Abstract
The family of aquaporins (AQPs), or major intrinsic proteins (MIPs), includes integral membrane proteins that function as transmembrane channels for water and other small molecules of physiological significance. MIPs are classified into five subfamilies in higher plants, including plasma membrane (PIPs), tonoplast (TIPs), NOD26-like (NIPs), small basic (SIPs) and unclassified X (XIPs) intrinsic proteins. This study reports a genome-wide survey of MIP encoding genes in sweet orange (Citrus sinensis L. Osb.), the most widely cultivated Citrus spp. A total of 34 different genes encoding C. sinensis MIPs (CsMIPs) were identified and assigned into five subfamilies (CsPIPs, CsTIPs, CsNIPs, CsSIPs and CsXIPs) based on sequence analysis and also on their phylogenetic relationships with clearly classified MIPs of Arabidopsis thaliana. Analysis of key amino acid residues allowed the assessment of the substrate specificity of each CsMIP. Gene structure analysis revealed that the CsMIPs possess an exon-intron organization that is highly conserved within each subfamily. CsMIP loci were precisely mapped on every sweet orange chromosome, indicating a wide distribution of the gene family in the sweet orange genome. Investigation of their expression patterns in different tissues and upon drought and salt stress treatments, as well as with ‘Candidatus Liberibacter asiaticus’ infection, revealed a tissue-specific and coordinated regulation of the different CsMIP isoforms, consistent with the organization of the stress-responsive cis-acting regulatory elements observed in their promoter regions. A special role in regulating the flow of water and nutrients is proposed for CsTIPs and CsXIPs during drought stress, and for most CsMIPs during salt stress and the development of HLB disease. These results provide a valuable reference for further exploration of the CsMIPs functions and applications to the genetic improvement of both abiotic and biotic stress tolerance in citrus.
Introduction
Aquaporins (AQPs) are integral membrane proteins that assist the rapid movement of water as well as other low molecular weight molecules across cellular membranes [1–3]. AQPs belong to the ancient family of major intrinsic proteins (MIPs) found in microorganisms, plants and animals. While a small number of different AQPs have been identified (2 in E. coli, 9 in S. cerevisiae, 11 in C. elegans and 13 in mammals [4]), a surprisingly large number of MIP homologues have been found in plants; for example, 35 AQPs were found in Arabidopsis [5], 36 in Zea mays [6], 33 in Oryza sativa [7], 28 in Vitis vinifera [8], 55 in Populus trichocarpa [9], 71 in Gossypium hirsutum [10], 47 in Solanum lycopersicum [11] and 66 in Glycine max [12]. These observations highlight a major role for plant MIPs as key regulators of the intricate flows of water and solutes required for growth and adaptive responses to the ever-changing environment.
Plant MIPs were originally categorized into four subfamilies on the basis of sequence homologies and subcellular localization: plasma membrane (PIP), tonoplast (TIP), nodulin-like (NIP) and small basic (SIP) intrinsic proteins [2,13]. More recently, studies in the moss Physcomitrella patens revealed the presence of novel AQP isoforms in addition to the four conserved plant AQP subfamilies: a homologue of the Escherichia coli intrinsic protein GlpF (GIPs), intrinsic hybrid proteins (HIPs) and unclassified X intrinsic proteins (XIPs) [9,13–17]. XIP homologues have also been identified in some higher plants, such as Solanum lycopersicum, Populus trichocarpa and Glycine max [9,11,12,15]. These findings suggest that the family of plant MIPs is larger and much more complex than previously anticipated and, hence, may play critical roles in a wide range of biological processes that go far beyond the current knowledge.
AQP-mediated water transport in plants has been implicated to play a central regulatory step in diverse biological processes, including cell elongation, seed germination and osmoregulation [18]. In addition, AQPs facilitate the transport of small uncharged molecules of physiological significance like glycerol, urea, boric acid, silicic acid, hydrogen peroxide (H2O2), ammonia (NH3) and carbon dioxide (CO2) through the plant cell membranes [1,2] and also regulate phloem sap loading and unloading, stomatal and leaf movement, and cytoplasmic homeostasis [1,2,13,19]. Therefore, it is not surprising that their expression and biological activities have been shown to be developmentally and differentially regulated in a cell-specific manner, via phytohormones such as abscisic acid (ABA), gibberellins and possibly brassinosteroids, and by environmental signals such as light, water stress, nematode infection, low temperature, and salinity [4]. However, a general expression pattern among the distinct MIP isoforms cannot be distinguished, as they are either up- or downregulated depending on the stimulus and/or the cell-type studied [4,19,20]. This difference in transcriptional regulation suggests that each MIP isoform may play a distinct role in plant growth, development and stress response [4].
As a major horticultural crop, the cultivated Citrus spp. face constant biotic and abiotic constraints in the main regions of production, including drought, salinity, extreme temperatures and serious diseases like Huanglongbing (HLB, or citrus greening), which are predicted to increase in intensity, frequency, and geographic extent as a consequence of global climate change. Despite the highlighted importance of AQPs, there are only a few studies to date on citrus MIPs and their predicted role in the transport of water and solutes required for plant growth, development and adaptive responses to the environment. The expression of two MIP genes, PIP1 and PIP2, has been investigated in roots of Poncirus trifoliata (L.) Raf., Cleopatra mandarin (C. reshni Hort exTan.) and one of their hybrids, subjected to moderate water deficit [21], and in roots of P. trifoliata, Cleopatra mandarin and Carrizo citrange (C. sinensis [L.] Osb.×P. trifoliata [L.] Raf.), subjected to salt treatment [22]. PIP1 and PIP2 gene expression differences were correlated with alterations in root hydraulic conductance (Kr) and chloride (Cl-) exclusion from leaves and, hence, tolerance to water and salt stresses, respectively. With the recent completion and publication of the genome sequences of sweet orange [23–25], it is now possible to identify and characterize the complete repertoire of MIPs in citrus, as well as to carry out comparative genome analysis in order to understand their evolutionary history. Therefore, the objective of the present study was to identify sweet orange MIP genes through a genome-wide analysis and to characterize their sequences, evolutionary relationships, putative functions and expression patterns in various tissues and in response to abiotic and biotic stresses. This is the first comprehensive study of the MIP gene family in sweet orange, providing valuable information for further exploration of the functions of this important gene family in citrus.
Materials and Methods
Plant materials and stress treatments
Two-year-old sweet orange plants grafted on Rangpur lime (C. limonia Osbeck), a rootstock highly resistant to drought, were used in the drought stress experiment. Plants were first pruned and acclimatized to greenhouse conditions (25±4°C, 16 h of light and relative humidity oscillating between 80 and 95%) during 90 days to obtain adequate root development and uniform leaf flushes. During acclimatization, plants were grown in plastic pots of 45-L, containing a mixture of soil and sand (ratio 3:1) and micronutrient mix FTE (50g per pot), irrigated with tap water twice a week, and fertilized weekly with 1 liter of the following nutrient solution: 1.0g l-1 Ca(NO3)2, 0.4g l-1 KNO3, 0.6g l-1 MgSO4 and 0.4g l-1 NH4H2P04 (MAP). Thereafter, the pots were closed with aluminum foil to prevent water loss by evaporation, and a set of 10 plants was randomized over the experimental area and subjected to the following treatments: (i) 5 plants in control, in which plants were maintained at leaf predawn water potential values of -0.2 to -0.4 MPa by daily irrigation and (ii) 5 plants in drought, in which the plants were exposed to a progressive soil water deficit until their leaves reach predawn water potential values of -1.5 MPa. The leaf predawn water potential was recorded on the third fully expanded mature leaf from the apex of each plant, between 2 AM and 4 AM, using a Scholander-type pressure pump (m670, Pms Instrument Co., Albany, USA).
For salt treatment, sweet orange seeds were germinated in vitro as described by de Oliveira et al. [26]. Twenty-day-old seedlings of nucellar origin were selected based on their uniformity, and transferred to MS medium alone (control) or containing 150 mM NaCl (Merck, Darmstadt, Germany). Each treatment consisted of 15 nucellar plants (biological replicates). Leaves and roots were harvested 20 days after the treatments and immediately frozen in liquid nitrogen and stored at -80°C.
Plants were infected with ‘Candidatus Liberibacter asiaticus’ as described in Fan et al. [27]. Briefly, two-year-old seedlings of rough lemon (C. jambhiri Lush.) and sweet orange (C. sinensis L. Osbeck) were graft-inoculated with bud wood from ‘Ca. L. asiaticus’ infected ‘Carrizo’ citrange trees kept under greenhouse conditions. For controls, the plants were grafted with bud wood from healthy Carrizo trees. All these plants were kept in a United States Department of Agriculture Animal and Plant Health Inspection Service and Center for Disease Control-approved and secured greenhouse at the University of Florida, Citrus Research and Education Center, Lake Alfred. Three biological replicates were produced for each citrus species in each treatment. Quantitative real-time PCR was performed to confirm the presence of ‘Ca. L. asiaticus’ in the inoculum source and inoculated plants as described in Li et al. [28]. Four fully expanded leaves were sampled separately from ‘Ca. L. asiaticus’ inoculated plants and mock-inoculated plants (used as controls) of each citrus species at 0, 7, 17, and 34 weeks after inoculation (WAI). Leaves were immediately frozen in liquid nitrogen and stored at -80°C until use. Three biological replicates were produced for each condition. In total, 12 plants with 48 leaf samples were collected (2 species x 2 treatments x 3 replicates x 4 time points).
Identification and classification of CsMIPs
The Hidden Markov Model profile of the PFAM (http://pfam.sanger.ac.uk/) [29] motif PF00230 (major intrinsic protein) was used as a keyword to search the sweet orange genome sequence database (http://www.phytozome.org/citrus/) [25]. The KEGG Orthology (KO) terms K09872 (aquaporin PIP), K09873 (aquaporin TIP), K09874 (aquaporin NIP) and K09875 (aquaporin SIP) were also used as keywords to search the sweet orange genome sequence at Phytozome. To avoid the deficiencies of the automatic annotation, the 35 Arabidopsis thaliana MIP protein sequences were retrieved from TAIR (http://www.arabidopsis.org/), according to previous reports [5,30], and also used to align with the sweet orange genome sequence assembly available at Phytozome using the TBLASTN tool. After merging the results from all these strategies, unique entries (with unique locus ID) were identified to remove the redundancy. The resulting sequences were manually inspected for the presence of characteristic and functionally important MIP amino acids and motifs.
The sweet orange MIPs were classified in different isoforms based on sequence analysis of the multiple alignments and on their phylogenetic relationship with those clearly classified MIPs of Arabidopsis thaliana, Ricinus communis and Nicotiana benthamiana, downloaded from the TAIR and NCBI databases. Multiple sequence alignments of the deduced amino acid sequences of CsMIPs and those of A. thaliana, R. communis and N. benthamiana were performed using the default parameters of ClustalW [31]. The dendrogram was generated by the MEGA 6 program [32] using the Neighbor-Joining (NJ) method [33] and bootstrap analysis (1,000 replications).
Analysis of CsMIP protein properties and conserved amino acid residues
Information about coding sequence (CDS), full-length sequence and predicted amino acid sequence was obtained for each sweet orange MIP gene from the Phytozome database. The GRAVY (grand average of hydropathy), molecular weight and isoelectric point (pI) of the deduced amino acid sequences were predicted by the PROTPARAM tool available on the Expert Protein Analysis System (ExPASy) proteomics server (www.expasy.ch/tools/protparam.html). The subcellular localization of MIP proteins was predicted using the WoLF PSORT tool available at http://www.genscript.com/psort/wolf_psort.html. Careful visual inspection of amino acid sequence alignments were performed to identify the characteristic MIP amino acids and motifs and the residues in seven key positions that have been reported to be specific for each functional subgroup [12,30,34,35].
Analysis of promoter sequences and chromosomal locations of CsMIPs
One kb upstream region from the translation start site was extracted from all the sweet orange MIP genes and subsequently analyzed in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) to identify the presence of the stress-responsive cis-acting regulatory elements ABRE (ABA-responsive element; ACGTG), DRE/CRT (dehydration responsive element/C-repeat; G/ACCGAC), MYBS (MYB binding site; TAACTG) and LTRE (low-temperature-responsive element; CCGAC) in their promoters. The physical locations of CsMIPs were determined by confirming the starting position of all genes on each chromosome, using BLASTN searching against the local database of the Citrus sinensis Annotation Project (CAP; http://citrus.hzau.edu.cn/orange/). MapChart software was used to plot the gene loci on the sweet orange chromosomes [36].
Expression analysis of CsMIPs
Total RNA isolation, cDNA synthesis and quantitative real-time RT-PCR (qPCR) analysis were performed as described previously [26]. qPCR primers were designed appropriately to avoid the conserved regions. Primer sequences are shown in detail in S1 Table. Glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2) was used as an internal reference gene to normalize expression among the different samples [37]. Data were obtained from a pool of three biological replicates that were individually validated.
RNA-seq data were downloaded from CAP [24] and used to analyze the expression patterns of CsMIPs in different tissues, namely callus (C), flower (Fl), leaf (L), fruit (Fr), and mixed tissues from fruit at three developmental stages (Mix.1, Mix.2, and Mix.3). The heatmap was generated using R 3.1.0 software.
Results and Discussion
MIP encoding genes in the sweet orange genome
Searches in the sweet orange genome sequence database at Phytozome using annotation information, as well as the 35 protein sequences of the complete set of A. thaliana MIPs (AtMIPs) as query sequences, resulted in the identification of 34 different genes encoding C. sinensis MIPs (CsMIPs) (Table 1). The retrieved sequences were manually inspected for the presence of characteristic and functionally important MIP domains and motifs, such as the highly conserved NPA (Asn-Pro-Ala) motifs, and considered to be correct. The number of MIP genes described in this study is similar to that found in the genomes of Arabidopsis [5], maize [6], rice [7] and grape [8], but significantly lower than that identified in the genomes of poplar [9], cotton [10], tomato [11] and soybean [12]. The absence of recent whole-genome duplication (WGD) events in the sweet orange genome, as described by Xu et al. [23], could account for the relatively small size of the MIP family in the citrus genome.
Table 1. Genes and encoded MIP proteins in sweet orange.
| Gene name | Locus | Chromosome location | Group | Kegg Orth ID | Polypeptide length (MW) | pI | GRAVY | Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CsPIP1;1 | orange1.1g018895m | chr7: 31,253,722…31,256,103 | PIP | K09872 | 349 (37.56kDa) | 9.39 | 0.397 | plasma membrane |
| CsPIP1;2 | orange1.1g023021m | chr7: 31,247,369…31,248,902 | 288 (30.67kDa) | 7.71 | 0.414 | plasma membrane | ||
| CsPIP1;3 | orange1.1g023107m | chr5: 1,804,896…1,807,677 | 287 (30.70kDa) | 8.97 | 0.344 | plasma membrane | ||
| CsPIP1;4 | orange1.1g023069m | chr6: 9,907,825…9,909,541 | 287 (30.82kDa) | 8.96 | 0.343 | plasma membrane | ||
| CsPIP2;1 | orange1.1g023108m | chr6: 12,833,884…12,835,042 | 287 (30.67kDa) | 8.74 | 0.376 | plasma membrane | ||
| CsPIP2;2 | orange1.1g022966m | chr8: 19,657,287…19,659,497 | 289 (31.05kDa) | 7.62 | 0.392 | plasma membrane | ||
| CsPIP2;3 | orange1.1g019681m | chr7: 26,202,220…26,205,546 | 337 (36.34KDa) | 9.77 | 0.403 | plasma membrane | ||
| CsPIP2;4 | orange1.1g023370m | chr8: 981,841…983,571 | 283 (30.14kDa) | 8.99 | 0.431 | plasma membrane | ||
| CsTIP1;1 | orange1.1g025548m | chrUn: 46,663,407…46,665,011 | TIP | K09873 | 251 (26.06kDa) | 6.12 | 0.675 | vacuole |
| CsTIP1;2 | orange1.1g025600m | chr8: 20,659,157…20,660,437 | 250 (25.65kDa) | 5.32 | 0.841 | cytosol | ||
| CsTIP1;3 | orange1.1g037978m | chr8: 20,659,157…20,660,437 | 124 (12.92kDa) | 4.37 | 0.734 | cytosol | ||
| CsTIP1;4 | orange1.1g025464m | chr7: 29,135,182…29,136,531 | 252 (26.01kDa) | 5,69 | 0,786 | vacuole | ||
| CsTIP2;1 | orange1.1g025817m | chr1: 18,627,617…18,629,472 | 247 (25.15kDa) | 5.59 | 0.894 | vacuole | ||
| CsTIP2;2 | orange1.1g025865m | chr1: 18,627,617…18,629,472 | 247 (25.10kDa) | 5.59 | 0.902 | vacuole | ||
| CsTIP2;3 | orange1.1g038895m | chr5: 5,749,487…5,750,939 | 206 (20.55kDa) | 4.72 | 0.979 | vacuole | ||
| CsTIP3;1 | orange1.1g025197m | chr5: 16,938,542…16,940,192 | 256 (26.99kDa) | 7.07 | 0.626 | cytosol | ||
| CsTIP4;1 | orange1.1g025864m | chr4: 19,032,254…19,033,990 | 247 (16.27kDa) | 6.27 | 0.825 | vacuole | ||
| CsTIP5;1 | orange1.1g046726m | chr9: 14,144,215…14,145,199 | 161 (16.86kDa) | 9.00 | 0.770 | cytosol | ||
| CsTIP6;1 | orange1.1g042738m | chr9: 14,144,215…14,145,199 | 107 (11.11kDa) | 4.54 | 0.636 | secreted | ||
| CsNIP1;1 | orange1.1g023184m | chr2: 2,151,220…2,153,387 | NIP | K09874 | 286 (30.45kDa) | 8.64 | 0.434 | vacuole |
| CsNIP2;1 | orange1.1g036721m | chr6: 18,134,848…18,136,228 | 223 (23.70kDa) | 9.69 | 0.889 | cytosol | ||
| CsNIP2;2 | orange1.1g040981m | chr6: 18,134,848…18,136,228 | 211 (22.32kDa) | 9.39 | 0.967 | cytosol | ||
| CsNIP2;3 | orange1.1g040755m | chr2: 13,464,261…13,465,771 | 275 (29.39kDa) | 8.88 | 0.628 | plasma membrane | ||
| CsNIP3;1 | orange1.1g023102m | chr6: 20,482,599…20,486,038 | 287 (30.30kDa) | 8.40 | 0.387 | plasma membrane | ||
| CsNIP4;1 | orange1.1g046511m | chr3: 23,770,831…23,774,009 | 282 (29.28kDa) | 8.84 | 0.372 | vacuole | ||
| CsNIP5;1 | orange1.1g035030m | chr1: 13,680,675…13,682,992 | 75 (7.75kDa) | 8.98 | 0.291 | chloroplast | ||
| CsNIP5;2 | orange1.1g027840m | chr1: 13,678,241…13,681,090 | 218 (22.51kDa) | 7.75 | 0.462 | plasma membrane | ||
| CsNIP6;1 | orange1.1g039196m | chr9: 3,798,017…3,800,780 | 288 (30.20kDa) | 7.53 | 0.718 | plasma membrane | ||
| CsSIP1;1 | orange1.1g026039m | chr5: 28,968,880…28,972,401 | SIP | K09875 | 244 (25.92kDa) | 9.35 | 0.727 | plasma membrane |
| CsSIP1;2 | orange1.1g026082m | chr3: 1,234,876…1,236,556 | 244 (26.17kDa) | 9.83 | 0.749 | plasma membrane | ||
| CsSIP2;1 | orange1.1g026600m | chr6: 17,078,102…17,081,323 | 236 (25.57kDa) | 9.70 | 0.600 | chloroplast | ||
| CsXIP1;1 | orange1.1g036381m | chr8: 7,139,938…7,141,114 | XIP | - | 235 (25.09kDa) | 8.70 | 0.821 | plasma membrane |
| CsXIP1;2 | orange1.1g040654m | chr8: 7,131,064…7,132,799 | 302 (32.68kDa) | 8.74 | 0.573 | plasma membrane | ||
| CsXIP2;1 | orange1.1g045670m | chr8: 7,128,184…7,129,448 | 319 (34.58kDa) | 8.32 | 0.681 | plasma membrane |
The CsMIPs were classified in five different subfamilies, PIPs, TIPs, NIPs, SIPs and XIPs, based on analysis of the amino acid residues located in seven key positions (P1 to P7) that were previously proposed [12,34,35] to discriminate the different subfamilies (S2 Table), as well as on their phylogenetic relationships with the well classified MIPs of A. thaliana and XIPs of R. communis and N. benthamiana (S1 Fig). Our analysis revealed the presence of 8 PIPs, 11 TIPs, 9 NIPs, 3 SIPs and 3 XIPs in the sweet orange genome (Table 1). The CsMIPs were named according to the nomenclature proposed in classification of the MIPs of A. thaliana. This nomenclature was based on phylogenetic analyses and where the names in a systematic way reflect distinct clades that are evolutionarily stable [5]. PIPs, TIPs, NIPs and SIPs from sweet orange grouped with their respective Arabidopsis counterparts, indicating the large extent of conservation between the sweet orange and Arabidopsis MIP gene families (S1 Fig). The only exception was CsTIP6;1, which was found to encode a N- and C-terminally truncated TIP protein compared to the rest of the subfamily.
To examine whether the number of MIP genes found in the diploid sweet orange is comparable to that of the dihaploid sweet orange and haploid Clementine mandarin, we also performed homology searches against the dihaploid sweet orange draft genome available at the Citrus sinensis Annotation Project (CAP) and the reference haploid Clementine mandarin (C. clementina) genome available at Phytozome (S3 Table). Although the total number of MIP genes was roughly similar among the different citrus genomes, significant differences were observed in the number of members within the subfamilies (Table 2). BLAST similarity analysis revealed that the dihaploid sweet orange and haploid Clementine contained two additional PIP isoforms closely related to the CsPIP2;1 (S3 Table). Clementine also contained one additional PIP isoform closely related to the CsPIP1;2 and one PIP, TIP and NIP isoform without homology to any MIP sequence from the diploid sweet orange (S3 Table). CsTIP1;2 and CsTIP1;3, CsTIP2;1 and CsTIP2;2, CsTIP5;1 and CsTIP6;1, and CsNIP2;1 and CsNIP2;2 exhibited significant hits to the same MIP isoforms of the dihaploid sweet orange and haploid Clementine (S3 Table). These observed variations in the size of the MIP subfamilies may be a consequence of the different sequencing depth and assembly quality between the diploid and dihaploid sweet orange genomes [23,25] and the evolutionary origin of Clementine, which is a hybrid of Willowleaf mandarin and sweet-orange [25] and, thereby, it contains more C. reticulata haplotype regions than found in sweet orange.
Table 2. Comparison of the number of the different MIP family members in diploid (Phytozome) and dihaploid (CAP) sweet oranges (C. sinensis) and haploid Clementine (C. clementina).
| Subfamily | Diploid C. sinensis | Dihaploid C. sinensis | C. clementina |
|---|---|---|---|
| PIP | 8 | 10 | 11 |
| TIP | 11 | 8 | 9 |
| NIP | 9 | 8 | 8 |
| SIP | 3 | 3 | 3 |
| XIP | 3 | 3 | 2 |
| Total | 34 | 32 | 33 |
CsMIP protein properties and conserved amino acid residues
The CsMIPs encode proteins ranging from 75 (7.7 kDa) to 349 (37.6 kDa) amino acids in length, and pI values ranging from 4.37 to 9.77 (Table 1). The average protein length of PIPs, TIPs, NIPs, SIPs and XIPs were 300.8 (32.2 kDa), 213.4 (21.1 kDa), 238.3 (25.1 kDa), 241.3 (25.9 kDa) and 285.3 (30.8 kDa) amino acids, respectively. The average pI of PIPs, TIPs, NIPs, SIPs and XIPs were 8.77, 5.86, 8.68, 9.63, and 8.59, respectively. These data reveal that CsTIPs are not only smaller, but most of them are also much more acidic than the other CsMIPs, as reported for Arabidopsis MIPs [38]. The cause of these large differences in TIPs has been attributed to the smaller amount of basic residues found at the carboxyl termini of TIPs compared with the other MIPs [5]. All the CsMIPs had a positive GRAVY score (Table 1), suggesting that they are hydrophobic proteins, which is a necessary characteristic for AQPs [1]. Analysis of the predicted subcellular localization showed that all CsPIPs and CsXIPs were localized to plasma membrane (Table 1). The predicted localization of CsTIPs, CsNIPs and CsSIPs was more diverse, including vacuole (CsTIP1;1, CsTIP1;4, CsTIP2;1, CsTIP2;2, CsTIP2;3, CsTIP4;1, CsNIP1;1 and CsNIP4;1), cytosol (CsTIP1;2, CsTIP1;3, CsTIP3;1, CsTIP5;1, CsNIP2;1 and CsNIP2;2), plasma membrane (CsNIP2;3, CsNIP3;1, CsNIP5;2, CsNIP6;1, CsSIP1;1 and CsSIP1;2), chloroplast (CsNIP5;1 and CsSIP2;1) and secreted (CsTIP6;1) (Table 1). These results seem to be in agreement with the experimentally determined localizations of MIPs reported in the literature [13,39].
MIP folding is characterized by six transmembrane α-helices (H1 to H6) that are connected by five loops (loops A-E), forming an aqueous transmembrane pore that constitutes the functional core of MIPs [35]. Loops B (LB) and E (LE) contain two highly conserved NPA (Asn-Pro-Ala) motifs that are considered to be critical for the substrate selectivity of MIPs [40,41]. Another set of four conserved residues forms the aromatic/Arginine selectivity filter (ar/R filter), which has been proposed to act as a size exclusion barrier for substrate molecules [42]. The first two residues are located in H2 and H5, while the latter two are found in LE (LE1 and LE2). Finally, seven key amino acid residues (named positions P1 to P7) have been proposed to discriminate the five subfamilies [12,34]. P1 is located in the terminal part of H3, while P2 and P3 are located in LE, just behind the second NPA motif (2nd and 6th residues after 2nd NPA, respectively). P4 and P5 correspond to two consecutive amino acids located in H6, while P6 and P7 also correspond to two consecutive amino acids located in H3. The multiple sequence alignments were carefully analyzed and all these conserved motifs and amino acid residues were identified in most CsMIPs, indicating that they are functional channels for water and other solutes (S2 Table). All the CsPIPs showed the dual typical NPA motifs and an ar/R filter configuration characteristic for a water-transporting MIP (F,H,T,R). Additional presence of the S-A-F-W residues at P2-P5 positions, as observed in all CsPIPs, except for CsPIP1;1, has been interpreted as a signature of CO2 transporter PIPs [35]. All the CsTIPs also had the two canonical NPA motifs, except CsTIP1;3, CsTIP5;1 and CsTIP6;1, which were found to encode truncated proteins lacking either the first (CsTIP1;3) or the second NPA motif (CsTIP5;1 and CsTIP6;1), as well as other conserved amino acid residues of the ar/R filter region (S2 Table). TIPs containing the H-I-A-V or H-I-G-R residues in the ar/R filter and T-A-A-Y-W or T-S-A-Y-W residues at P1-P5 positions, like CsTIP1s, CsTIP2s and CsTIP3, have been shown to transport urea and H2O2 [35]. The CsNIP1;1, CsNIP2;1, CsNIP2;2 and CsNIP2;3 showed an ar/R filter configuration identical to that of soybean Nodulin 26, indicating that they are also able to facilitate water and solute transport capability [30]. The residue at the H5 position of the ar/R filter of AtNIP5;1 was shown to play a key role in the membrane permeability to water, silicic acid (Si) and boric acid (B) [42]. AtNIP5;1 with AIGR residues for the ar/R filter was shown to transport water, B and arsenite (As), but not Si [42]. CsNIP3;1, with GSGR residues for ar/R filter, can be expected to transport water, Si and B, while CsNIP4;1 (AIGR residues) may transport water, B and As. The CsSIPs showed a less conserved first NPA motif, while the second NPA motif was perfectly conserved in all members (S2 Table). AtSIP1 isoforms, but not AtSIP2;1, were shown to be functional water channels [43], suggesting that the latter may be involved in the transport of solutes. However, SIP transport function and structural organization still await biochemical characterization. The CsXIPs also showed a modified first NPA motif, NPL (CsXIP1s) or SPV (CsXIP2), and a conserved second NPA motif (S2 Table). The four positions in the ar/R filter region contained amino acid residues that were strictly conserved among the CsXIPs. CsXIP1;1 was observed to contain an internal deletion of 13 amino acid residues in the H2 region that abolished the conserved amino acid V at position H2 of the ar/R selective filter. Since the first three amino acid of the ar/R filter have rather hydrophobic residues (VVAR or VVVR), the CsXIPs might be involved in the transport of molecules other than water [35]. In fact, a recent study has indicated that the Solanaceae XIPs are plasma membrane aquaporins involved in the transport of many uncharged substrates, such as urea, H2O2 and B [44].
Genomic organization of CsMIPs
The exon-intron structure of all 34 CsMIP genes was analyzed using the sweet orange gene models annotated in Phytozome. With a few exceptions, the number and size of the exons, but not of the introns, were observed to be conserved within each CsMIP subfamily (S2 Fig). All the CsPIPs presented three introns and four exons, as reported for all Arabidopsis [5], poplar [9], tomato [11] and soybean PIPs [12]. The majority of CsTIPs contained two introns and three exons, with exception of CsTIP1;1, CsTIP2;3 and CsTIP6;1, which showed one intron and two exons, and the truncated CsTIP1;3 (one exon) gene. Such a more varied pattern of exon-intron structure has been also observed in Arabidopsis [5], poplar [9], tomato [11] and soybean TIPs [12]. Most CsNIPs contained four introns and five exons, like all Arabidopsis [5] and most poplar [9], tomato [11] and soybean NIPs [12]. The exceptions were CsNIP2;2 (three introns and four exons), CsNIP5;2 (two introns and three exons) and the truncated CsNIP5;1 (one intron and two exons) genes. The CsSIPs featured two introns and three exons, except CsSIP1;2 (one exon and one intron in the 3’-UTR region). Similar patterns of exon-intron structure were also reported for tomato SIPs [11], while all the Arabidopsis [5] and poplar SIPs [9] had two introns and three exons. The gene structure of CsXIPs varied among all members, which contained two introns and three exons (CsXIP1;2), either one intron and two exons (CsXIP2;1) or only one exon (CsXIP1;1). The pattern of two intron and three exons has been reported for most poplar [9] and tomato XIPs [11], while all soybean XIPs contained a single intron and two exons [12].
The positions of all 34 CsMIPs were mapped on the sweet orange chromosomes by homology searches against the full-length sweet orange genome assembly available at the CAP database (Table 1 and S3 Table). Except for CsTIP1;1, which was not exactly located on any chromosome because of an incomplete physical map for sweet orange, all the CsMIP loci were precisely mapped on every sweet orange chromosome, indicating a wide distribution of the gene family in the sweet orange genome (Table 1 and S3 Fig). The closely related CsMIP isoforms CsTIP1;2 and CsTIP1;3, CsTIP2;1 and CsTIP2;2, CsTIP5;1 and CsTIP6;1, and CsNIP2;1 and CsNIP2;2 were respectively mapped on identical chromosome positions since they showed significant hits to the same genes in the CAP database (S3 Table). Seven CsMIPs were found to be tandem duplicated genes according to the criteria of Hanada et al. [45], which defined tandem duplicates as genes in any gene pair, T1 and T2, that (1) belong to the same gene family, (2) are located within 100 kb each other, and (3) are separated by 10 or fewer nonhomologous spacer genes. These were CsNIP5;1 and CsNIP5;2 on chromosome 1, CsPIP1;1 and CsPIP1;2 on chromosome 7, and CsXIP1;1, CsXIP1;2 and CsXIP2;1 on chromosome 8 (Table 1 and S3 Fig). These results suggest that all these CsMIPs may have evolved from tandem duplication events, as also recently proposed for the tomato XIPs [11].
Analysis of previously mapped traits revealed that the 282-kb region surrounding the Citrus Tristeza Virus resistance (Ctv) locus is physically linked (~40-kb) to the CsNIP5;1 and CsNIP5;2 genes, on the chromosome 1 [46]. This region was also reported to contain Tyr1, the major locus controlling citrus nematode (Tylenchulus semipenetrans) resistance [47,48].
Expression patterns of CsMIP genes in different tissues
To investigate the expression patterns of CsMIPs in different tissues, RNA-seq data were downloaded from CAP [24]. The heatmap generated showed a differential transcript abundance of the 34 CsMIPs in four major tissues, namely callus, flower, leaf, fruit, and mixed fruit tissues at three developmental stages (S4 Fig). Some of the CsMIPs (CsPIP1;1, CsPIP1;3, CsPIP2;2, CsPIP2;3 and CsSIP1;1) showed higher expression in all the seven tissues, indicating a role in constitutive transport processes throughout the plant. Others genes were found to have a low expression in all the tissues (CsTIP1;1, CsTIP1;2, CsTIP1;3, CsTIP2;3, CsTIP5;1, CsTIP6;1, CsNIP2;1, CsNIP2;2, CsNIP2;3, CsNIP5;1, CsNIP5;2, CsXIP1;1 and CsXIP2;1). The putative tandem duplicated CsMIP genes were observed to have divergent expression profiles, which probably has contributed to their maintenance through regulatory subfunctionalization and neofunctionalization [49]. CsPIP1;1 showed a higher expression than CsPIP1;2 in all the seven tissues analyzed. CsXIP1;1 and CsXIP2;1 showed low expression in all the seven tissues analyzed, while CsXIP1;2 exhibited a high expression in flower, leaf and mixed fruit tissue (Mix.3).
The cell type localization of aquaporin expression can also provide clues about their physiological roles. For instance, expression of PIP aquaporins is generally localized in organs and tissues characterized by large fluxes of water, such as vascular tissues, guard cells, flowers and fruits [4]. Their expression in roots and leaves has been also correlated with the presence of apoplastic barriers, the exodermis and endodermis in roots or in suberized bundle sheath cells in leaves, suggesting their essential role in the transmembrane water diffusion when its movement is hindered [19,20,50–55]. Except for CsPIP2;1, all the CsPIPs were found to be highly expressed in flower, leaf, fruit and mixed fruit tissues (S4 Fig), supporting their active role in the transport of water and solutes across these tissues. By contrast, the expression of TIP isoforms has been more related to developmental stages and/or organ specificity [56]. For instance, the expression of AtTIP2;1 is especially high in the vascular system of the shoot but is barely detectable in the root [57]. AtTIP3;1 is highly expressed in cotyledons and associated with the membrane of protein storage vacuoles [4]. TIPs are also differentially expressed during fruit maturation, e.g., the TIP1;1 homolog in pear is highly expressed in the young fruit, whereas TIP proteins levels in grape gradually increase along with ripening [58,59]. Vacuoles participate in cell expansion and, thus, their enlargement by water compartmentation is essential to provoke the rapid fruit growth that is characteristic of the ripening process. The lack of specific regulation observed along fruit ripening for PIPs isoforms points out the essential role of TIPs in this process [60]. The differential expression of CsTIP1;3, CsTIP1;4, CsTIP2;1, CsTIP2;2, CsTIP3;1, CsTIP4;1 and CsTIP5;1 highlights the functional importance of these CsMIPs on each tissue and stage of fruit development analyzed (S4 Fig). The overall level of NIP expression is usually lower than the expression of PIPs and TIPs, and it is usually associated with specialized organs and cells [10]. For instance, AtNIP2;1 is specifically expressed in the endoplasmic reticulum (ER) of roots, whereas AtNIP5;1 is a plasma membrane MIP mainly expressed in root elongation zones [39,61]. Our analysis showed that the CsNIPs had preferential expression either in flower (CsNIP6;1), leaf (CsNIP2;2 and CsNIP3;1) or mixed fruit tissues at different developmental stages (CsNIP1;1, CsNIP2;2, CsNIP3;1, CsNIP4;1 and CsNIP6;1) (S4 Fig). SIPs seem to be expressed in a range of tissues in Arabidopsis, including young roots, flowers and pollen [62]. It is remarkable that SIPs are also strongly expressed in suspension cultured cells compared to other MIPs [62]. CsSIP1;1 was observed to be constitutively expressed in all the seven tissues analyzes, while the others were preferentially expressed in flower and fruit tissues (S4 Fig). XIPs were reported to be expressed in almost all the poplar tissues [9]. By contrast, CsXIPs showed a low expression in all tissues analyzed, except CsXIP1;2, which show a relatively high expression in flower, leaf and mixed fruit tissues at third developmental stage (Mix.3) (S4 Fig).
Expression patterns of CsMIP genes under abiotic and biotic stresses
To identify CsMIPs with a potential role in abiotic and biotic stress response of sweet orange, the expression patterns of all the 34 sweet orange MIPs were investigated in plants exposed to drought, high salinity and ‘Ca. L. asiaticus’ (HLB) infection, by qPCR. Considering the log2 fold change (LFC) of ≥1.00 or ≤-1.00 as cutoff threshold between stressed and control plants, the qPCR analyses showed that all the CsMIPs were differentially expressed in at least one stress condition and tissue analyzed (Figs 1–3). Twelve CsMIPs (CsPIP1;1, CsPIP2;4, CsTIP1;3, CsTIP2;1, CsTIP2;2, CsTIP3;1, CsTIP4;1, CsNIP1;1, CsSIP1;2, CsXIP1;1, CsXIP1;2 and CsXIP2;1) were observed to be differentially expressed in response to all the three stress conditions analyzed, in at least one tissue studied. Interestingly, only CsTIP1;1 showed differential expression exclusively in response to the abiotic stress treatments, while six CsMIPs (CsPIP1;2, CsPIP2;2, CsNIP2;2, CsNIP5;2, CsNIP6;1 and CsSIP1;1) were differentially expressed exclusively under the biotic stress treatment. These results seem to be consistent with the respective organization of the stress-responsive cis-acting regulatory elements observed in the CsMIPs promoters (S5 Fig). CsTIP1;1 was observed to contain the highest number of ABRE copies in the promotor region among the sweet orange MIP genes, while no or a low number of ABRE (less than 2 copies) and other stress-responsive cis-acting regulatory elements was detected in the promoter regions of the CsMIPs that were not induced by the abiotic stress treatments (S5 Fig). A single copy of DRE/CRT has been observed to be sufficient for ABA-independent stress-responsive gene expression, while more than two ABRE sequences are usually required for the ABA-responsive transcription [63].
Fig 1. Expression analysis of the complete set of sweet orange MIPs in response to drought treatment.
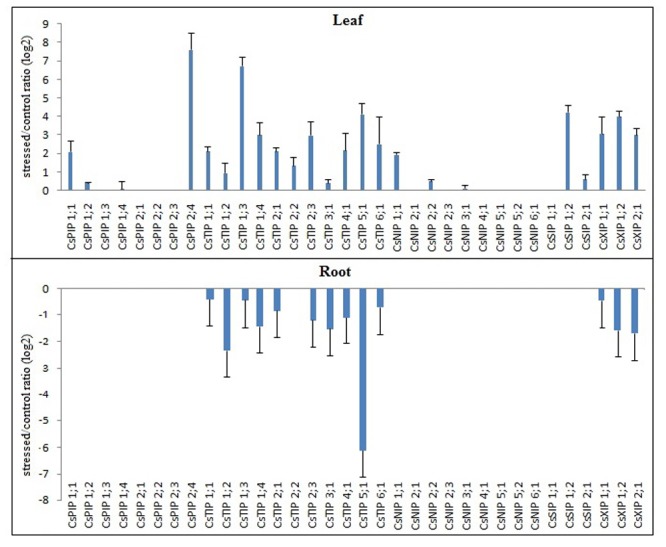
Ratios (log2) of relative mRNA levels between stressed and control plants for all 34 CsMIPs in leaves and roots, as measured by qPCR. GAPC2 was used as an endogenous control. The bars show means ± SE from three biological replicates.
Fig 3. Expression analysis of the complete set of sweet orange MIPs in response to ‘Ca. L. asiaticus’ infection in rough lemon (LEM) and sweet orange (SWO).

Ratios (log2) of relative mRNA levels between infected and control plants at 0, 7, 17, and 34 WAI for all 34 CsMIPs, as measured by qPCR. GAPC2 was used as an endogenous control.
All the CsTIPs and CsXIPs were upregulated in leaves but downregulated in roots by drought treatment, while only two CsPIPs (CsPIP1;1 and CsPIP2;4) and one CsNIP (CsNIP1;1) and CsSIP (CsSIP1;2) were differentially upregulated by drought treatment in leaves (Fig 1). Most CsMIPs were upregulated by salt treatment in roots, and either upregulated or downregulated by this treatment in leaves, depending on the isoform (Fig 2). A coordinated up- and downregulation, depending on the MIP gene and organ examined, has been described as a general pattern of MIP regulation during drought and salt stresses in Arabidopsis [64–67], soybean [12] and rice [68]. It has been proposed that a general downregulation of MIPs might be a way for the plant to minimize water loss and the hence loss of turgor in specific organs, and that the upregulation of individual MIPs could be a way for the plant to direct water flow to certain organs or cells that are crucial for plant survival during stress, or necessary for its fast recovery upon rehydration [65]. Thus, the concomitant upregulation in leaves and downregulation in roots of the CsTIPs and CsXIPs by drought treatment likely reveal a coordinated regulation of these MIP isoforms to direct the water flow through the leaf plasma membrane and tonoplast, while avoiding the water loss in roots. On the other hand, the upregulation of most CsMIPs in roots by salt treatment suggests their coordinated regulation to increase the overall water flow into this organ, since it is well known that salt stress reduces the hydraulic conductivity in roots, resulting in decreases of water flow from root to shoot. Those CsMIPs that were upregulated in leaves by salt treatment (i.e., CsPIP1;4, CsTIP1;1, CsTIP2;1, CsTIP2;2, CsTIP3;1, CsNIP2;3, CsNIP4;1, CsXIP1;1 and CsXIP1;2) may also contribute to increase the water flow into this organ.
Fig 2. Expression analysis of the complete set of sweet orange MIPs in response to salt treatment.
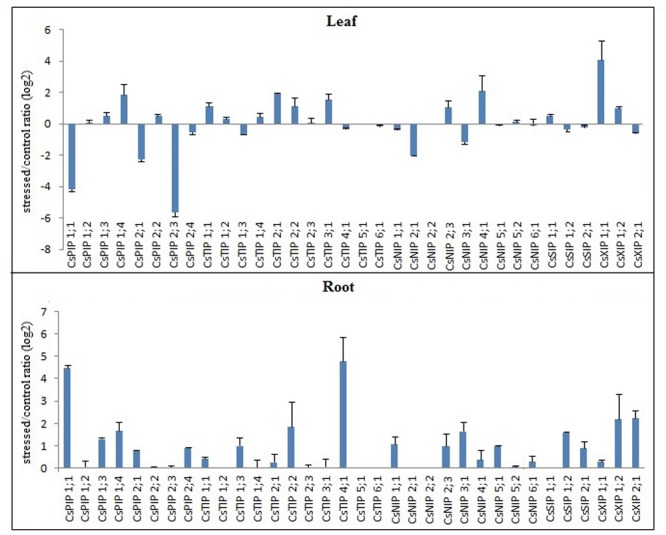
Ratios (log2) of relative mRNA levels between stressed and control plants for all 34 CsMIPs in leaves and roots, as measured by qPCR. GAPC2 was used as an endogenous control. The bars show means ± SE from three biological replicates.
The time-course transcriptional analysis in response to ‘Ca. L. asiaticus’ infection showed that 32 out of 34 CsMIPs were strongly downregulated at the early stage (7 weeks) in the highly susceptible sweet orange, but not in the tolerant rough lemon, whose expression levels of all CsMIPs were either upregulated or unchanged at this stage (Fig 3). At the later time points (17 and 34 weeks), significantly differential expression levels between the susceptible and tolerant citrus species were essentially limited to CsPIP2;2, CsTIP1;2, CsTIP2;1, CsTIP2;2 and CsNIP5;1. ‘Ca. L. asiaticus’ is the most widely distributed of the three species of the phloem-limited α-proteobacterium, designated as Candidatus Liberibacter (i.e., ‘Ca. L. asiaticus’, ‘Ca. L. africanus’ and ‘Ca. L. americanus’), which has been associated with the most destructive disease affecting citrus worldwide, Huanglongbing (HLB) [69]. Leaf symptoms of HLB disease have been well documented [69]. These include vein yellowing and blotchy mottle, reduced leaf size and premature leaf abscission. Anatomical alterations caused by the disease in leaves include the excessive accumulation of starch, callose depositions, phloem plugging, necrosis and collapse, swelling of sieve elements and companion cell walls, and the disruption of chloroplast inner grana structures [27,69–74]. Significant differences on phloem ultrastructure and phloem loading activity were also observed between HLB-infected sweet orange and rough lemon plants, highlighting some underlying differences between tolerance and susceptibility mechanisms to HLB disease [27]. While the phloem transport activity in the midribs of leaves was considerably impaired in diseased sweet orange, it was much less affected in infected rough lemon [27]. The downregulation of most CsMIPs as observed 7 WAI in the susceptible sweet orange, but not in the tolerant rough lemon, correlates with the onset of disease symptoms, such as blotchy leaf mottle and yellowing [27]. The continued contrasting expression patterns of CsPIP2;2, CsTIP1;2, CsTIP2;1, CsTIP2;2 and CsNIP5;1 between the tolerant and susceptible citrus species throughout the 17 and/or 34 WAI suggests an involvement of these CsMIP genes in further symptom development, such as growth inhibition and impaired phloem loading [27]. Taken together, these results indicate that MIP genes may play an active role in the pathogenesis of HLB disease by regulating the flow of water and nutrients required for the normal growth and development of citrus plants. The recent finding in a microarray experiment that CsPIPs were observed to be downregulated in stems of HLB-symptomatic sweet orange trees, 16 months after inoculation of ‘Ca. L. asiaticus’, is consistent with this interpretation [74].
Conclusions
This study presented a genome-wide survey of the MIP gene family in sweet orange. A total of 34 open reading frames (ORFs) encoding MIP proteins were identified and characterized as to their sequences, phylogenetic relationships, genomic organization, tissue-specific gene expression, and expression profiles upon abiotic and biotic stresses. Our results allow us to assess the relative contribution of each CsMIP member to water and solute transport in different tissues and in response to drought, salinity and ‘Ca. L. asiaticus’ infection. These results suggest a special role for CsTIPs and CsXIPs in delivering water to the leaves while preventing root tissue dehydration under drought stress, and for most CsMIPs in increasing the overall water flow into the roots during salt stress and also regulating the flow of water and nutrients during the development of HLB disease. Taken together, our results support the idea that these CsMIP genes represent an important genetic resource for improving citrus tolerance or resistance to both abiotic and biotic stresses.
Supporting Information
The deduced amino acid sequences were aligned using ClustalW2 and the phylogenetic tree was generated using Bootstrap N-J tree (1,000 resamplings) method and MEGA program (v6.0.5). Numbers at internal nodes denotes the results of bootstrapping analysis (n = 1000). Black diamonds indicate MIP gene from sweet orange. Cs, Citrus sinensis; At, Arabidopsis thaliana; Rc, Ricinus communis; Nb, Nicotiana benthamiana.
(TIF)
NOI denotes the number of introns, E the exon and I the intron.
(TIF)
The chromosomal position of each CsMIP was mapped according to the Citrus sinensis Annotation Project (CAP). The scale is in Mb. CsSIP (circle), CsPIP (star), CsNIP (square), CsTIP (triangle), CsXIP (diamond).
(TIF)
Mix.1, Mix.2 and Mix.3 indicate mixed fruit tissues from different developmental stages. The heatmap was generated using R 3.1.0 software. The color scale shown represents RPKM-normalized log2-transformed counts.
(TIF)
The cis-elements were analyzed in the 1 kb upstream promoter region of translation start site of all CsMIPs using the PLACE database.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors are grateful to Dr. Eduardo A. Girardi (Embrapa Cassava & Fruits, Cruz das Almas, Bahia, Brazil) for kindly providing the grafted citrus plant material used in this study. We gratefully acknowledge the Ph.D. scholarship to C.P.S.M. by the CAPES Foundation (Brasília, Brazil).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by research grants from CNPq (Brasília, Brazil), CAPES (Brasília, Brazil), FAPESP (São Paulo, Brazil) and Florida Citrus Research and Development Foundation (USA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Maurel C, Verdoucq L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol. 2008;59: 595–624. 10.1146/annurev.arplant.59.032607.092734 [DOI] [PubMed] [Google Scholar]
- 2. Maurel C, Santoni V, Luu DT, Wudick MM, Verdoucq L. The cellular dynamics of plant aquaporin expression and functions. Plant Biol. 2009;12: 690–698. [DOI] [PubMed] [Google Scholar]
- 3. Sutka M, Li G, Boudet J, Boursiac Y, Doumas P, Maurel C. Natural variation of root hydraulics in Arabidopsis grown in normal and salt stressed conditions. Plant Physiol. 2011;155: 1264–1276. 10.1104/pp.110.163113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Acta Biochim Biophys. 2009;1788: 1213–1228. [DOI] [PubMed] [Google Scholar]
- 5. Johanson U, Karlsson M, Jahansson I, Gustavsson S, Sjovall S, Fraysse L, et al. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126: 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125: 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 rice aquaporin genes and analyses of their expression and function. Plant Cell Physiol. 2005;45: 1568–1577. [DOI] [PubMed] [Google Scholar]
- 8. Fouquet R, Leon C, Ollat N, Barrieu F. Identification of grapevine aquaporins and expression analysis in developing berries. Plant Cell Rep. 2008;27: 1541–1550. 10.1007/s00299-008-0566-1 [DOI] [PubMed] [Google Scholar]
- 9. Gupta AB, Sankararamakrishnan R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009;9: 134 10.1186/1471-2229-9-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park W, Scheffler BE, Bauer PJ, Campbell BT. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2010;10: 142 10.1186/1471-2229-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reuscher S, Akiyama M, Mori C, Aoki K, Shibata D, Shiratake K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS One. 2013;8: e79052 10.1371/journal.pone.0079052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, et al. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS One. 2013;8: e56312 10.1371/journal.pone.0056312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wudick MM, Luu D, Maurel C. A look inside: localization patterns and functions of intracellular plant aquaporins. New Phytol. 2009;184: 289–302. 10.1111/j.1469-8137.2009.02985.x [DOI] [PubMed] [Google Scholar]
- 14. Gustavsson S, Lebrun AS, Nordén K, Chamont F, Johanson U. A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiol. 2005;139: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Danielson JAH, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens . BMC Plant Biol. 2008;8: 45 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liénard D, Durambur G, Kiefer-Meyer MC, Nogue F, Menu-Bouaouiche L, Charlot F, et al. Water transport by aquaporins in the extant plant Physcomitrella patens . Plant Physiol. 2008;146: 1207–1218. 10.1104/pp.107.111351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Postaire L, Verdoucq L, Maurel C. Aquaporins in plants: from molecular structures to integrated functions. Adv Bot Res. 2008;46: 75–136. [Google Scholar]
- 18. Maurel C. Plant aquaporins: novel functions and regulation properties. FEBS Lett. 2007;581: 2227–2236. [DOI] [PubMed] [Google Scholar]
- 19. Prado K, Maurel C. Regulation of leaf hydraulics: from molecular to whole plant levels. Front Plant Sci. 2013;4: 255 10.3389/fpls.2013.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hachez C, Zelazny E, Chaumont F. Modulating the expression of aquaporin genes in planta: a key to understand their physiological functions. Acta Biochim Biophys. 2006;1758: 1142–1156. [DOI] [PubMed] [Google Scholar]
- 21. Rodríguez-Gamir J, Ancillo G, Aparicio F, Bordas M, Primo-Millo E, Forner-Giner MA. Water-deficit tolerance in citrus is mediated by the down regulation of PIP gene expression in the roots. Plant Soil. 2011;347: 91–104. [Google Scholar]
- 22. Rodríguez-Gamir J, Ancillo G, Legaz F, Primo-Millo E, Forner-Giner MA. Influence of salinity on PIP gene expression in citrus roots and its relationship with root hydraulic conductance, transpiration and chloride exclusion from leaves. Environ Exp Bot. 2012;78: 163–166. [Google Scholar]
- 23. Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 2013;45: 59–66. 10.1038/ng.2472 [DOI] [PubMed] [Google Scholar]
- 24. Wang J, Chen D, Lei Y, Chang J-W, Hao B-H, Xing F, et al. Citrus sinensis Annotation Project (CAP): a comprehensive database for sweet orange genome. PLoS One. 2014;9: e87723 10.1371/journal.pone.0087723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 2014;32: 656–662. 10.1038/nbt.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Oliveira TM, Cidade LC, Gesteira AS, Coelho-Filho MA, Soares-Filho WS, Costa MGC. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet Genom. 2011;7: 1123–1134. [Google Scholar]
- 27. Fan J, Chen C, Yu Q, Khalaf A, Achor DS, Brlansky RH, et al. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to 'Candidatus Liberibacter asiaticus' infection. Mol Plant Microbe Interact. 2012;25: 1396–1407. 10.1094/MPMI-06-12-0150-R [DOI] [PubMed] [Google Scholar]
- 28. Li WB, Hartung JS, Levy L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J Microbiol Meth. 2006;66: 104–115. [DOI] [PubMed] [Google Scholar]
- 29. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40: D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallace IS, Roberts DM. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 2004;135: 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson JD, Higgens DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saitou N, Nei M. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4: 406–425. [DOI] [PubMed] [Google Scholar]
- 34. Froger A, Tallur B, Thomas D, Delamarche C. Prediction of functional residues in water channels and related proteins. Protein Sci. 1998;7: 1458–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hove RM, Bhave M. Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol. 2011;75: 413–430. 10.1007/s11103-011-9737-5 [DOI] [PubMed] [Google Scholar]
- 36. Voorrips R. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93: 77–78. [DOI] [PubMed] [Google Scholar]
- 37. Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One. 2012;7: e31263 10.1371/journal.pone.0031263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. The role of aquaporins in cellular and whole plant water balance. Acta Biochim Biophys. 2000;1465: 324–342. [DOI] [PubMed] [Google Scholar]
- 39. Takano J, Wada M, Ludewig U, Schaaf G, von Wirén N, Fujiwara T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell. 2006;18: 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathai JC, Agre P. Hourglass pore-forming domains restrict aquaporin-1 tetramer assembly. Biochemistry. 1999;38: 923–928. [DOI] [PubMed] [Google Scholar]
- 41. Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407: 599–605. [DOI] [PubMed] [Google Scholar]
- 42. Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot. 2011;62: 4391–4398. 10.1093/jxb/err158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell specific expression in Arabidopsis thaliana . FEBS Lett. 2005;579: 5814–5820. [DOI] [PubMed] [Google Scholar]
- 44. Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011;66: 306–317. 10.1111/j.1365-313X.2011.04496.x [DOI] [PubMed] [Google Scholar]
- 45. Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148: 993–1003. 10.1104/pp.108.122457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z-N, Ye X-R, Molina J, Roose ML, Mirkov TE. Sequence analysis of a 282-kilobase region surrounding the Citrus Tristeza Virus resistance gene (Ctv) locus in Poncirus trifoliata L. Raf. Plant Physiol. 2003;131: 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, et al. Cloning and characterization of NBS-LRR class resistance-gene candidate sequences in citrus. Theor Appl Genet. 2000;101: 814–822. [Google Scholar]
- 48. Deng Z, Tao Q, Chang Y-L, Huang S, Ling P, Yu C, et al. Construction of a bacterial artificial chromosome (BAC) library for citrus and identification of BAC contigs containing resistance gene candidates. Theor Appl Genet. 2001;102: 1177–1184. [Google Scholar]
- 49. Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis . Mol Biol Evol. 2006;23: 469–478. [DOI] [PubMed] [Google Scholar]
- 50. Schäffner AR. Aquaporin function, structure, and expression: are there more surprises to surface in water relations? Planta. 1998;204: 131–139. [DOI] [PubMed] [Google Scholar]
- 51. Suga S, Murai M, Kuwagata T, Maeshima M. Differences in aquaporin levels among cell types of radish and measurement of osmotic water permeability of individual protoplasts. Plant Cell Physiol. 2003;44: 277–286. [DOI] [PubMed] [Google Scholar]
- 52. Hachez C, Heinen RB, Draye X, Chaumont F. The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Mol Biol. 2008;68: 337–353. 10.1007/s11103-008-9373-x [DOI] [PubMed] [Google Scholar]
- 53. Hachez C, Veselov D, Ye Q, Reinhardt H, Knipfer T, Fricke W, et al. Short-term control of maize cell and root water permeability through plasma membrane aquaporin isoforms. Plant Cell Environ. 2012;35: 185–198. 10.1111/j.1365-3040.2011.02429.x [DOI] [PubMed] [Google Scholar]
- 54. Vandeleur RK, Mayo G, Shelden MC, Gilliham M, Kaiser BN, Tyerman SD. The role of plasma membrane intrinsic protein aquaporins in water transport through roots: diurnal and drought stress responses reveal different strategies between isohydric and anisohydric cultivars of grapevine. Plant Physiol. 2009;149: 445–460. 10.1104/pp.108.128645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shatil-Cohen A, Attia Z, Moshelion M. Bundle-sheath cell regulation of xylem-mesophyll water transport via aquaporins under drought stress: a target of xylem-borne ABA? Plant Mol Biol. 2011;67: 72–80. [DOI] [PubMed] [Google Scholar]
- 56. Jauh GY, Fischer AM, Grimes HD, Ryan CA, Rogers JC. Delta-tonoplast intrinsic protein defines unique plant vacuole functions. Proc Natl Acad Sci USA. 1998;95: 12995–12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ. Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell. 1996;8: 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shiratake K, Kanayama Y, Maeshima M, Yamaki S. Changes in H(+)-pumps and a tonoplast intrinsic protein of vacuolar membranes during the development of pear fruit. Plant Cell Physiol. 1997;38: 1039–1045. [DOI] [PubMed] [Google Scholar]
- 59. Shiratake K, Goto H, Maeshima M, Yamaki S. Fluctuations of the protein levels of H+-pumps and water channels of tonoplast and plasma membrane grape berry development. J. Japan Soc Hort Sci. 2001;70: 287–293. [Google Scholar]
- 60. Shiratake K, Martinoia E. Transporters in fruit vacuoles. Plant Biotechnol. 2007;24: 127–133. [Google Scholar]
- 61. Mizutani M, Watanabe S, Nakagawa T, Maeshima M. Aquaporin NIP2;1 is mainly localized to the ER membrane and shows root-specific accumulation in Arabidopsis thaliana . Plant Cell Physiol. 2006;47: 1420–1426. [DOI] [PubMed] [Google Scholar]
- 62. Maeshima M, Ishikawa F. ER membrane aquaporins in plants. Pflugers Arch, Eur J Physiol. 2008;456: 709–716. [DOI] [PubMed] [Google Scholar]
- 63. Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10: 88–94. [DOI] [PubMed] [Google Scholar]
- 64. Jang JY, Kim DG, Kim YO, Kim JS, Kang H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana . Plant Mol Biol. 2004;54: 713–725. [DOI] [PubMed] [Google Scholar]
- 65. Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, et al. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol. 2005;59: 469–484. [DOI] [PubMed] [Google Scholar]
- 66. Alexandersson E, Danielson JAH, Rade J, Moparthi VK, Fontes M, Kjellbom P, et al. Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 2010;61: 650–660. 10.1111/j.1365-313X.2009.04087.x [DOI] [PubMed] [Google Scholar]
- 67. Boursiac Y, Chen S, Luu DT, Sorieul M, Dries NVD, Maurel C. Early effects of salinity on water transport in Arabidopsis roots molecular and cellular features of aquaporin expression. Plant Physiol. 2005;139: 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li G-W, Peng Y-H, Yu X, Zhang M-H, Cai W-M, Sun W-N, et al. Transport functions and expression analysis of vacuolar membrane aquaporins in response to various stresses in rice. J Plant Physiol. 2008;165: 1879–1888. 10.1016/j.jplph.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 69. Bove JM. Huanglongbing: A destructive, newly emerging, century-old disease of citrus. J Plant Pathol. 2006;88: 7–37. [Google Scholar]
- 70. Etxeberria E, Gonzalez P, Achor D, Albrigo G. Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol Mol Plant Pathol. 2009;74: 76–83. [Google Scholar]
- 71. Kim JS, Sagaram US, Burns JK, Li JL, Wang N. Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: microscopy and microarray analyses. Phytopathology. 2009;99: 50–57. 10.1094/PHYTO-99-1-0050 [DOI] [PubMed] [Google Scholar]
- 72. Achor DS, Etxeberria E, Wang N, Folimonova SY, Chung KR, Albrigo LG. Sequence of anatomical symptom observations in citrus affected with Huanglongbing disease. Plant Pathol J. 2010;9: 56–64. [Google Scholar]
- 73. Folimonova SY, Achor DS. Early events of citrus greening (Huanglongbing) disease development at the ultrastructural level. Phytopathology. 2010;100: 949–958. 10.1094/PHYTO-100-9-0949 [DOI] [PubMed] [Google Scholar]
- 74. Aritua V, Achor D, Gmitter FG, Albrigo G, Wang N. Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus infection. PLoS One. 2013;8: e73742 10.1371/journal.pone.0073742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The deduced amino acid sequences were aligned using ClustalW2 and the phylogenetic tree was generated using Bootstrap N-J tree (1,000 resamplings) method and MEGA program (v6.0.5). Numbers at internal nodes denotes the results of bootstrapping analysis (n = 1000). Black diamonds indicate MIP gene from sweet orange. Cs, Citrus sinensis; At, Arabidopsis thaliana; Rc, Ricinus communis; Nb, Nicotiana benthamiana.
(TIF)
NOI denotes the number of introns, E the exon and I the intron.
(TIF)
The chromosomal position of each CsMIP was mapped according to the Citrus sinensis Annotation Project (CAP). The scale is in Mb. CsSIP (circle), CsPIP (star), CsNIP (square), CsTIP (triangle), CsXIP (diamond).
(TIF)
Mix.1, Mix.2 and Mix.3 indicate mixed fruit tissues from different developmental stages. The heatmap was generated using R 3.1.0 software. The color scale shown represents RPKM-normalized log2-transformed counts.
(TIF)
The cis-elements were analyzed in the 1 kb upstream promoter region of translation start site of all CsMIPs using the PLACE database.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


