Metabolomics Facility for analysis of cellular metabolites; Dr Richard Higashi and Sathyaramya Balasubramaniam in CREAM and Dr Sengodagounder Arumugam in the NMR Facility of the J.G. Brown Cancer Center at University of Louisville for processing extracts for SIRM analysis and for SIRM measurement by NMR, respectively; Dr David Weinstein, Director of the Glycogen Storage Disease Program at the University of Florida, for GSD III patient data from the Faroe Islands; and Drs Christiaan P. Sentner and G. Peter A. Smit at the University Medical Center Groningen for GSD III patient data from Groningen. DT conceived the study and formulated the hypothesis. CO aided in the in vitro and in vivo phenotypic experiments. YR and GD performed statistical analysis on the in vivo xenograft, microarray data, and IHC data. SK and AS conducted the initial shRNA pool library screen and developed the UMUC3 shCTL and shAGL lines. SGup and DH performed IHC analysis on the human normal, primary tumor, and metastatic tissue specimens. SGui and CP developed T24T and T24 shCTL and shAGL cell lines, validated AGL KD through western blot and/or real-time PCR, conducted in vitro anchorage-dependent and -independent growth assays, conducted in vivo tumor formation assays, initial NMR samples, and glucose deprivation experiments. SGui investigated the role of glycogen phosphorylase and AGL enzymatic activity on bladder cancer cell proliferation. SGui and CRL prepared samples for 13C-glucose-based SIRM experiments (NMR/GC-MS) and investigated the role of glycine synthesis under AGL loss. CRL helped analyze the NMR/GC-MS data and prepare the manuscript. MH, PL, ANL, and TW-MF carried out the design and NMR/GC-MS profiling and data analysis. SGui, CP, JED, HH, MH, ANL, TW-MF, and DT interpreted data and wrote the paper.
References
- 1. Gregory MA, Phang TL, Neviani P, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010;18(1):74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coussens MJ, Corman C, Fischer AL, et al. MISSION LentiPlex pooled shRNA library screening in mammalian cells. J Vis Exp. 2011;58. 10.3791/3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. [DOI] [PubMed] [Google Scholar]
- 4. Gildea JJ, Golden WL, Harding MA, et al. Genetic and phenotypic changes associated with the acquisition of tumorigenicity in human bladder cancer. Genes Chromosomes Cancer. 2000;27(3):252–263. [DOI] [PubMed] [Google Scholar]
- 5. Nitz MD, Harding MA, Smith SC, et al. RREB1 transcription factor splice variants in urologic cancer. Am J Pathol. 2011;179(1):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang H, Owens C, Chandra N, et al. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70(21):8760–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansel DE, Platt E, Orloff M, et al. Mammalian target of rapamycin (mTOR) regulates cellular proliferation and tumor growth in urothelial carcinoma. Am J Pathol. 2010;176(6):3062–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38(12):1386–1396. [DOI] [PubMed] [Google Scholar]
- 9. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 10. Kim W-J, Kim E-J, Kim S-K, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanchez-Carbayo M, Socci ND, Lozano J, et al. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24(5):778–789. [DOI] [PubMed] [Google Scholar]
- 12. Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11(11):4044–4055. [DOI] [PubMed] [Google Scholar]
- 13. Pinheiro J, Bates D, DebRoy S, et al. nlme: Linear and Nonlinear Mixed Effects Models. In: Team RDC, (ed). R package version 3.1–102; 2011. [Google Scholar]
- 14. Illa-Bochaca I, Fernandez-Gonzalez R, Shelton DN, et al. Limiting-dilution transplantation assays in mammary stem cell studies. Methods Mol Biol. 2010;621:29–47. [DOI] [PubMed] [Google Scholar]
- 15. Demo E, Frush D, Gottfried M, et al. Glycogen storage disease type III-hepatocellular carcinoma a long-term complication? J Hepatol. 2007;46(3):492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dagli A, Sentner CP, Weinstein WA. Glycogen Storage Disease Type III. Seattle: University of Washington; 2011. [Google Scholar]
- 17. Hicks J, Wartchow E, Mierau G. Glycogen storage diseases: a brief review and update on clinical features, genetic abnormalities, pathologic features, and treatment. Ultrastruct Pathol. 2011;35(5):183–196. [DOI] [PubMed] [Google Scholar]
- 18. Roach PJ, Depaoli-Roach AA, Hurley TD, et al. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441(3):763–787. [DOI] [PMC free article] [PubMed]
- 19. Jain M, Nilsson R, Sharma S, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng A, Zhang M, Okubo M, et al. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet. 2009;18(11):2045–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng A, Zhang M, Gentry MS, et al. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21(19):2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 24. Brown J. Effects of 2-deoxyglucose on carbohydrate metablism: review of the literature and studies in the rat. Metabolism. 1962;11:1098–1112. [PubMed] [Google Scholar]
- 25. Fan TW, Lane AN, Higashi RM, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer. 2009;8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan TW-M, Yuan P, Lane AN, et al. Stable isotope-resolved metabolomic analysis of lithium effects on glial-neuronal metabolism and interactions. Metabolomics. 2010;6(2):165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fan T, Lane A, Higashi R, et al. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics 2011;7(2):257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane AN, Fan TW, Bousamra M, et al. Stable isotope-resolved metabolomics (SIRM) in cancer research with clinical application to nonsmall cell lung cancer. OMICS. 2011;15(3):173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane AN, Fan TW-M, Higashi RM, et al. Stable isotope tracing in metabolic pathways. Presentation given at JG Brown Cancer Center, University of Louisville, KY. 7–11 April 2008. [Google Scholar]
- 30. Moseley H, Lane A, Belshoff A, et al. A novel deconvolution method for modeling UDP-GlcNAc biosynthetic pathways based on 13C mass isotopologue profiles under non steady-state conditions. BMC Biol. 2011;9(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fan TW-M, Tan JL, McKinney MM, et al. Stable isotope resolved metabolomics analysis of ribonucleotide and rna metabolism in human lung cancer cells. Metabolomics. 2012;8(3):517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lorkiewicz P, Higashi RM, Lane AN, et al. High information throughput analysis of nucleotides and their isotopically enriched isotopologues by direct-infusion FTICR-MS. Metabolomics. 2012;8(5):930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. 2nd ed. New York: W. H. Freeman; 1993. [Google Scholar]
- 34. Murray RK, Mayes PA, Rodwell VW, Granner DK. Harper’s Illustrated Biochemistry. 26th ed. New York: McGraw-Hill; 2003. [Google Scholar]
- 35. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino acid. J Biol Chem 2012;287(24):19786–19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hebbring SJ, Chai Y, Ji Y, et al. Serine hydroxymethyltransferase 1 and 2: gene sequence variation and functional genomic characterization. J Neurochem. 2012;120(6):881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen YT, Burhchell A, Scriver CR, et al. The Metabolomic and Molecular Basis of Inherited Disease. New York: McGraw-Hill. [Google Scholar]
- 39. Greenberg CC, Jurczak MJ, Danos AM, et al. Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways. Am J Physiol Endocrinol Metab. 2006;291(1):E1–E8. [DOI] [PubMed] [Google Scholar]
- 40. Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Cancer. 2006;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ritterson Lew C, Tolan DR. Targeting of several glycolytic enzymes using RNA interference reveals aldolase affects cancer cell proliferation through a non-glycolytic mechanism. J Biol Chem. 2012;287(51):42554–42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furuta E, Okuda H, Kobayashi A, et al. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805(2):141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.