Abstract
Epigenetic modifications by DNA methylation are associated with a wide range of diseases. Previous studies in psoriasis have concentrated on epigenetic changes in immune cells or in total skin biopsies that include stromal-associated changes. In order to improve our understanding of the role of DNA methylation in psoriasis, we sought to obtain a comprehensive DNA methylation signature specific for the epidermal component of psoriasis and to analyze methylation changes during therapy. Genome-wide DNA methylation profiling of epidermal cells from 12 patients undergoing narrow-band UVB phototherapy and 12 corresponding healthy controls revealed a distinct DNA methylation pattern in psoriasis compared with controls. A total of 3,665 methylation variable positions (MVPs) were identified with an overall hypomethylation in psoriasis patient samples. DNA methylation pattern was reversed at the end of phototherapy in patients showing excellent clinical improvement. Only 7% of phototherapy-affected MVPs (150 out of 2,108) correlate with nearby gene expression. Enrichment of MVPs in enhancers indicates tissue-specific modulation of the transcriptional regulatory machinery in psoriasis. Our study identified key epigenetic events associated with psoriasis pathogenesis and helps understand the dynamic DNA methylation landscape in the human genome.
Introduction
Psoriasis is a common chronic inflammatory skin disease that also can affect nails and joints (Perera et al., 2012). Approximately 2–3% of the world's population have psoriasis, with rates varying between countries and races (Crow, 2012). Psoriasis is characterized by altered keratinocyte differentiation and an exaggerated inflammatory response with both genetic and environmental etiologies (Perera et al., 2012). Several susceptibility loci and distinct gene expression patterns have been identified, and genes related to cell differentiation, immune response, metabolism, and oxidation reduction are implicated in disease pathogenesis (Mitsui et al., 2012; Tian et al., 2012; Tsoi et al., 2012; Gu et al., 2015).
Recently, epigenetic regulations such as abnormal DNA methylation were reported in psoriasis (Roberson et al., 2012; Hou et al., 2013; Zhang et al., 2013; Park et al., 2014). DNA methylation at the 5-position of cytosine in mammals is the most studied epigenetic mark that is dynamic across lifetime, affected by environmental insults, and essential for several developmental and cellular processes (Smith and Meissner, 2013; Ziller et al., 2013). Aberrant DNA methylation has been recognized in several developmental disorders and cancer (Feinberg, 2007; Jones, 2012). In psoriasis, altered global DNA methylation status in CD4+ T cells (Park et al., 2014) and dermal mesenchymal stem cells (Hou et al., 2013) has been shown. Changes in DNA methylation have also been seen in psoriatic lesions (Roberson et al., 2012; Zhang et al., 2013) and could be reverted back to baseline after 1 month of anti-TNF–α therapy (Roberson et al., 2012).
Psoriasis is a lifelong systemic disorder that is temporarily quenched when appropriately treated. Therefore, psoriasis has long served as a model for the reciprocity between disease and treatment (Keaney and Kirsner, 2010). A variety of options are available for treatment of psoriasis, including topical agents, systemic agents, phototherapy, combination therapies, and biological therapies (Rahman et al., 2012). Insights into the therapeutic mechanisms for psoriasis have improved our understanding of its pathogenesis (Piskin et al., 2004; Johnson-Huang et al., 2010; Racz et al., 2011; Gu et al., 2015). In order to gain further insight, we sought to obtain a more comprehensive and specific DNA methylation signature of psoriatic epidermis following narrow-band UVB phototherapy, a well-established first-line treatment for psoriasis, which is effective for about 70% of patients (Kirke et al., 2007). By the use of the HumanMethylation450 BeadChip to examine genome-wide methylation status of epidermal cells from patients undergoing phototherapy, we here provide an in-depth DNA methylation signature of psoriasis and have identified key methylation events involved in psoriasis pathogenesis.
Results
Identification of DNA methylation changes in psoriatic epidermis
Epidermis-specific DNA methylation profiles in psoriasis and matched controls were measured using the Illumina Infinium HumanMethylation450 BeadChip platform (450K) (Illumina, San Diego, CA), which evaluates methylation status of 482,421 CpG sites covering key features of the human genome (Bibikova et al., 2011). The ChIP analysis methylation pipeline package was applied for data analysis (Morris et al., 2014). The beta-value was chosen as a measure of the methylation level, which ranges from 0 (no methylation) to 1 (complete methylation). On the basis of the methylation level of 470,903 sites, similar global methylation status was seen between psoriasis and controls, with a median beta-value of 0.590 in controls and 0.554 in psoriasis (PRE-UV).
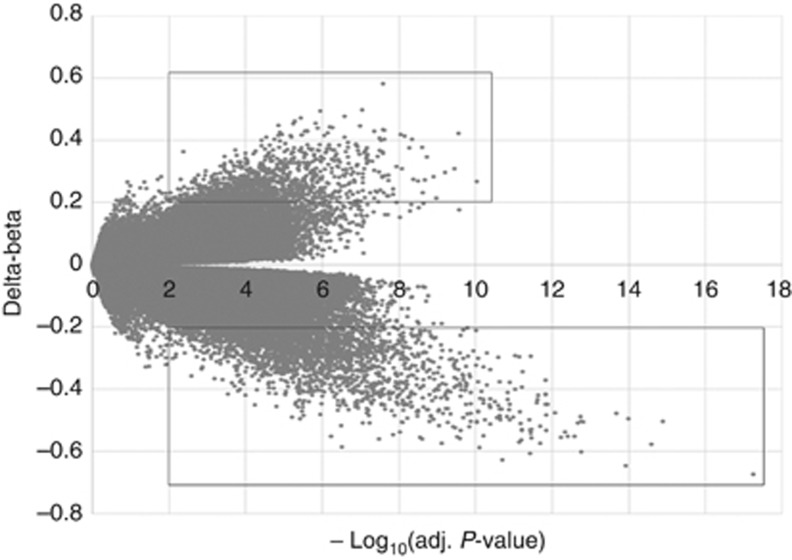
Methylation differences between psoriasis and controls were calculated as delta-beta and plotted against corresponding −log10 (false-discovery rate (FDR) adjusted P-value), as shown in Figure 1. We defined CpG sites with delta-beta>0.2 and FDR-adjusted P-value<0.01 as methylation variable positions (MVPs). From the 470,903 informative probes, 3,665 MVPs were identified, representing <1% of the total sites surveyed. There were more hypomethylated CpG sites in psoriasis (2,542 MVPs, delta-beta<−0.2; bottom square in Figure 1) than hypermethylated CpG sites (1,123 MVPs, delta-beta>0.2; upper square), and the hypomethylated CpG sites displayed larger degrees of methylation changes.
Figure 1.
Identification of differences in DNA methylation between psoriasis and controls. Scatter plot between methylation changes (delta-beta, psoriasis PRE-UV vs. controls) and corresponding −log10(false-discovery rate (FDR) adj. P-value) for total assessed 470,903 sites was shown. CpG sites with delta-beta>0.2 and −log10(FDR adj. P-value)>2 were defined as methylation variable positions (MVPs). The upper square indicates hypermethylated MVPs, and the bottom square indicates hypomethylated MVPs in psoriasis compared with controls.
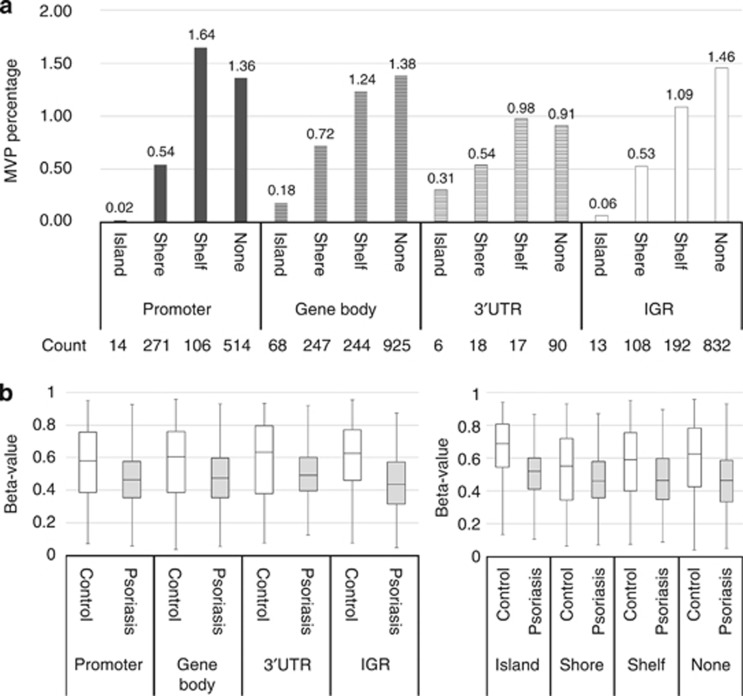
All variant probes were annotated with respect to location and then categorized into groups based on chromosome (1–22), enhancer (true or none), genomic location (promoter region, gene body, 3′-UTR or IGR), and CpG density (island, shore, shelf, none). We found that MVPs appeared within all autosomal chromosomes, ranging from 0.39% of assessed sites in chromosome 19 to 1.03% in chromosome 1. Psoriasis MVPs were preferentially located at the enhancers, with 1.84% of enhancer-located sites differentially methylated, whereas only 0.49% of non-enhancer-related sites were differentially methylated. When probes were grouped into 16 classes based on genomic position and CpG density, profound differences in MVP percentage were found across different groups. Only 0.02% of sites located in CpG islands of the promoter region were differentially methylated. The region having the highest percentage of CpG sites as MVPs (1.64%) was distal to CpG island (shelf) in the promoter region (Figure 2a). The DNA methylation status of 3,665 MVPs was compared between psoriasis and controls. The medium methylation level in controls is 0.61, whereas 0.46 in psoriasis. Figure 2b shows that, regardless of genomic region or CpG density, significant changes in DNA methylation lead to an overall hypomethylated status in psoriasis.
Figure 2.
Distribution of psoriasis methylation variable positions (MVPs) across genomic regions and related to CpG density. (a) Total accessed CpG sites (470,903) were divided into 16 groups according to genomic location (promoter region, gene body, 3′-UTR or IGR) and CpG density (island, shore, shelf, none). The promoter region refers to TSS1,500, TSS200, 5′-UTR, and 1stExon. MVP percentage in each group is shown in the bar graph. Count of MVPs in each group is also shown in the bottom. (b) Overall hypomethylation in psoriasis. Total MVPs (3,665) were divided into four groups based on genomic location or CpG density. Box plots showed the distribution of beta-value for corresponding MVPs in each group. White boxes represent controls, and gray boxes represent psoriasis (PRE-UV samples).
Effect of phototherapy on psoriasis and DNA methylation
Differential analysis using significance analysis of microarrays (SAM) was performed to identify significantly affected MVPs by phototherapy in paired samples. A methylation change with delta-beta>0.1 and FDR<0.05 was considered significant. After 1 month of treatment (MID-UV vs. PRE-UV), the methylation level of 73 CpG sites was significantly modified. At the end of phototherapy (POST-UV vs. PRE-UV), a total of 2,108 CpG sites were significantly affected. Top 50 hyper and 50 hypomethylated MVPs are listed in Supplementary Table S1 online, ordered by methylation changes following phototherapy (POST-UV vs. PRE-UV). DAVID (Database for Annotation, Visualization, and Integrated Discovery) functional annotation analysis was performed for 1,082 known genes that are proximal to the 2,108 MVPs. Top five enriched annotation clusters in biological processes relate to inflammatory response, cytoskeleton organization, response to hormone stimulus, regulation of cell motion, and regulation of programmed cell death (Supplementary Table S2 online).
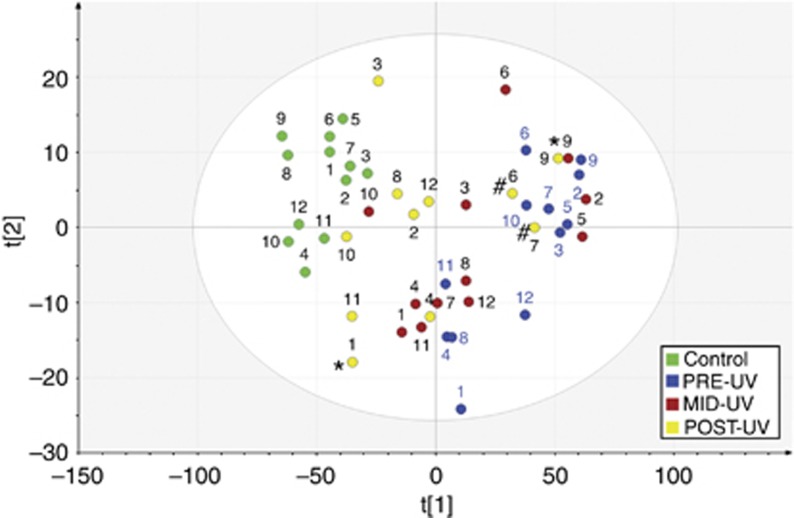
Principle component analysis (PCA) was performed to study sample similarities based on 2,108 MVPs. Figure 3 displays the score plot from principle component analysis showing first and second component that explains 75.1 and 4.8% of the variance, respectively. Samples close to each other have similar methylation profiles. We can see that control samples position closer to each other in the upper left-hand corner, whereas psoriasis samples before treatment are located most distal to the controls. It is also visualized that DNA methylation profiles in psoriasis changed following phototherapy to become more similar to the control group. Three POST-UV samples from patient 6, 7, and 9 are positioned in the upper right-hand corner, showing distinct methylation patterns compared with other POST-UV samples. Response assessment on the 11 patients that fulfilled treatment showed 7 patients with excellent clinical response, 2 with good response (patient 1, 9), and 2 with unsatisfactory response (patient 6, 7). When connecting clinical data to DNA methylation, we found that the DNA methylation status was associated with clinical outcome. All seven POST-UV samples from patients with excellent clinical improvement are positioned closer to the controls. The three particular POST-UV samples are from patients with unsatisfactory response (patient 6, 7) or good rather than excellent clinical improvement (patient 9). For the other POST-UV sample with good clinical improvement (patient 1), although appearing more similar to the controls following phototherapy, was positioned most distal to the controls in the lower left-hand corner.
Figure 3.
Score plot from PCA analysis for 12 psoriasis patients undergoing phototherapy and 12 corresponding controls. PCA score plot of the two first PCs of the methylation data set providing a map of how the samples relate to each other. The samples are colored by the sample group. Assessment of clinical response was given in three grades: Excellent, good, or unsatisfactory. POST-UV samples from patients with good or unsatisfactory response to phototherapy are indicated with symbol * or #, respectively.
For each patient, we further calculated the overall improvement of DNA methylation at the end of phototherapy (percentage increase or decrease), as well as the overall methylation difference between POST-UV and healthy controls (Supplementary Table S3 online). It is clear that, at the end of treatment, patients with excellent and good clinical outcome presented the highest methylation improvements (except patient 4 and 9), as well as the least methylation differences to healthy controls (except patient 9).
DNA methylation and gene expression
The role of DNA methylation was studied by examining their impact on the expression of proximal genes. Epidermal-specific gene expression data from the same patients and controls were obtained from our previously reported gene expression profiling study (Gu et al., 2015). The relationships between proximal gene expression and DNA methylation (Pearson's correlation coefficient (r)) were determined. For 2,108 MVPs, only 150 MVPs were moderately/strongly correlated with proximal gene expression (moderate correlation (r=0.4–0.6), strong correlation (r=0.7–0.9)). The strongest correlation (r=−0.89) was seen for S100A9 (S100 Calcium Binding Protein A9), a regulator of inflammatory processes and immune response (Kerkhoff et al., 2012). Expression of several other S100 genes and epithelial cell differentiation genes also strongly correlated with DNA methylation (Supplementary Table S4 online). On average, 7% of MVPs showed a correlation between methylation and expression levels. When allocating MVPs to different genomic locations as shown in Supplementary Table S5 online, we found that MVPs located in the promoter regions tend to correlate to nearby gene expression (14.12%) compared with MVPs located in other genomic regions (0–8.77%).
When annotating probes according to enhancer, we found that 1,098 out of 2,108 MVPs were located in enhancers. Hypergeometric test indicated that enhancer-located MVPs are enriched in this list (P (x ≥1,098)=0). When analyzing associations between DNA methylation and gene expression for enhancer-located MVPs (Supplementary Table S5 online), less correlations were found. Enhancers are remote regulatory elements that can locate at great distances (as far as 1 Mb) from the genes they control, and the gene most proximal to an enhancer is not necessarily its target (Visel et al., 2009). In order to connect MVPs to transcriptional regulation of distal genes, we started to investigate whether or not expression of genes situated distal to MVPs tend to be modified by phototherapy. A total of 9,447 genes are with a distance of up to 500 kb to 1,098 enhancer-located MVPs. However, expression of only 250 genes was significantly affected by phototherapy, most likely due to selection by chance (Fisher's exact test, P>0.05).
DNase I hypersensitive sites (DHSs) are markers of regulating DNA and have been used to map regulatory DNA regions including enhancer, promoter, insulators, silencers, and locus control regions (Thurman et al., 2012). Recently, cell type–specific DHSs were identified (Sheffield et al., 2013), allowing us to investigate whether psoriasis MVPs co-localize with keratinocyte-specific regulatory elements. A total of 3,115 keratinocyte-specific DHSs were extracted from the Regulatory Elements Database (available from: http://dnase.genome.duke.edu), and we found that only eight MVPs position in keratinocyte-specific DHSs.
450K data validation
Validation of BeadChip measurements was carried out on selected CpG sites using OneStep qMethyl (Zymo Research, Irvine, CA). For all the genes tested, qMethyl data were highly correlated with array data, with Pearson's correlation coefficient (r) of 0.96 for HN1L and IRF2, 0.85 for IFI27, and 0.90 for PDK2 (P<0.001; Supplementary Figure S1 online).
Discussion
We performed a genome-wide study of epidermis-specific methylation changes in psoriasis, before, during, and after phototherapy. As the DNA methylation profile varies between cell types, it is important to consider that the infiltration of inflammatory cells in psoriatic skin alters the cell composition, as compared with normal tissue (Jaffe and Irizarry, 2014). In this study, in order to minimize the influence of inflammatory cells and to obtain the most comprehensive data set possible in the genome-scale methylation examination, we separated the epidermis from the dermis and used the most comprehensive array platform currently available (Bibikova et al., 2011). Nevertheless, some methylation alterations might arise from differences in inflammatory infiltration into the epidermis and/or from differences in epidermal cell composition with age, rather than be true psoriasis-related keratinocyte defects. It is also possible that keratinocyte-specific methylation is regulated by immune or other stromal cells, so that therapy-related changes in methylation profiles may be indirectly due to therapeutic effects on immune cells rather than direct effects on keratinocytes.
Global hypomethylation and promoter-specific hypermethylation have been reported in cancer (Berman et al., 2012). We also found an overall hypomethylation in psoriasis that is in contrast to previous results from other psoriasis studies (Roberson et al., 2012; Zhang et al., 2013), probably due to the limitation of using whole-biopsy DNA and/or targeted arrays of promoter regions in these studies. The tendency of differences toward DNA hypermethylation in the CD4+ T cells of psoriasis patients (Park et al., 2014) could explain the hypermethylation pattern in psoriatic lesions where both keratinocytes and inflammatory cells are dominant. Variant methylation pattern in different cell types from psoriatic lesions was also suggested by showing more promoter hypomethylated genes than hypermethylated genes in mesenchymal stem cells (Hou et al., 2013).
Among 3,665 identified MVPs, only 14 positions were located at promoter CpG islands. CpG islands are generally unmethylated in normal cells and less dynamic and less tissue-specific compared with non-CpG island methylation (Jones, 2012). Our finding that the methylation status barely changed within CpG islands in the promoters supports that methylation status in these regions is relatively stable. On the other hand, it has been shown that enhancers tend to be CpG-poor and have incomplete and dynamic methylation (Jones, 2012). Consistent with this, we found most DNA methylation changes to be located in enhancers.
DNA methylation is an inherently reversible change, and, our study shows that following phototherapy, altered methylation status in the epidermis could be reversed back toward that observed in normal tissue. It has been shown previously that, after 1 month of anti-TNF–α therapy, DNA methylation in several assessed CpG sites was partially reversed in responders, which thus might be useful for predicting response early in treatment (Roberson et al., 2012). However, in our study by examining genome-wide methylation changes after 1 month of phototherapy (at the middle stage of treatment), we found that only 73 out of 3,665 MVPs were significantly affected by phototherapy. More importantly, at this stage, no obvious difference in DNA methylation pattern could be seen between patients with different clinical outcome. Therefore, even though reversal of DNA methylation is associated with clinical outcome at the end of phototherapy, detecting DNA methylation changes at the early stage of phototherapy cannot predict outcome of treatment. Nevertheless, it is possible that DNA methylation processes offer targets for therapeutic intervention if it can be verified that changes in the DNA methylation pattern by itself has a positive effect on the clinical symptoms.
DNA methylation is acknowledged to have a key role in gene regulation and disease susceptibility, whereas the relationship between DNA methylation and transcription is not fully dissected (Jones, 2012). We believe that CpG sites that are dysregulated in psoriasis and respond to treatment represent key methylation events during the development of psoriasis. A total of 2,108 phototherapy-responding MVPs were identified in this study, and, their nearby genes were significantly enriched in biological processes known for driving psoriasis, such as inflammatory response and cytoskeleton organization. However, altered DNA methylation status is barely correlated to expression of nearly genes. Although we found that MVPs are enriched in enhancers, we could not show their connection with distal genes. Therefore, the impact of DNA methylation on transcriptional regulation is far from understood. Regulatory DNA variations associated with common human disease have been shown (Maurano et al., 2012). As regulatory elements are not organized in a gene-centric manner but could form large regulatory networks with great tissue specificity (Maurano et al., 2012; Symmons and Spitz, 2013), it is possible that modifications in DNA methylation could affect the interplay between different regulatory elements and lead to altered gene expression that is tissue specific. It is worth noting that keratinocyte-specific regulatory DNA marked by DHSs is depleted of methylation variation in psoriasis, indicating that DNA methylation provides additional signals for tissue-specific transcriptional regulation.
A total of 32 genes showed a strong correlation between DNA methylation and gene expression, and, many of these genes/gene families are represented by multiple probes (Supplementary Table S4 online), suggesting involvement of epigenetic modifications for regulation of these genes. Interestingly, several psoriasis susceptibility genes were found, encoding key components of the skin barrier, such as small proline-rich proteins (Kainu et al., 2009), late cornified envelope protein 3D (Tsoi et al., 2012), and gap junction protein connexin 26 (Liu et al., 2012; Tang et al., 2014). Genetic and/or epigenetic variance in these genes might have a role in psoriasis susceptibility.
In summary, genome-scale epidermis-specific DNA methylation profiles were obtained for 12 psoriasis patients before, during, and at the end of phototherapy. Altered DNA methylation was seen in psoriasis compared with healthy controls, and reversion of abnormal methylation was observed following phototherapy in patients with clinical improvement. As part of the coordinated regulatory programs, epigenetic alterations in the human genome are important events during development of psoriasis.
Materials and Methods
Patients and tissue samples
Twelve patients diagnosed with plaque-type psoriasis were recruited to the study, which was approved by the Regional Ethics Review Board, Umeå, Sweden (Dnr 08-108M), and performed in accordance with the Declaration of Helsinki Principles. Written informed consent was obtained from all subjects. narrow-band UVB irradiation was administered to the whole body using a cabinet (PCL 8000, Puva Combi Light—ARKADE, Heverlee, Belgium) equipped with fluorescent lamps (UVB TL100W/01, Philips, Eindhoven, The Netherlands). Treatment comprising ~24 sessions was given during 2 to 3 months. A total of six 4 mm diameter punch biopsies were taken from lesions on each patient: two prior to treatment (PRE-UV), two at the middle stage of treatment (after 1 month of treatment, MID-UV), and two before the last treatment session (POST-UV). One patient (patient 5) left at the middle stage of phototherapy; thus, only PRE-UV and MID-UV samples could be collected. All patients had moderate–severe psoriasis and a history stating improvement by sun exposure. Clinical improvement was assessed by evaluation of erythema, desquamation, and induration following phototherapy. Three grade evaluation (excellent, good, and unsatisfactory) was performed due to clinical routine by an experienced nurse and included the patient's statement of improvement. Clinical data on patients are shown in Supplementary Table S6 online. Twelve healthy age–, sex–, and skin type–matched volunteers were also recruited, and two punch biopsies were taken from the buttocks.
Epidermal DNA isolation and bisulfite treatment
Skin biopsies were fresh frozen in liquid nitrogen and stored at −80°C until DNA extraction. For epidermis and dermis separation, biopsies were incubated in 3.8% ammonium thiocyanate (Sigma-Aldrich, St Louis, MO) in Dulbecco's phosphate-buffered saline, pH 7.4, at room temperature for 20 minutes (Trost et al., 2007). Separated epidermis was processed for DNA isolation using the PureLink Genomic DNA Kit (Life Technologies, Carlsbad, CA). The quantity and purity of DNA were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). DNA quality was confirmed by gel electrophoresis. A measure of 500 ng of genomic DNA was used for bisulfite conversion using the Zymo EZ DNA Methylation kit (Zymo Research).
Genome-scale DNA methylation array
Bisulfite-converted DNA was used for methylation profiling using the HumanMethylation450 platform. iScan Reader was used to image BeadChips. The ChIP analysis methylation pipeline package was applied for data analysis (Morris et al., 2014). By default, ChIP analysis methylation pipeline filtered the data for detection P-value<0.01. Probes with <3 beads in at least 5% of samples per probe were filtered out and probes from X and Y chromosomes were removed, resulting in 470,903 probes. Intra-array data normalization using the BMIQ (Beta MIxture Quantile dilation) method (Teschendorff et al., 2013) was performed for correcting bias introduced by the Infinium type 2 probe design. Singular value decomposition method (Teschendorff et al., 2011) was used to assess the magnitude of batch effects in relation to biological variation.
The beta-value was chosen as a measure of the methylation level and was transformed to M-value for differential analysis (Du et al., 2010). The limma package implemented in ChIP analysis methylation pipeline was used to calculate the Benjamini and Hochberg FDR-adjusted P-value for differential methylation between psoriasis (PRE-UV) and controls. To identify significantly modified CpG sites following phototherapy, significance analysis of microarrays was performed in Mev 4.8.1 (Dana-Farber Cancer Institute, Boston, MA) (Saeed et al., 2003). Functional annotation analysis was performed using DAVID (Huang da et al., 2009a, b). Principle component analysis was performed using SIMCA software (v. 13, MKS Umetrics AB, Sweden).
Validation of 450K data
Validation of BeadChip measurements was carried out using OneStep qMethyl (Zymo Research). The list of top differentially methylated CpG sites was analyzed, and primers for four CpG sites, cg27431500 (HN1L), cg20161089 (IFI27), cg11802666 (IRF2), and cg04091816 (PDK2), were designed (Supplementary Table S7 online). Sufficient DNA was available from only four patients for validation. Methylation levels in four healthy controls were also determined. Real-time PCR was performed using 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA).
Statistics
All statistical tests were conducted in R (version 3.1.1) (The R Foundation for Statistical Computing, Austria). Fisher's exact test was performed to evaluate the significance of overlap between two lists of genes. The Hypergeometric test was used to evaluate the significance of enrichment for a given data set. Pearson's correlation coefficient (r) was calculated to evaluate the strength of correlation between DNA methylation and gene expression levels, as well as the correlation between array and qMethyl data.
Data access
The DNA methylation data have been deposited in the NCBI Gene Expression Omnibus database (GEO) under access number GSE63315. Gene expression data related to this study are also available at GEO (GSE53431).
Acknowledgments
This study was supported by grants from Hudfonden, Lion's Cancer Research Foundation, Umeå University, and The Swedish Cancer Society—contract number 140752. We are grateful to Maria Östman for her help with patient recruitment and collection of biopsies.
Glossary
- DHS
DNase I hypersensitive site
- FDR
false-discovery rate
- MVP
methylation variable position
The authors state no conflict of interest.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Berman BP, Weisenberger DJ, Aman JF et al. (2012) Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics 98:288–295 [DOI] [PubMed] [Google Scholar]
- Crow JM. (2012) Psoriasis uncovered. Nature 492:S50–S51 [DOI] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC et al. (2010) Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformat 11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440 [DOI] [PubMed] [Google Scholar]
- Gu X, Nylander E, Coates PJ et al. (2015) Oxidation reduction is a key process for successful treatment of psoriasis by narrow-band UVB phototherapy. Acta Derm Venereol 95:140–146 [DOI] [PubMed] [Google Scholar]
- Hou R, Yin G, An P et al. (2013) DNA methylation of dermal MSCs in psoriasis: identification of epigenetically dysregulated genes. J Dermatol Sci 72:103–109 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. (2009. a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. (2009. b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA. (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genom Biol 15:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Huang LM, Suarez-Farinas M, Sullivan-Whalen M et al. (2010) Effective narrow-band UVB radiation therapy suppresses the IL-23/IL-17 axis in normalized psoriasis plaques. J Investig Dermatol Symp Proc 130:2654–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492 [DOI] [PubMed] [Google Scholar]
- Kainu K, Kivinen K, Zucchelli M et al. (2009) Association of psoriasis to PGLYRP and SPRR genes at PSORS4 locus on 1q shows heterogeneity between Finnish, Swedish and Irish families. Exp Dermatol 18:109–115 [DOI] [PubMed] [Google Scholar]
- Keaney TC, Kirsner RS. (2010) New insights into the mechanism of narrow-band UVB therapy for psoriasis. J Investig Dermatol Symp Proc 130:2534. [DOI] [PubMed] [Google Scholar]
- Kerkhoff C, Voss A, Scholzen TE et al. (2012) Novel insights into the role of S100A8/A9 in skin biology. Exp Dermatol 21:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirke SM, Lowder S, Lloyd JJ et al. (2007) A randomized comparison of selective broadband UVB and narrowband UVB in the treatment of psoriasis. J Investig Dermatol Symp Proc 127:1641–1646 [DOI] [PubMed] [Google Scholar]
- Liu QP, Wu LS, Li FF et al. (2012) The association between GJB2 gene polymorphism and psoriasis: a verification study. Arch Dermatol Res 304:769–772 [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337:1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Suarez-Farinas M, Belkin DA et al. (2012) Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Investig Dermatol Symp Proc 132:1615–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A et al. (2014) ChAMP: 450k chip analysis methylation pipeline. Bioinformatics 30:428–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GT, Han J, Park SG et al. (2014) DNA methylation analysis of CD4+ T cells in patients with psoriasis. Arch Dermatol Res 306:259–268 [DOI] [PubMed] [Google Scholar]
- Perera GK, Di Meglio P, Nestle FO. (2012) Psoriasis. Annu Rev Pathol 7:385–422 [DOI] [PubMed] [Google Scholar]
- Piskin G, Tursen U, Sylva-Steenland RM et al. (2004) Clinical improvement in chronic plaque-type psoriasis lesions after narrow-band UVB therapy is accompanied by a decrease in the expression of IFN-gamma inducers—IL-12, IL-18 and IL-23. Exp Dermatol 13:764–772 [DOI] [PubMed] [Google Scholar]
- Racz E, Prens EP, Kurek D et al. (2011) Effective treatment of psoriasis with narrow-band UVB phototherapy is linked to suppression of the IFN and Th17 pathways. J Investig Dermatol Symp Proc 131:1547–1558 [DOI] [PubMed] [Google Scholar]
- Rahman M, Alam K, Ahmad MZ et al. (2012) Classical to current approach for treatment of psoriasis: a review. Endocr Metab Immune Disord Drug Targets 12:287–302 [DOI] [PubMed] [Google Scholar]
- Roberson ED, Liu Y, Ryan C et al. (2012) A subset of methylated CpG sites differentiate psoriatic from normal skin. J Investig Dermatol Symp Proc 132:583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J et al. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Thurman RE, Song L et al. (2013) Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genom Res 23:777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14:204–220 [DOI] [PubMed] [Google Scholar]
- Symmons O, Spitz F. (2013) From remote enhancers to gene regulation: charting the genome's regulatory landscapes. Philos Trans R Soc Lond B Biol Sci 368:20120358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Jin X, Li Y et al. (2014) A large-scale screen for coding variants predisposing to psoriasis. Nat Genet 46:45–50 [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M et al. (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Zhuang J, Widschwendter M. (2011) Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics 27:1496–1505 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R et al. (2012) The accessible chromatin landscape of the human genome. Nature 489:75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Krueger JG, Li K et al. (2012) Meta-analysis derived (MAD) transcriptome of psoriasis defines the "core" pathogenesis of disease. PLoS One 7:e44274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost A, Bauer JW, Lanschutzer C et al. (2007) Rapid, high-quality and epidermal-specific isolation of RNA from human skin. Exp Dermatol 16:185–190 [DOI] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J et al. (2012) Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44:1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. (2009) Genomic views of distant-acting enhancers. Nature 461:199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao M, Liang G et al. (2013) Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun 41:17–24 [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F et al. (2013) Charting a dynamic DNA methylation landscape of the human genome. Nature 500:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.