Abstract
Sports related traumatic brain injury (TBI) is a worldwide public health issue, and damage to the corpus callosum (CC) has been considered as an important indicator of TBI. However, contact sports players suffer repeated hits to the head during the course of a season even in the absence of diagnosed concussion, and less is known about their effect on callosal anatomy. In addition, T1-weighted and diffusion tensor brain magnetic resonance images (DTI) have been analyzed separately, but a joint analysis of both types of data may increase statistical power and give a more complete understanding of anatomical correlates of subclinical concussions in these athletes. Here, for the first time, we fuse T1 surface-based morphometry and a new DTI analysis on 3D surface representations of the CCs into a single statistical analysis on these subjects. Our new combined method successfully increases detection power in detecting differences between pre- vs. post-season contact sports players. Alterations are found in the ventral genu, isthmus, and splenium of CC. Our findings may inform future health assessments in contact sports players. The new method here is also the first truly multimodal diffusion and T1-weighted analysis of the CC in TBI, and may be useful to detect anatomical changes in the corpus callosum in other multimodal datasets.
2 Description of purpose
Sports related traumatic brain injury (TBI) has been drawing broad attention in the last few years as short and long term consequences on athletes' health have come to light. In paticular, regional corpus callosum (CC) damage has been detected in moderate to severe TBI cases in MRI based studies [6, 9]. However, less is known about the consequences of repeated head blows sustained by contact sports players even in the absence of diagnosed concussion. Finding sensitive indicators of CC alterations is of great importance for athletes' health assessment and to the design of sports safety rules, with the aim of reducing long-term brain damage.
To capture the potential brain microstructure changes resulted from repeated sport-associated hits, a better understanding of the alteration in fiber structure of the CC is much needed. However, current analysis methods that zoom in on specific brain white matter tracts may discard some of the information in the CC. This is because popular methods such as Tract-Based Spatial Statistics (TBSS) [14] and Tract Specific Analysis (TSA) [19] project the CC into midline or mid-plane surface. Previous postmortem and probabilistic tractography studies have shown that the CC is not a homogenous structure, in terms of fiber composition [1] and topographical distribution[13]. Thus, 3D representations may better localize injury, and may have higher statistical detection power to identify neuro-circuits alterations underling the observed changes in behavior.
In addition to the potential value of using 3D surface representations to investigate diffusivity changes in the CC, morphological alterations may provide complementary information in deciphering brain alterations due to trauma. Impaired neurological abilities in TBI subjects have been attributed to both parenchyma and diffuse injuries, and the former is more easily seen on T1 images [3, 7], while the latter is better detected using DTI. For instance, in moderate to severe TBI studies, T1-based analyses found morphometric changes of several subcortical structures [4, 16]. However, parenchyma and diffuse injuries often occur concomitantly in white matter structures such as the CC. As a result, a joint analysis of diffusion and T1-weighted data may therefore provide a more complete picture of CC changes brought on by brain injury, and is likely to yield higher detection power.
Here we perform the first ever joint analysis of structural and diffusion MRI data on the CC of contact sports players scanned pre- vs. post-season. As only one of our subjects is known to have had a concussion during that particular season, we test whether repeated subclinical injury may lead to detectable changes in callosal anatomy.
3 Method
3.1 Data and Preprocessing
T1-weighted MRI and DTI scans of male collegiate contact sports athletes (19 set of scans in total:10 pre- and 9 post-season scans) were acquired on a 3T GE HDxT scanner. 8 subjects were scanned both pre- and post-season, while 2 subjects were scanned pre-season only, and one subject was scanned post-season only. Each of the DTI scans was obtained using a single shot echo-planar imaging sequence with a b-value of 1000s/mm2 and 25 gradient directions.
T1 images for each of the subjects were first skull stripped and bias corrected, and were then registered to a common template space through linear registration [5]. The CCs were then manually traced on the linear registered T1 images, and the intra-rater percentage overlap was 0.937, in four participants at two different times. To combine the information from DTI and T1, we need to integrate the information in the same space. After preprocessing of DTI data, we first transformed DT images from each subject to their corresponding T1 space, then from each of their own T1 space to the T1 common template space. Both of the transformation were performed using linear registration between the b0-weighted and corresponding T1 images. After each of the linear registrations, the diffusion tensors were resampled using the b0 transformation matrices, and rotated according to the underling anatomy. These steps are all achieved using MedINRIA [17].
3.2 3D Representations and Registration
Based on previously obtained CC segmentations on T1 images, 3D surface representations and conformal mesh grids of the CC were constructed. Subsequently, one-to-one correspondences between vertices were obtained through a constrained harmonic algorithm [18]. After registration, a deformation tensor - where J is the Jacobian of the transformation from the registration - was computed at each vertex on the surface. Its determinant (det J, the difference in surface area) and projection on the log-Euclidean space (log ) [2] were used in later statistical analysis [8, 18].
3.3 Statistical Analysis
Our statistical analysis is performed using either the morphometry information only, the diffusion information only, or a combination of both. To project diffusion indices of each of the CCs onto its surface, we first calculated midlines of all the 3D CCs, and then collected diffusion parameters to each surface vertex along its corresponding radius to the midline. To make sure each of the vertices has some voxels assigned, and to minimize overlap with neighboring vertices, here we chose R = 0.6mm3.
Vertex-wise univariate analysis or multivariate analysis are performed based on the following variables:
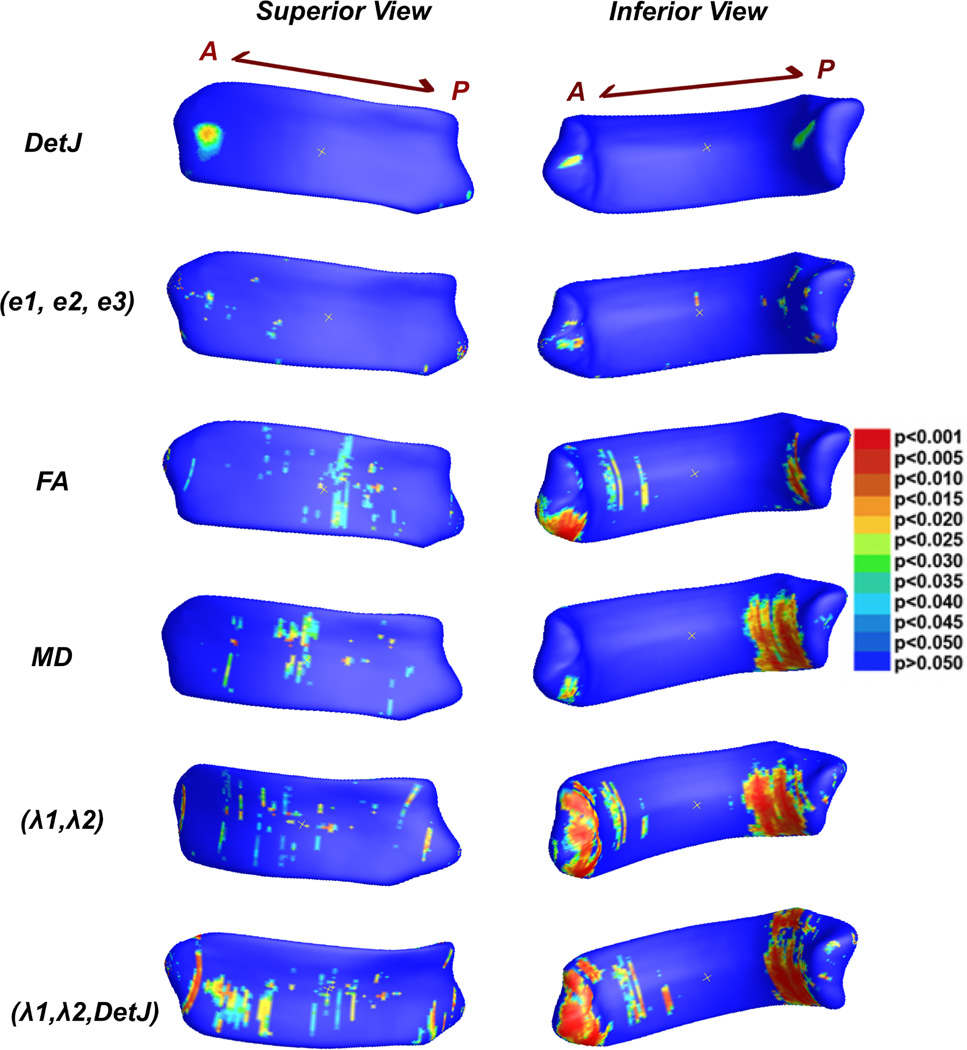
Morphometry information: univariate det J and multivariate (e1, e2, e3) from the logged deformation tensors (Fig. 1, 1st and 2nd row).
Diffusion information: mean FA and MD along the radius of CC for each vertex (Fig. 1, 3th and 4th row).
Diffusion information: multivariate λ1 and λ2 (Fig. 1, 5th row). Note: we did not include λ3. Being small, this value is susceptible to noise and may reduce detection power.
A fusion of morphometry (detJ) and diffusion indices (λ1 and λ2) (Fig.1, 6th row).
Fig. 1.
pre- versus post-season contact sports players using different measures. Vertex-wise corresponding p–values are displayed. In addition, whole structure corrected p–values are p=0.5061 for det J, p= 0.2253 for (e1,e2,e3), p= 0.0797 for FA, p= 0.1181 for MD, p= 0.0352 for (λ1, λ2), p= 0.0136 for (λ1, λ2, det J).
Two types of permutation tests were performed: a vertex-based one to avoid the normal distribution assumption and one over the whole segmented image to correct for multiple comparisons [11, 8].
4 Results
Vertex-wise group differences results based on different methods are displayed in Fig. 1. All methods give consistent results, with the main clusters located in the ventral genu, isthmus, and splenium. For statistics based on morphometry only (row 1st to 2nd), neither univariate det J or multivariate (e1, e2, e3) are able to detect significant whole structure differences between pre-season and post-season contract sport players. For statistics based on diffusion information only (row 3rd to 5th), the multivariate test of (λ1, λ2) shows a significant overall p-value (p=0.0352), while the univariate FA (p=0.0797) or MD (p=0.1181) only display trends. For the T1 and DTI fusion method, the multivariate analysis of (λ1, λ2, det J) shows consistent regions of significant group differences (row 6th), and as expected, outperforms all the other univariate or multivariate methods in detection power (p=0.0136).
5 New or breakthrough work to be presented
Group differences of brain white matter (WM), including CC, are typically analyzed based on voxels, midlines, or midplanes. However, voxel-based methods give poor localization of differences in anatomical regions compared to surface-based ones and may be contaminated by differently oriented tracts [12], while midline- or midplane-based methods rely on assumptions that WM perpendicular to the mid-line or the mid-plane are uniformly distributed. The method introduced in this paper uses clearly defined CC regions traced in T1 images, and largely preserved within tract information projected onto the surface of the CCs. In our dataset, our new method outperforms previous methods in specificity and detection power.
6 Discussion and Conclusion
The ventral genu is neuroanatomically connected to lateral prefrontal cortex, which mediates executive functions and attention. Damage to this cortex region has been linked to impaired learning ability [10]. Posterior CC regions including the ventral isthmus and splenium are connected with the occipital, temporal and the posterior parietal lobe, and in particular are in charge of visual related tasks, including visual memory. The significantly altered ventral anterior and posterior regions detected by our fusion method are interestingly consistent with cognitive analyses on mild TBI contact sports players [15], in which impairments of visual working memory and altered dorsal lateral prefrontal cortex activation are observed.
In this paper, for the first time, we fuse the T1 based morphometry information and DTI based diffusion information on 3D CCs, and successfully detect significant differences between pre- vs. post-season contact sports players. The fused method also outperforms univariate and multivariate 3D analyses based on the structural information only, or the diffusion information only in terms of detection power. Our findings provide new neuroanatomic evidence for the diagnosis of mTBI, and may enhance our understanding of the neuroanatomical consequences mTBI related pathologies.
References
- 1.Aboitiz F, et al. Fiber composition of the human corpus callosum. Brain research. 1992;598(1):143–153. doi: 10.1016/0006-8993(92)90178-c. [DOI] [PubMed] [Google Scholar]
- 2.Arsigny V, et al. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2006. Springer; 2006. A log-euclidean framework for statistics on diffeomorphisms; pp. 924–931. [DOI] [PubMed] [Google Scholar]
- 3.Granacher RP., Jr . Traumatic brain injury: Methods for clinical and forensic neuropsychiatric assessment. CRC Press; 2007. [Google Scholar]
- 4.Holli KK, et al. Texture analysis of mr images of patients with mild traumatic brain injury. BMC medical imaging. 2010;10(1):8. doi: 10.1186/1471-2342-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkinson M, et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 6.Kraus MF, et al. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain. 2007;130(10):2508–2519. doi: 10.1093/brain/awm216. [DOI] [PubMed] [Google Scholar]
- 7.Lebby PC. Brain Imaging: A Guide for Clinicians. Oxford University Press; 2013. [Google Scholar]
- 8.Lepore F, et al. Generalized tensor-based morphometry of hiv/aids using multivariate statistics on deformation tensors. Medical Imaging, IEEE Transactions on. 2008;27(1):129–141. doi: 10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer A, et al. A prospective diffusion tensor imaging study in mild traumatic brain injury. Neurology. 2010;74(8):643–650. doi: 10.1212/WNL.0b013e3181d0ccdd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mega MS, et al. Frontal-subcortical circuits and neuropsychiatric disorders. The Journal of Neuropsychiatry and Clinical Neurosciences. 1994 doi: 10.1176/jnp.6.4.358. [DOI] [PubMed] [Google Scholar]
- 11.Nichols TE, et al. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human brain mapping. 2001;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell LJ, et al. Tract-based morphometry for white matter group analysis. Neuroimage. 2009;45(3):832–844. doi: 10.1016/j.neuroimage.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park HJ, et al. Corpus callosal connection mapping using cortical gray matter parcellation and dt-mri. Human brain mapping. 2008;29(5):503–516. doi: 10.1002/hbm.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith SM, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Talavage T, et al. Functionally-detected cognitive impairment in high school football players without clinically-diagnosed concussion. Journal of neurotrauma. 2013 doi: 10.1089/neu.2010.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomaiuolo F, et al. Gross morphology and morphometric sequelae in the hippocampus, fornix, and corpus callosum of patients with severe non-missile traumatic brain injury without macroscopically detectable lesions: a t1 weighted mri study. Journal of Neurology, Neurosurgery & Psychiatry. 2004;75(9):1314–1322. doi: 10.1136/jnnp.2003.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toussaint N, et al. Medinria: Medical image navigation and research tool by inria. Proc. of MICCAI'07 Workshop on Interaction in medical image analysis and visualization. 2007 [Google Scholar]
- 18.Wang Y, et al. Surface-based tbm boosts power to detect disease effects on the brain: An n= 804 adni study. Neuroimage. 2011;56(4):1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. A tract-specific framework for white matter morphometry combining macroscopic and microscopic tract features. Medical image analysis. 2010;14(5):666–673. doi: 10.1016/j.media.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]