Abstract
Interleukin-17 (IL-17) belongs to a relatively new family of cytokines that has garnered attention as the signature cytokine of Th17 cells. This cytokine family consists of 6 ligands, which bind to 5 receptor subtypes and induce downstream signaling. Although the receptors are ubiquitously expressed, cellular responses to ligands vary across tissues. The cytokine family is associated with various autoimmune disorders including rheumatoid arthritis, multiple sclerosis, inflammatory bowel disease, asthma and psoriasis in addition to being implicated in the pathogenesis of cancer. In addition, this family plays a role in host defense against bacterial and fungal infections. The signaling mechanisms of the IL-17 family of proinflammatory cytokines are not well explored. In this study, we present a resource of literature-annotated reactions induced by IL-17. The reactions are catalogued under 5 categories, namely; molecular association, catalysis, transport, activation/inhibition and gene regulation. A total of 93 molecules and 122 reactions have been annotated. The IL-17 pathway is freely available through NetPath, a resource of signal transduction pathways previously developed by our group.
Keywords: Activation, Differential expression, Inhibition, NetPath, Post-translational modifications, Protein-protein interaction, Translocation
Introduction
Cytokines are small proteins that mediate intercellular communication. Interleukin-17 (IL-17 also known as IL17A or CTLA8) was first identified from murine T cell hybridoma cDNA library as a virus-captured cellular gene related to the immune system (Rouvier et al. 1993). While the majority of the cytokines belong to the Th1/Th2 paradigm, IL-17 served as a signature cytokine which led to the identification of a new subset of T helper cells, Th17 cells (Rouvier et al. 1993; Yao et al. 1995b; Korn et al. 2009; Weaver et al. 2007). Based on the amino acid sequence homology, five other members of this family IL-17B, IL-17C (Li et al. 2000), IL-17D (Starnes et al. 2002), IL-17E (also known as IL-25) (Lee et al. 2001), and IL-17F have been identified. In humans, all members of the IL-17 family show sequence similarity to IL-17A. While IL-17F is its closest homolog with 55 % identity; IL-17E is the most distant homolog with only 17 % identity (Moseley et al. 2003). IL-17A and IL-17F are expressed in NK, NKT, γδ T cells and LTi cells (Cua and Tato 2010). Expression of IL-17 and its subtypes is reported in diverse cells and tissues as listed in Table 1.
Table 1.
IL‐17 ligand family members and their expression
| Ligand | Alternate Name(s) | Cell/tissue Expression |
|---|---|---|
| IL-17A | CTLA-8 IL-17 |
Th17, NK, NKT, γδ T cells and LTi |
| IL-17B | NIRF ZCYTO7 IL-20 |
Intestine, pancreas and neurons |
| IL-17C | CX2 | Prostate and fetal kidney |
| IL-17D | IL-27 | Adipose tissue, brain, heart, lung, pancreas and spleen |
| IL-17E | IL-25 | Th2 cells, eosinophils, basophils, lung epithelial cells, mast cells, intestine, lung and uterus |
| IL-17F | ML-1 CANDF6 |
Th17, NK, NKT, γδ T cells and LTi |
IL-17 binds to IL-17R, which is its cognate receptor. IL-17R family consists of 5 subtypes of IL-17 receptors known as IL-17RA, IL-17RB, IL-17RC, IL-17RD and IL-17RE. IL-17RA is ubiquitously expressed in tissues with a relatively higher expression in haematopoietic tissues (Yao et al. 1995a). IL-17RB receptors are expressed in various endocrine tissues, kidney, liver and Th2 cells (Lee et al. 2001). IL-17RC is expressed in non-immune cells such as prostate, thyroid and joints (Haudenschild et al. 2002; Kuestner et al. 2007). IL-17RD is expressed in epithelial cells of breast, prostate, thyroid gland, ovarian surface (Zisman-Rozen et al. 2007) and endothelial cells (Yang et al. 2003). Expression of IL-17RE is not well defined. IL-17Rs are single pass transmembrane glycoprotein receptors. The subtypes of IL-17R contain conserved SEFIR (similar expression to FGF receptor/IL-17R) domain in the cytoplasm and an extracellular fibronectin III like domain (Yao et al. 1997). IL-17RA also has a specialized motif called the TILL (TIR-like loop) which is a crucial mediator of IL-17 family signaling (Maitra et al. 2007).
IL-17 is predominantly a proinflammatory cytokine and mediates multiple cell type specific functions. It induces the expression of a number of inflammatory effectors that includes cytokines and chemokines. It may also act synergistically with other cytokines like TNF-α and IFN-γ to amplify inflammation (Onishi and Gaffen 2010). Recent studies have implicated a role for IL-17 in the development of breast (Huang et al. 2013) and colorectal (Song et al. 2014) cancers. It has been shown that IL-17 promotes the self-renewal of ovarian CD133(+) cancer stem-like cells (Xiang et al. 2015). A significant number of studies have implicated increased expression of IL-17 in inflammatory autoimmune disorders which are multifactorial. These disorders include rheumatoid arthritis (RA), psoriasis, multiple sclerosis (MS), inflammatory bowel disease and systemic lupus erythematosus (Firestein 2003; Miossec 2009; Qian et al. 2010). Significantly elevated levels of IL-17 have been observed in the synovial fluid of patients with RA (Gaffen 2009). Mouse model of RA have shown excess IL-17 in exacerbated collagen induced arthritis (CIA) (Lubberts et al. 2002) and silencing of IL-17 or IL-17R in CIA mice reduced the symptoms of CIA (Lubberts et al. 2004). Similarly, mice deficient in IL-17 were resistant to the induction of experimental autoimmune encephalomyelitis, a mouse model of MS (Yang et al. 2008). Despite such observations, the details of signaling and the intracellular changes elicited by IL-17 and its cognate receptors are not available in the public domain. In this study, we performed a literature-based survey to systematically compile the molecular reactions orchestrated by stimulation of IL-17R with IL-17.
Materials and methods
We performed an extensive search of published literature using PubMed and iHOP (Information Hyperlinked Over Proteins). The articles were screened for information pertaining to molecular association (protein-protein interactions), catalysis (post-translational modifications), transport (translocation of proteins between sub-cellular compartments), activation/inhibition and gene regulation events that have been reported under stimulation by IL-17 and its subtypes. The pathway data were annotated using PathBuilder, a curation tool developed by our group (Kandasamy et al. 2009). The annotation criteria were followed as described in IL-11 (Balakrishnan et al. 2013), RANKL (Raju et al. 2011) and TSLP (Zhong et al. 2014) signaling pathways. A hyperlink has been provided for each of the PubMed identifier from which the molecular information was annotated. Further, reactions were categorized based on the ligands and ligand-specific pathway maps were generated using PathVisio, a freely available pathway drawing tool (van Iersel et al. 2008). The reactions were exported to an in-house pathway resource known as NetPath database (Kandasamy et al. 2010).
Results and discussion
IL-17 signaling pathway resource contains 93 molecules that are involved in IL-17 family signaling events. We cataloged 43 molecular associations, 73 catalytic reactions, 6 transportation events, 9 activation/inhibition reactions and 154 gene regulations. The IL-17 pathway web page in the NetPath database (http://www.netpath.org/pathways?path_id=NetPath_128) includes a description of the pathway, statistics of the number of molecules and reactions involved in the pathway. The pathway reactions are provided in various community standard data formats including Biological Pathway Exchange (BioPAX level 3) (Demir et al. 2010), Proteomics Standards Initiative for Molecular Interaction (PSI-MI version 2.5) (Orchard and Kerrien 2009) and Systems Biology Markup Language (SBML version 2.1) (Hucka et al. 2003). Users can download these files from the database to visualize the pathway reactions using various freely available software such as Cytoscape (Shannon et al. 2003) and VISIBIOweb (Dilek et al. 2010). The data is also accessible in tab delimited and Microsoft Excel formats. The IL-17 signaling pathway will be periodically updated as and when more signaling events will be available. All the pathway reactions have been reviewed by the pathway authority (SS).
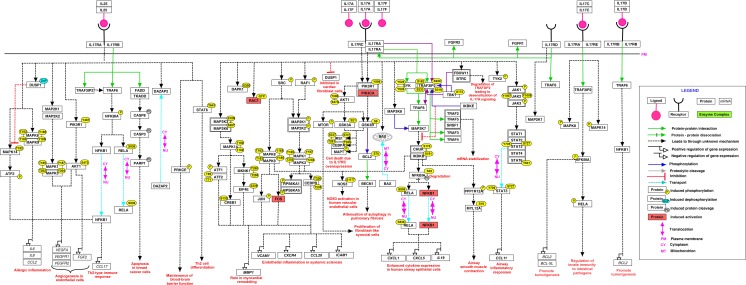
The signaling events triggered by IL-17 are shown in Fig. 1. IL-17A is the most studied member of the IL-17 family of cytokines. IL-17A and its closest homolog IL-17F, signal through a heterodimeric complex consisting of IL-17RA and IL-17RC (Toy et al. 2006). IL-17A and IL-17F can form homodimers (IL-17A/A and IL-17F/F) and heterodimer (IL17A/F) (Hymowitz et al. 2001; Wright et al. 2008). IL-17A induces TRAF6 dependent Nuclear factor kappa B (NFκB) activation, which leads to enhanced cytokine expression in human airway epithelial cells (Huang et al. 2007). Attenuation of autophagy in pulmonary fibrosis was observed through PI3K-GSK3B signaling pathway (Liu et al. 2013). The secretion of matrix metalloproteinases (MMP1 and MMP3) is also enhanced under the influence of IL-17A and IL-17F (Yagi et al. 2007). IL-17B signals through an IL-17RB receptor and promotes tumorigenesis through NFκB dependent BCL2 expression in breast cancer cells (Huang et al. 2013). IL-17C signals through a heterodimeric receptor complex consisting of IL-17RA and IL-17RE (Chang et al. 2011; Song et al. 2011). It regulates innate immunity via TRAF3IP2 dependent activation of NFκB (Song et al. 2011) as shown in Fig. 1. IL-17C also induces the phosphorylation of MAPK8/9 and MAPK14. IL-17C induces BCL-2 and BCL-XL expression in intestinal epithelial cells to promote cell survival and tumorigenesis in both chemically induced and spontaneous intestinal tumor models (Song et al. 2014). The receptor of IL-17D is unknown. An orphan receptor IL-17RD interacts with IL-17RA to regulate IL-17A signaling (Mellett et al. 2012; Rong et al. 2009). It also interacts with FGFR1 and FGFR2 and is involved in cell differentiation (Xiong et al. 2003).
Fig. 1.
A schematic representation of reactions induced by IL-17. The pathway reaction map depicts molecules involved in molecular associations, catalysis, and translocation events induced upon treatment with IL-17 ligand family. Site and residues of post-translational modifications are also mentioned, wherever available. Upon stimulation of IL-17, important signaling pathways including PI3K-AKT, Ras-MAPK, and NFκB are found to be activated
IL-25 is associated with Th2 responses and binds to IL-17RA and IL-17RB (Ely et al. 2009) to trigger downstream signaling events (Fig. 1). TRAF3IP2-TRAF6 dependent NFκB activation is responsible for Th2-type immune response (Maezawa et al. 2006). IL-25 induces the expression of FGF2 through PIK3R1 (Wang et al. 2012). STAT6 is responsible for the differentiation of Th2 cells (Angkasekwinai et al. 2007) and PRKCE is involved in the maintenance of blood–brain barrier (Sonobe et al. 2009).
Conclusions
Availability of IL-17 signaling reactions in a centralized resource will accelarate the understanding of the role of various molecules in the biology of this pathway. These data have been submitted to the NetPath resource and are made available in diverse community standard data exchange formats so that they can be easily visualized and analyzed. This resource will be useful to understand IL-17 signaling in normal and disease conditions.
Acknowledgments
We thank the Department of Biotechnology, Government of India for research support to the Institute of Bioinformatics, Bangalore. We thank the Infosys Foundation for research support to the Institute of Bioinformatics. JS is a recipient of Senior Research Fellowship from the Council of Scientific and Industrial Research, Government of India. KKD and AS are recipients of Junior Research Fellowship from the University Grants Commission, Government of India. AAK is a recipient of Senior Research Fellowship from Indian Council of Medical Research, Government of India. HG is a Wellcome Trust/DBT India Alliance Early Career Fellow.
Conflict of interests
The authors declare no conflicts of interest.
Abbreviations
- IL-17
Interleukin- 17
- IL-25
Interleukin- 25
- IL-17R
Interleukin- 17 receptor
- PPIs
Protein-protein interactions
- PTMs
Post-translational modifications
- BioPAX
Biological Pathway Exchange
- SBML
Systems Biology Markup Language
- PSI-MI
Proteomics Standards Initiative for Molecular Interaction
Contributor Information
Jyoti Sharma, Email: jyoti@ibioinformatics.org.
Lavanya Balakrishnan, Email: lavanya@ibioinformatics.org.
Keshava K. Datta, Email: keshavadatta@ibioinformatics.org
Nandini A. Sahasrabuddhe, Email: nandini.jhmi@gmail.com
Aafaque Ahmad Khan, Email: aafaque@ibioinformatics.org.
Apeksha Sahu, Email: apeksha@ibioinformatics.org.
Anish Singhal, Email: asksinghal@gmail.com.
Derese Getnet, Email: get.derese@gmail.com.
Rajesh Raju, Email: rajrrnbt@gmail.com.
Aditi Chatterjee, Email: aditi@ibioinformatics.org.
Harsha Gowda, Email: harsha@ibioinformatics.org.
T. S. Keshava Prasad, Email: keshav@ibioinformatics.org
Subramanian Shankar, Phone: +91-8971880033, Email: shankar@ibioinformatics.org.
Akhilesh Pandey, Phone: +1-410-502-6662, Email: pandey@jhmi.edu.
References
- Angkasekwinai P, Park H, Wang YH, Chang SH, Corry DB, Liu YJ, Zhu Z, Dong C. Interleukin 25 promotes the initiation of proallergic type 2 responses. J Exp Med. 2007;204:1509–1517. doi: 10.1084/jem.20061675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L, Soman S, Patil YB, Advani J, Thomas JK, Desai DV, Kulkarni-Kale U, Harsha HC, Prasad TS, Raju R, Pandey A, Dimitriadis E, Chatterjee A. IL-11/IL11RA receptor mediated signaling: a web accessible knowledgebase. Cell Commun Adhes. 2013;20:81–86. doi: 10.3109/15419061.2013.791683. [DOI] [PubMed] [Google Scholar]
- Chang SH, Reynolds JM, Pappu BP, Chen G, Martinez GJ, Dong C. Interleukin-17C promotes Th17 cell responses and autoimmune disease via interleukin-17 receptor E. Immunity. 2011;35:611–621. doi: 10.1016/j.immuni.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Reubenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novere N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilek A, Belviranli ME, Dogrusoz U. VISIBIOweb: visualization and layout services for BioPAX pathway models. Nucleic Acids Res. 2010;38:W150–W154. doi: 10.1093/nar/gkq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely LK, Fischer S, Garcia KC. Structural basis of receptor sharing by interleukin 17 cytokines. Nat Immunol. 2009;10:1245–1251. doi: 10.1038/ni.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- Gaffen SL. The role of interleukin-17 in the pathogenesis of rheumatoid arthritis. Curr Rheumatol Rep. 2009;11:365–370. doi: 10.1007/s11926-009-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudenschild D, Moseley T, Rose L, Reddi AH. Soluble and transmembrane isoforms of novel interleukin-17 receptor-like protein by RNA splicing and expression in prostate cancer. J Biol Chem. 2002;277:4309–4316. doi: 10.1074/jbc.M109372200. [DOI] [PubMed] [Google Scholar]
- Huang F, Kao CY, Wachi S, Thai P, Ryu J, Wu R. Requirement for both JAK-mediated PI3K signaling and ACT1/TRAF6/TAK1-dependent NF-kappaB activation by IL-17A in enhancing cytokine expression in human airway epithelial cells. J Immunol. 2007;179:6504–6513. doi: 10.4049/jimmunol.179.10.6504. [DOI] [PubMed] [Google Scholar]
- Huang CK, Yang CY, Jeng YM, Chen CL, Wu HH, Chang YC, Ma C, Kuo WH, Chang KJ, Shew JY, Lee WH. Autocrine/paracrine mechanism of interleukin-17B receptor promotes breast tumorigenesis through NF-kappaB-mediated antiapoptotic pathway. Oncogene. 2013;33:2968–2977. doi: 10.1038/onc.2013.268. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, Filvaroff EH, Yin JP, Lee J, Cai L, Risser P, Maruoka M, Mao W, Foster J, Kelley RF, Pan G, Gurney AL, de Vos AM, Starovasnik MA. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 2001;20:5332–5341. doi: 10.1093/emboj/20.19.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Keerthikumar S, Raju R, Keshava Prasad TS, Ramachandra YL, Mohan S, Pandey A. PathBuilder–open source software for annotating and developing pathway resources. Bioinformatics. 2009;25:2860–2862. doi: 10.1093/bioinformatics/btp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, Aujla SJ, Reardon B, Moore M, Shea P, Schreckhise R, Bukowski TR, Presnell S, Guerra-Lewis P, Parrish-Novak J, Ellsworth JL, Jaspers S, Lewis KE, Appleby M, Kolls JK, Rixon M, West JW, Gao Z, Levin SD. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J Immunol. 2007;179:5462–5473. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ho WH, Maruoka M, Corpuz RT, Baldwin DT, Foster JS, Goddard AD, Yansura DG, Vandlen RL, Wood WI, Gurney AL. IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J Biol Chem. 2001;276:1660–1664. doi: 10.1074/jbc.M008289200. [DOI] [PubMed] [Google Scholar]
- Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, Dowd P, Gurney AL, Wood WI. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc Natl Acad Sci U S A. 2000;97:773–778. doi: 10.1073/pnas.97.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mi S, Li Z, Hua F, Hu ZW. Interleukin 17A inhibits autophagy through activation of PIK3CA to interrupt the GSK3B-mediated degradation of BCL2 in lung epithelial cells. Autophagy. 2013;9:730–742. doi: 10.4161/auto.24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberts E, Joosten LA, van de Loo FA, Schwarzenberger P, Kolls J, van den Berg WB. Overexpression of IL-17 in the knee joint of collagen type II immunized mice promotes collagen arthritis and aggravates joint destruction. Inflamm Res. 2002;51:102–104. doi: 10.1007/BF02684010. [DOI] [PubMed] [Google Scholar]
- Lubberts E, Koenders MI, Oppers-Walgreen B, van den Bersselaar L, Coenen-de Roo CJ, Joosten LA, van den Berg WB. Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of collagen-induced arthritis reduces joint inflammation, cartilage destruction, and bone erosion. Arthritis Rheum. 2004;50:650–659. doi: 10.1002/art.20001. [DOI] [PubMed] [Google Scholar]
- Maezawa Y, Nakajima H, Suzuki K, Tamachi T, Ikeda K, Inoue J, Saito Y, Iwamoto I. Involvement of TNF receptor-associated factor 6 in IL-25 receptor signaling. J Immunol. 2006;176:1013–1018. doi: 10.4049/jimmunol.176.2.1013. [DOI] [PubMed] [Google Scholar]
- Maitra A, Shen F, Hanel W, Mossman K, Tocker J, Swart D, Gaffen SL. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc Natl Acad Sci U S A. 2007;104:7506–7511. doi: 10.1073/pnas.0611589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellett M, Atzei P, Horgan A, Hams E, Floss T, Wurst W, Fallon PG, Moynagh PN. Orphan receptor IL-17RD tunes IL-17A signalling and is required for neutrophilia. Nat Commun. 2012;3:1119. doi: 10.1038/ncomms2127. [DOI] [PubMed] [Google Scholar]
- Miossec P. IL-17 and Th17 cells in human inflammatory diseases. Microbes Infect. 2009;11:625–630. doi: 10.1016/j.micinf.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Moseley TA, Haudenschild DR, Rose L, Reddi AH (2003) Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev 14:155–174 [DOI] [PubMed]
- Onishi RM, Gaffen SL (2010) Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 129:311–321 [DOI] [PMC free article] [PubMed]
- Orchard S, Kerrien S. Molecular interactions and data standardisation. Methods Mol Biol. 2009;604:309–318. doi: 10.1007/978-1-60761-444-9_21. [DOI] [PubMed] [Google Scholar]
- Qian Y, Kang Z, Liu C, Li X. IL-17 signaling in host defense and inflammatory diseases. Cell Mol Immunol. 2010;7:328–333. doi: 10.1038/cmi.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Balakrishnan L, Nanjappa V, Bhattacharjee M, Getnet D, Muthusamy B, Kurian Thomas J, Sharma J, Rahiman BA, Harsha HC, Shankar S, Prasad TS, Mohan SS, Bader GD, Wani MR, Pandey A (2011) A comprehensive manually curated reaction map of RANKL/RANK-signaling pathway. Database (Oxford) 2011:bar021 [DOI] [PMC free article] [PubMed]
- Rong Z, Wang A, Li Z, Ren Y, Cheng L, Li Y, Wang Y, Ren F, Zhang X, Hu J, Chang Z. IL-17RD (Sef or IL-17RLM) interacts with IL-17 receptor and mediates IL-17 signaling. Cell Res. 2009;19:208–215. doi: 10.1038/cr.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvier E, Luciani MF, Mattei MG, Denizot F, Golstein P. CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol. 1993;150:5445–5456. [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, Qian Y. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat Immunol. 2011;12:1151–1158. doi: 10.1038/ni.2155. [DOI] [PubMed] [Google Scholar]
- Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang J, Liu Y, Yao X, Meng G, Shen N, Shi Y, Iwakura Y, Qian Y. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40:140–152. doi: 10.1016/j.immuni.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Sonobe Y, Takeuchi H, Kataoka K, Li H, Jin S, Mimuro M, Hashizume Y, Sano Y, Kanda T, Mizuno T, Suzumura A. Interleukin-25 expressed by brain capillary endothelial cells maintains blood–brain barrier function in a protein kinase Cepsilon-dependent manner. J Biol Chem. 2009;284:31834–31842. doi: 10.1074/jbc.M109.025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol. 2002;169:642–646. doi: 10.4049/jimmunol.169.2.642. [DOI] [PubMed] [Google Scholar]
- Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, Tocker J, Peschon J. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J Immunol. 2006;177:36–39. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinforma. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fan YQ, Lv Z, Yao XJ, Huang KW, Meng Q, Fang CL, Lee TH, Corrigan CJ, An YQ, Ying S. Interleukin-25 promotes basic fibroblast growth factor expression by human endothelial cells through interaction with IL-17RB, but not IL-17RA. Clin Exp Allergy. 2012;42:1604–1614. doi: 10.1111/j.1365-2222.2012.04062.x. [DOI] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Wright JF, Bennett F, Li B, Brooks J, Luxenberg DP, Whitters MJ, Tomkinson KN, Fitz LJ, Wolfman NM, Collins M, Dunussi-Joannopoulos K, Chatterjee-Kishore M, Carreno BM. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J Immunol. 2008;181:2799–2805. doi: 10.4049/jimmunol.181.4.2799. [DOI] [PubMed] [Google Scholar]
- Xiang T, Long H, He L, Han X, Lin K, Liang Z, Zhuo W, Xie R, Zhu B. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133(+) cancer stem-like cells in ovarian cancer. Oncogene. 2015;34:165–176. doi: 10.1038/onc.2013.537. [DOI] [PubMed] [Google Scholar]
- Xiong S, Zhao Q, Rong Z, Huang G, Huang Y, Chen P, Zhang S, Liu L, Chang Z. hSef inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. J Biol Chem. 2003;278:50273–50282. doi: 10.1074/jbc.M306936200. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Andoh A, Inatomi O, Tsujikawa T, Fujiyama Y. Inflammatory responses induced by interleukin-17 family members in human colonic subepithelial myofibroblasts. J Gastroenterol. 2007;42:746–753. doi: 10.1007/s00535-007-2091-3. [DOI] [PubMed] [Google Scholar]
- Yang RB, Ng CK, Wasserman SM, Komuves LG, Gerritsen ME, Topper JN. A novel interleukin-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J Biol Chem. 2003;278:33232–33238. doi: 10.1074/jbc.M305022200. [DOI] [PubMed] [Google Scholar]
- Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- Yao Z, Painter SL, Fanslow WC, Ulrich D, Macduff BM, Spriggs MK, Armitage RJ. Human IL-17: a novel cytokine derived from T cells. J Immunol. 1995;155:5483–5486. [PubMed] [Google Scholar]
- Yao Z, Spriggs MK, Derry JM, Strockbine L, Park LS, VandenBos T, Zappone JD, Painter SL, Armitage RJ. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine. 1997;9:794–800. doi: 10.1006/cyto.1997.0240. [DOI] [PubMed] [Google Scholar]
- Zhong J, Sharma J, Raju R, Palapetta SM, Prasad TS, Huang TC, Yoda A, Tyner JW, van Bodegom D, Weinstock DM, Ziegler SF, Pandey A (2014) TSLP signaling pathway map: a platform for analysis of TSLP-mediated signaling. Database (Oxford) 2014:bau007 [DOI] [PMC free article] [PubMed]
- Zisman-Rozen S, Fink D, Ben-Izhak O, Fuchs Y, Brodski A, Kraus MH, Bejar J, Ron D. Downregulation of Sef, an inhibitor of receptor tyrosine kinase signaling, is common to a variety of human carcinomas. Oncogene. 2007;26:6093–6098. doi: 10.1038/sj.onc.1210424. [DOI] [PubMed] [Google Scholar]