Abstract
In North American swine, there are numerous antigenically distinct H1 influenza A virus (IAV) variants currently circulating, making vaccine development difficult due to the inability to formulate a vaccine that provides broad cross-protection. Experimentally, live-attenuated influenza virus (LAIV) vaccines demonstrate increased cross-protection compared to inactivated vaccines. However, there is no standardized assay to predict cross-protection following LAIV vaccination. Hemagglutination-inhibiting (HI) antibody in serum is the gold standard correlate of protection following IAV vaccination. LAIV vaccination does not induce a robust serum HI antibody titer; however, a local mucosal antibody response is elicited. Thus, a live-animal sample source that could be used to evaluate LAIV immunogenicity and cross-protection is needed. Here, we evaluated the use of oral fluids (OF) and nasal wash (NW) collected after IAV inoculation as a live-animal sample source in an enzyme-linked immunosorbent assay (ELISA) to predict cross-protection in comparison to traditional serology. Both live-virus exposure and LAIV vaccination provided heterologous protection, though protection was greatest against more closely phylogenetically related viruses. IAV-specific IgA was detected in NW and OF samples and was cross-reactive to representative IAV from each H1 cluster. Endpoint titers of cross-reactive IgA in OF from pigs exposed to live virus was associated with heterologous protection. While LAIV vaccination provided significant protection, LAIV immunogenicity was reduced compared to live-virus exposure. These data suggest that OF from pigs inoculated with wild-type IAV, with surface genes that match the LAIV seed strain, could be used in an ELISA to assess cross-protection and the antigenic relatedness of circulating and emerging IAV in swine.
INTRODUCTION
Influenza A virus (IAV), of the family Orthomyxoviridae, infects many species, including humans, pigs, horses, sea mammals, and birds. The structural proteins hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2), along with the cellular lipid bilayer, form the envelope of IAV. The HA and NA proteins play essential roles in virus entry and release of progeny, respectively, and are primary immunogens involved in a protective antibody response (reviewed in reference 1), while internal genes such as nucleoprotein (NP), polymerase subunit PB1, and matrix protein (M)1 are also important for a protective immune response (2).
Pigs are possible “mixing vessels” for the emergence of antigenically distinct IAV isolates since the respiratory tracts of pigs contain receptors for both avian and mammalian IAV (3, 4). Interspecies transmission of IAV is a real concern and has been well documented, with transmission between pigs and humans (5–7) as well as between pigs and birds or poultry (8–10). The relationship between human IAV and swine IAV is an animal and public health concern because of noted spillover events. Routine IAV vaccination is an important tool in the management of animal health to reduce economic loss associated with IAV infection (11) and for pigs displayed at agricultural fairs (12) in terms of both public and animal health.
One of the main roadblocks for swine IAV vaccine development is the presence of multiple antigenic variants within H1 and H3 subtypes, which results in the need for vaccines that provide cross-protection. Classical H1N1 (cH1N1) was the predominant IAV in U.S. swine until the introduction of the novel H3N2 strain in 1998 (13). H3N2 quickly became endemic in U.S. swine and reassorted with extant cH1N1, resulting in genes from human, avian, and swine IAV combining into a single IAV strain (13, 14). Through introductions of human seasonal IAV into swine and genetic evolution of existing strains, there are currently six phylogenetic clusters of H1 IAV circulating in U.S. herds that are designated beta (β), gamma (γ), γ-2, delta-1 (δ1), delta-2 (δ2), and pandemic (15–19). There is limited hemagglutination-inhibiting (HI) cross-reactivity between H1 clusters when using pig hyperimmune serum (derived through vaccination with adjuvanted, monovalent whole-inactivated virus [WIV] vaccine) to evaluate antigenic relatedness (17, 18).
The majority of currently licensed IAV vaccines for swine are adjuvanted, WIV vaccines given by the intramuscular (i.m.) route. This vaccine formulation induces IAV-specific antibody in serum as measured by the HI assay or a whole-virus enzyme-linked immunosorbent assay (ELISA) (20). WIV vaccines primarily provide protection against homologous or antigenically related strains but provide limited cross-protection against heterologous strains (i.e., strains with the same subtype but limited antigenic cross-reactivity) (21, 22). Notably, differences in the immune response following vaccination through the i.m. route compared to natural infection include minimal mucosal IAV-specific IgA and cell-mediated immune responses (21). To overcome limitations associated with WIV vaccines, several next-generation IAV vaccines, including live-attenuated influenza virus (LAIV) vaccines and replication-defective adenovirus vectors that encode IAV genes, have been experimentally tested for use in pigs (23). LAIV and adenovirus-vectored vaccines have been shown experimentally to provide better cross-protection than WIV vaccines (24–28) and to elicit cellular immunity (24, 29), and IAV-specific maternal immunity does not interfere with adenovirus-vectored or LAIV vaccine efficacy in piglets (29–31).
Nonstructural protein 1 (NS1) of IAV is a virulence factor as a consequence of its type I interferon (IFN) antagonist activity (32). Introducing nucleotide mutations into the NS1 gene causes the loss of type I IFN inhibition; thus, the host IFN response inhibits growth of the virus (33). Through truncation of the last 93 amino acids of the NS1 IAV protein, an LAIV was generated that was shown to induce a robust IFN response and to be attenuated in pigs (34). This truncated NS1 LAIV with H3 surface genes was further shown to elicit protection against homologous and heterologous H3 challenge viruses, as well as partial heterosubtypic protection when vaccinated pigs were challenged with H1 virus (35).
Human replication-defective adenovirus serotype 5 encoding HA (Ad5-HA) has been shown experimentally to provided homologous protection (24, 31, 36) when administered by either the intramuscular or intranasal (i.n.) route and to overcome the problem of interference by maternal-derived immunity (31). Additionally, Ad5-HA IAV vaccines in swine were shown to provide partial heterologous protection when administered intranasally (24).
Despite advantages of LAIV- and Ad5-based vaccines, these platforms have yet to come to market. A live-animal sample and assay to predict vaccine efficacy and cross-protection, as well as additional cross-protection studies using contemporary viruses, will aid in the continued development of these platforms. Serum HI titer is the established gold-standard method to predict IAV cross-reactivity and, subsequently, the cross-protective efficacy of a particular vaccine. A reciprocal serum HI titer of >40 is usually considered protective (correlated with a 50% reduction in the risk of IAV infection [37]), and a greater than 4-fold reduction in HI titer between viruses is considered to represent significant antigenic drift with a predicted loss in cross-protection (37). LAIV vaccination elicits modest serum HI antibody responses to homologous antigens, but LAIV also provides protection against heterologous IAV in which LAIV antibodies did not cross-react (25, 38). Thus, HI titers following LAIV vaccination may be useful for evaluating vaccine immunogenicity and predicting homologous protection, but predictions of cross-protection using this gold standard method are unreliable.
Since intranasal LAIV vaccination elicits IAV-specific antibody at mucosal surfaces (29, 39), it is possible that a live-animal mucosal sample would be better for evaluating intranasal IAV vaccine immunogenicity and cross-protective efficacy. To test this hypothesis, a study was completed in which groups of pigs were exposed to wild-type (WT) IAV, LAIV, or Ad5-HA, all with a pandemic lineage surface gene(s), for collection of mucosal and serum samples postexposure. Vaccinated pigs were then challenged with either a heterologous β-cluster or γ-cluster IAV to evaluate the cross-protective efficacy of the vaccines and to evaluate IAV-specific antibody levels in prechallenge samples in association with protective efficacy. Taking the results together, WT IAV inoculation or LAIV or Ad5-HA vaccination provided some level of cross-protection (depending on the challenge strain), and immunogenicity likely impacted the extent of cross-protective efficacy. IAV-specific IgA in oral fluids was found to be associated with cross-protective efficacy; the data may serve as predictive indices, and additional work is warranted to further validate these findings.
MATERIALS AND METHODS
Ethics statement.
Animal experiments were approved by the Institutional Animal Care and Use Committee of the National Animal Disease Center in Ames, IA. Animals were housed in animal biosafety level 2 (ABSL-2) conditions for the entirety of the study.
Vaccines and viruses.
The LAIV vaccine expressed the 2009 pandemic H1N1 (H1N1pdm09) (pdm) surface genes from A/New York/18/2009 (NY/09) with the A/turkey/Ohio/313053/2004 (H3N2) internal genes, in which a truncated NS1 gene (34) was carried (see Fig. S1 in the supplemental material). The reverse-engineered, genetically matched parent virus was derived by rescuing the HA and NA genes from A/New York/18/2009 with the A/turkey/Ohio/313053/2004 internal genes (no attenuation mutations introduced) as previously described (40, 41) and is referred to as RG NY/09. The replication-defective adenovirus type 5 vector HA (Ad5-HA) vaccine was derived using an AdEasy system (Agilent, Santa Clara, CA) by cloning the A/California/04/2009 HA gene into the pShuttle-cytomegalovirus (CMV) vector, and adenovirus rescue was performed as previously described (42). Heterologous challenge viruses were H1N1 β-cluster A/swine/Minnesota/03012/2010 virus (MN/10) and H1N2 γ-cluster A/swine/Illinois/3134/2010 IAV (IL/10) obtained through submission of clinical samples to the University of Minnesota Veterinary Diagnostic Laboratory (kindly provided by Marie Culhane). All vaccines and viruses were propagated in Madin-Darby canine kidney (MDCK) cells in serum-free Opti-MEM media (Gibco, Grand Island, NY) supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma, St. Louis, MO), l-glutamine, and antibiotics, with the exception of Ad5-HA, which was grown in AD-HEK293 cells with high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum. Ad5-HA was purified using a double discontinuous cesium chloride gradient per the manufacturer's instructions and dialyzed as previously described (42). All vaccines and viruses were diluted in phosphate-buffered saline (PBS) to the desired concentration immediately prior to administration.
Experimental design.
Three-week-old pigs were obtained from a high-health-status herd free from IAV and porcine reproductive and respiratory syndrome virus. Pigs were administered enrofloxacin (Baytril; Bayer, Pittsburgh, PA) and ceftiofur crystalline free acid (Excede; Zoetis, Florham Park, NJ) upon arrival according to the recommendations of the manufacturers. IAV antibody status was confirmed to be negative using the IDEXX Multi-Screen Ab test (IDEXX, Westbrook, ME). In part I of the experiment (attenuation), piglets were randomly assigned to groups of eight and immunized i.n. at 4 weeks of age. Groups received LAIV, WT NY/09, or RG NY/09 parent virus or were not vaccinated (NV). Pigs were humanely euthanized at 3 days postvaccination (dpv) to assess the attenuation and pathogenicity of the LAIV compared to the wild-type parent virus. In part II of the experiment (immune measures of cross-protection), piglets were randomly assigned to groups of eight, immunized i.n. at 4 weeks of age, and boosted 3 weeks later by the same route with the same dose (Table 1). LAIV vaccine and WT NY/09 virus were targeted at 106 50% tissue culture infective doses (TCID50)/ml and at 2 ml per dose. The back titer of the inoculum after administration indicated that the WT NY/09 dose was 105.5 TCID50 per pig, whereas the LAIV dose was 104.2 TCID50 per pig. Pigs in the Ad5-HA group received 1010 TCID50 in 2 ml i.n. Nonvaccinated (NV) controls received 2 ml PBS i.n. Three weeks following the boost, all pigs except for the nonvaccinated/nonchallenged (NV/NC) controls were challenged i.n. with 105.8 TCID50 MN/10(β) or IL/10(γ). Pigs were humanely euthanized at 5 days postinfection (dpi) for collection of samples to evaluate vaccine efficacy.
TABLE 1.
Experimental design for evaluation of IAV vaccine immunogenicity and cross-protective efficacya
| Group (n = 8) | Vaccine | Challenge virus |
|---|---|---|
| NV/NC | None | None |
| NV/CH | None | MN/10(β) |
| WT | NY/09 | MN/10(β) |
| LAIV | LAIV | MN/10(β) |
| Ad5-HA | Ad5-HA | MN/10(β) |
| NV/CH | None | IL/10(γ) |
| WT | NY/09 | IL/10(γ) |
| LAIV | LAIV | IL/10(γ) |
| Ad5-HA | Ad5-HA | IL/10(γ) |
n, number of pigs per group; NV, nonvaccinated; NC, nonchallenged; WT, wild type; LAIV, truncated nonstructural protein 1 live-attenuated influenza virus vaccine; MN/10(β), A/swine/MN/03012/2010; IL/10(γ), A/swine/IL/3134/2010.
Sample collection.
For postvaccination sample collection, nasal swabs (NS) were taken at 0 to 3 dpv for the subset of pigs necropsied on day 3 postpriming. Serum, nasal washes (NW), and oral fluids (OF) were collected from all remaining pigs every 7 days following primary immunization. Blood was collected by venous puncture and stored in BD Vacutainer serum separator tubes (BD, Franklin Lakes, NJ). Serum was collected by centrifugation at 800 × g for 20 min, divided into aliquots, and frozen at −80°C. NW were collected by instilling 5 ml PBS into a naris and collecting effluent as previously described (43), divided into aliquots, and frozen at −80°C. Typically, 0.5 to 2 ml of NW was collected. OF were obtained following previously described methods (44) with few modifications. Briefly, cotton ropes were hung in each room for approximately 30 min for the pigs to chew. Ropes from each group were placed into separate ziplock bags. The rope was manually squeezed inside the bag and liquid sample decanted into a 50-ml conical tube. The tubes were centrifuged at 800 × g for 20 min, and the supernatant was filtered through 0.45-μm-pore-size syringe filters and immediately frozen at −80°C. For postchallenge sample collection, nasal swabs were collected daily beginning on the day of challenge (0 dpi) through necropsy (5 dpi) using polyester-tipped swabs (Puritan, Guilford, ME) prewetted in 2 ml minimal essential media (MEM) and stored frozen. At necropsy, trachea wash was obtained by removing the trachea prior to lung lavage. The trachea, from 1 cm below the larynx to 2 cm above the bifurcation, was removed and submerged in 3 ml MEM and vigorously agitated for 15 s. The trachea was then removed, and the medium was aliquoted and frozen at −80°C. Lung lavage samples were obtained by lavage performed with 50 ml MEM as previously described (38). An aliquot of lung lavage fluid was plated on blood agar and Casman's agar plates containing 0.01% (wt/vol) NAD and 5% horse serum for routine aerobic culture to rule out bacterial infection. All postchallenge samples were stored on ice until they were divided into aliquots and frozen within 2 h of collection.
Pathological examination.
At necropsy, the percentage of lung afflicted with the purple-red consolidation typical of IAV infection was evaluated (45). The total percentage of pneumonia was calculated on the basis of the weighted proportions of each lobe with respect to the total lung volume (46). A portion of the right middle lobe was fixed in 10% buffered formalin for 48 h, processed by routine histopathologic procedures, and stained with hematoxylin and eosin. Microscopic lesions were evaluated and scored by a veterinary pathologist blinded to the treatment groups using parameters previously described (47).
Antibody evaluation.
The HI assay was performed as recommended in the WHO animal influenza training manual using turkey red blood cells and WT NY/09 virus or H1 challenge virus as the target antigen as previously described (41). The HI assay using OF and NW was performed following the same protocol as the serum HI assay, beginning with an initial dilution of 1:10. Data are expressed as the reciprocal titer. The serum neutralization assay was performed by generating serial 2-fold dilutions of heat-inactivated sera in MEM supplemented with 5% bovine serum albumin and antibiotics and by incubating with 100 TCID50 before inoculation of confluent MDCK monolayers in 96-well plates as previously described (48). Cells were incubated for 48 h, fixed, and stained for viral nucleoprotein (NP) antigen by immunocytochemistry (49). OF and NW neutralization assays were performed similarly except that the NW samples were not heat inactivated prior to use in the neutralization assay. Reciprocal titers were Log2 transformed and used for statistical analysis of HI and neutralization titer data. IAV-specific IgA and IgG levels in the OF and the NW were recorded as the optical density (OD) value using an indirect whole-virus ELISA and the viruses listed in Table 2 as previously described (38), with the exception that the NW and OF samples were diluted 1:2 in PBS for use in the assay. Antibody levels were reported as the mean OD at 405 nm for each vaccine group. Endpoint antibody titers were obtained by initially diluting OF and NW samples (pooled by treatment group) 1:2 in PBS and titrating 2-fold in duplicate before performing the ELISA. The resulting OD data were modeled as a nonlinear function of the Log10 dilution using the GraphPad Prism (GraphPad software Inc., La Jolla, CA) Log (agonist) versus response-variable slope four-parameter logistic model. Endpoints were interpolated by using 2× the average OD of the nonvaccinated control as the cutoff. Endpoints for each virus were organized by cluster, and average cluster endpoint dilutions are expressed for both OF and NW. Total IgA and IgG ELISAs were performed using a pig IgA or IgG quantification kit (Bethyl Laboratories, Montgomery, TX) following the manufacturer's protocol.
TABLE 2.
IAV isolates used to evaluate antibody responses following intranasal immunization
| Cluster | Virus name | Abbreviation | Subtype |
|---|---|---|---|
| Pandemic | A/NY/18/2009 | NY/09 | H1N1 |
| Pandemic | A/CA/04/2009 | CA/09 | H1N1 |
| Pandemic | A/swine/IL/5265/2010 | IL/10 | H1N1 |
| γ | A/swine/IL/3134/2010 | IL/10 | H1N2 |
| γ | A/swine/IN/3062/2010 | IN/10 | H1N1 |
| γ | A/swine/OH/511445/2007 | OH/07 | H1N1 |
| β | A/swine/MN/03012/2010 | MN/10 | H1N1 |
| δ-2 | A/swine/MO/03013/2010 | MO/10 | H1N2 |
| δ-1 | A/swine/MN/02011/2008 | MN/08 | H1N2 |
| H3 | A/turkey/OH/313053/2004 | OH/04 | H3N2 |
Virus titration.
To determine viral loads, nasal swab, trachea wash, and lung lavage samples were titrated on MDCK cells to determine the TCID50/ml as previously described (38). Briefly, frozen samples were thawed and filtered through 0.45-μm-pore-size syringe filters, and 10-fold serial titrations were made in triplicate in serum-free media containing TPCK-trypsin (Sigma, St. Louis, MO) (1 μg/ml) and added to confluent MDCK monolayers in 96-well plates. Cells were incubated for 48 h, fixed, and stained for NP by immunocytochemistry (49). For each titration, the Log10 transformed TCID50/ml was calculated for each sample using the method of Reed and Muench (50).
Statistical analysis.
Reciprocal HI and neutralization titers were Log2 transformed and viral titers were Log10 transformed for analysis and expressed as reciprocal titer values. ELISA data are presented as the average optical density at 405 nm. Statistical analysis was completed with Graph Pad prism version 6 using the Kruskal-Wallis test followed by Dunn's multiple comparison or using two-way analysis of variance (ANOVA) and Tukey's multiple comparison where appropriate.
RESULTS
Attenuation characteristics of H1 LAIV vaccine.
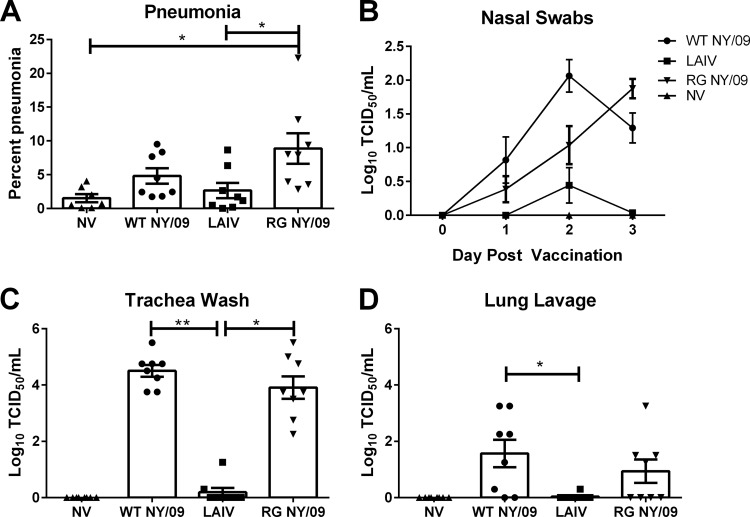
Pigs were i.n. inoculated with WT NY/09, H1 LAIV, or the reverse-genetics-rescued isogenic parent virus (RG NY/09) to assess the attenuation characteristics of the H1 LAIV vaccine. Macroscopic lung lesions (pneumonia) at 3 dpv were significantly reduced in pigs inoculated with LAIV compared to those in pigs inoculated with RG NY/09 (Fig. 1A). Amounts of WT NY/09 virus were detected in nasal swabs by 1 dpv, peaked at 2 dpv, and began to decline by 3 dpv. The amount of virus detected in nasal swabs from pigs inoculated with LAIV was significantly reduced at each time point tested compared to the amount of virus detected in nasal swabs from pigs exposed to WT NY/09 and was significantly reduced at 3 dpv compared to the amount detected in pigs exposed to RG NY/09 (average, 0.03 versus 1.8 Log10 TCID50/ml) (Fig. 1B). Pigs inoculated with LAIV had significantly reduced mean viral titers in the trachea (0.2 Log10 TCID50/ml) compared to animals exposed to WT NY/09 and RG NY/09 (4.5 and 3.9 Log10 TCID50/ml, respectively) (Fig. 1C). Additionally, LAIV pigs had reduced mean viral titers in the lung lavage fluid compared to animals exposed to WT NY/09 (P = 0.01) (Fig. 1D). The back titer of the challenge indicated that each pig in the RG NY/09 groups received 105.9 TCID50, each pig in the WT NY/09 group received 105.5 TCID50, and each pig in the LAIV group received 104.2 TCID50.
FIG 1.
Attenuation characteristics of H1 NS1-truncated LAIV vaccine. Groups of pigs were inoculated with WT NY/09 virus, LAIV, or the reverse-genetics (RG)-rescued NY/09 parent virus by the intranasal route or were left nonvaccinated (NV) and samples collected at 3 days postvaccination to evaluate attenuation. (A) Percent macroscopic lung lesions. (B) Viral shedding in nasal swabs collected 0 to 3 days postimmunization. (C and D) Viral load in trachea wash (C) and lung lavage (D) fluids. Each dot represents a single animal in that respective treatment group, with the means ± standard errors of the means (SEM) for n = 8 shown for the group indicated. Data were analyzed with the Kruskal-Wallis test and Dunn's posttest. P values of <0.05 were considered significant. *, P < 0.05; **, P < 0.01.
IAV-specific antibody in serum, nasal wash, and oral fluids.
Serum, NW, and OF samples were collected weekly following primary exposure (to WT NY/09, LAIV, and Ad5-HA) for evaluation of IAV-specific antibodies. At 42 dpv, serum HI antibodies against WT NY/09 virus were detected for all immunization groups, though there were differences in titers between treatment groups (Table 3). Average serum HI titers against homologous NY/09 virus were 180, 55, and 31 in animals immunized with WT NY/09, LAIV, and Ad5-HA, respectively. Average serum HI titers against the challenge viruses were below the limit of detection (<10), with the exception of the WT NY/09 group, which had a low but detectable (12 ± 2) serum HI titer against the β-cluster (MN/10) virus. NW and OF samples collected at 42 dpv were also evaluated as a sample source in the HI assay, but there was no measureable HI activity using these samples, even at a dilution of 1:10. All samples from nonvaccinated animals (NV) had no detectable HI titers (<10).
TABLE 3.
Serum hemagglutination inhibition and virus neutralization titers measured at 42 days postvaccination
| Vaccine | Serum titer of indicated target virusa |
|||||
|---|---|---|---|---|---|---|
| HI |
Neutralization |
|||||
| NY/09 (pdm) | IL/10(γ) | MN/10(β) | NY/09 (pdm) | IL/10(γ) | MN/10(β) | |
| WT NY/09 | 180 ± 32.9 | <10 | 12.5 ± 1.6 | 168 ± 26.8 | 152 ± 24 | 54 ± 6.7** |
| LAIV | 55 ± 7.3 | <10 | <10 | 74 ± 26.8 | 72 ± 17 | 25 ± 6.3* |
| Ad5-HA | 31 ± 10 | <10 | <10 | <2 | <2 | <2 |
| NV | <10 | <10 | <10 | <2 | <2 | <2 |
*, P = 0.03; **, P = 0.005 (compared to NY/09 neutralization). Statistical significance was measured by the Kruskal-Wallis test (n = 8).
Functional IAV-specific antibody levels were also measured using a virus neutralization assay (Table 3). WT NY/09 and LAIV exposure induced production of serum neutralizing antibody against NY/09 virus (titers, 168 ± 27 and 74 ± 27, respectively), and titers against IL/10(γ) virus were similar. However, serum neutralization titers against the MN/10(β) challenge virus compared to neutralization of NY/09 virus were significantly reduced (54 ± 6.7 [P = 0.005] for WT NY/09-immunized animals and 25 ± 6.3 [P = 0.03] for LAIV-immunized animals). Ad5-HA immunization did not result in detectable serum neutralization antibody. Moreover, OF samples from animals exposed to WT NY/09 were able to neutralize WT NY/09 virus (titer 8 ± 1) but not the heterologous challenge viruses, while NW samples were unable to neutralize any virus used in the assay.
NW and OF samples were used in an indirect whole-virus ELISA to detect antibody specific to H1N1pdm09 lineage virus throughout the course of the vaccine regimen (Fig. 2). By 14 dpv, significant levels of IAV-specific IgA were measured in the NW (Fig. 2A) and OF (Fig. 2B) from pigs in the WT NY/09 (NW and OF, P < 0.0001) and LAIV (NW, P < 0.001; OF, P < 0.0001) groups and were maintained throughout the experiment. H1N1pdm09 IAV-specific IgG was detected only in OF samples beginning at 28 dpv (1 week postboost) and was maintained above background levels (NV group) only in samples from pigs in the WT NY/09 group (Fig. 2C). The total IgA and IgG antibody levels in each sample were determined, and amounts never varied more than 2-fold between time points (data not shown), suggesting that sample collection was consistent throughout the experiment. The Ad5-HA vaccine did not elicit detectable levels of H1N1pdm09-specific IgA or IgG in NW or OF samples (see Fig. S2A in the supplemental material). Due to an inability to measure H1N1pdm09-specific IgA in NW or OF collected from Ad5-HA-vaccinated animals, additional heterologous viruses were not evaluated using samples from Ad5-HA vaccinates.
FIG 2.
Kinetics of IAV-specific mucosal IgA and IgG levels following immunization. Groups of pigs were intranasally inoculated with WT NY/09 or LAIV on day 0 and boosted on day 21. NW and OF were collected weekly to day 42 to evaluate IAV H1N1pdm09 (CA/09)-specific mucosal antibody by whole-virus ELISA compared to nonvaccinated (NV) animals. (A and B) IAV-specific IgA detected in nasal wash (A) and oral (B) fluids. (C) IAV-specific IgG detected in OF. Data are expressed as means ± SEM of OD determined for n = 8 for the indicated group.
Macroscopic and microscopic lung pathology.
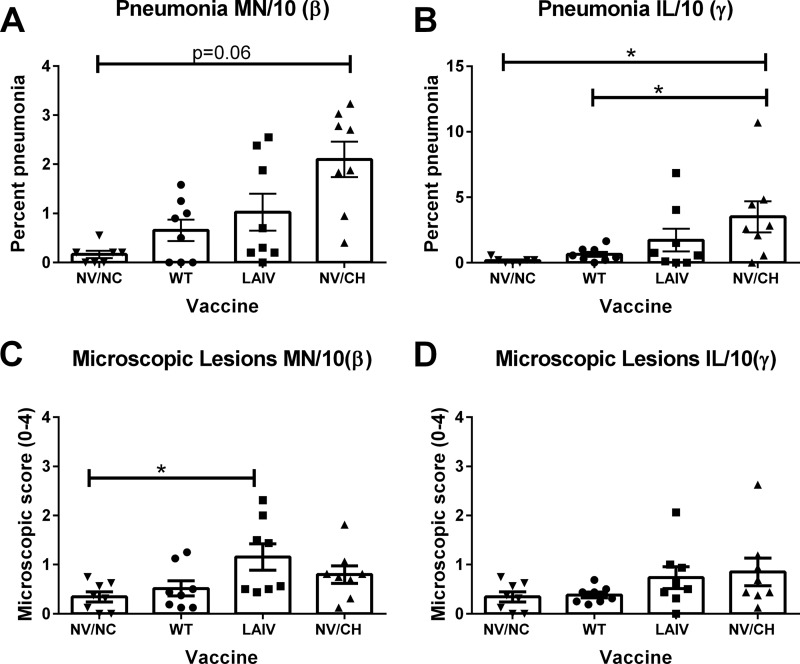
Macroscopic and microscopic lung lesions were evaluated at 5 dpi following challenge with heterologous H1 β-cluster MN/10 or γ-cluster IL/10 virus (Fig. 3). Pigs in the nonvaccinated, nonchallenged (NV/NC) control group had negligible macroscopic pneumonia (score, 0.16% ± 0.07). Pigs in the nonvaccinated challenged (NV/CH) groups challenged with IL/10(γ) or MN/10(β) had minimal macroscopic pneumonia (3.5% ± 1.1 and 2.1% ± 0.3, respectively). Pigs with prior exposure to WT NY/09 virus, challenged with MN/10(β), did not have reduced pneumonia compared to NV/CH pigs due to overall low scores at 5 dpi (Fig. 3A); however, these animals did not have significantly increased pneumonia scores compared to the NV/NC controls (P = 0.63). Prior WT NY/09 inoculation did reduce macroscopic lung lesions following IL/10(γ) challenge (0.63%, P = 0.001) (Fig. 3B). While LAIV vaccination did not statistically significantly reduce pneumonia following challenge with either virus, there was a trend for reduced macroscopic pneumonia following challenge and the percentage of pneumonia for the LAIV groups was not significantly increased over that seen with NV/NC pigs (P = 0.41). While LAIV-vaccinated pigs challenged with MN/10(β) had microscopic pathology scores greater than those seen with NV/NC controls (1.1 and 1.3 versus 0.3, respectively), these scores were not significantly increased over NV/CH pig scores (0.79) (Fig. 3C). Pigs challenged with IL/10(γ) demonstrated mild microscopic lesions and did not have scores greater than those determined for the NV/NC controls (Fig. 3D). Ad5-HA vaccination did not protect against the development of lung lesions following heterologous IL/10(γ) or MN/10(β) challenge (see Fig. S2B and C in the supplemental material).
FIG 3.
Pneumonia following heterologous IAV-challenged, IAV-vaccinated pigs. Groups of pigs were inoculated with the indicated vaccine and subsequently challenged intranasally with heterologous virus, and lung pathology was evaluated at 5 dpi. (A and B) Macroscopic lesion scores of animals following challenge with MN/10(β) (A) or IL/10(γ) (B) IAV compared to those calculated for nonvaccinated and challenged (NV/CH) or nonvaccinated and nonchallenged (NV/NC) animals. (C and D) Microscopic lesion scores following challenge with (C) MN/10(β) or (D) IL/10(γ) IAV. Each dot represents a single animal in that respective treatment group, with the mean ± SEM for n = 8 shown for the group indicated. Data were analyzed with the Kruskal-Wallis test and Dunn's posttest. P of <0.05 was considered significant. *, P < 0.05.
Viral load in the respiratory tract.
Following challenge, viral titers in nasal swabs (NS), trachea washes, and lung lavage fluid were measured to evaluate protection against virus replication. NS were collected daily from 0 dpi through necropsy at 5 dpi (Fig. 4A and D). Pigs exposed to WT NY/09 virus and subsequently challenged with MN/10(β) had reduced average viral titers in NS on dpi 1 through 4 (P < 0.0001), but the average viral titers in NS collected on dpi 5 were similar to the titers measured in NS collected from NV/CH animals (Fig. 4A). Virus was not recovered from any NS collected from pigs exposed to WT NY/09 and challenged with IL/10(γ) (Fig. 4D). LAIV vaccination limited the amount of MN/10(β) challenge virus in NS only at dpi 1 (P < 0.0001), and NS mean viral titers were similar to the titers measured in NS from NV/CH pigs on dpi 2 to 5. However, LAIV vaccination significantly limited the amount of IL/10(γ) challenge virus shed and the length of time during which the virus was shed in nasal passages (P = 0.005) (Fig. 4D). Pigs vaccinated with Ad5-HA also had reduced amounts of challenge virus in NS on dpi 1 to 2, but the average titers were the same as those in the respective NV/CH groups on dpi 3 to 5 (see Fig. S2D in the supplemental material).
FIG 4.
IAV vaccination impacted IAV burden in the respiratory tract following heterologous challenge. Groups of pigs were inoculated with the indicated vaccine and subsequently challenged intranasally with heterologous virus. Viral titers in NS (A and D), trachea wash fluid (B and E), and lung lavage fluid (C and E) of pigs challenged with MN/10(β) (A to C) or IL/10(γ) (D to F) are indicated. Each dot represents a single animal in that respective treatment group, with the mean ± SEM for n = 8 shown for the group indicated. The Kruskal-Wallis test and Dunn's multiple-comparison test were used for analysis. P of <0.05 was considered significant. ***, P < 0.001; ****, P < 0.0001.
Mean titers of the MN/10(β) challenge virus were significantly reduced in trachea washes from pigs previously exposed to WT NY/09 virus compared to NV/CH pigs (1.5 Log10 TCID50/ml and 4.6 Log10 TCID50/ml, respectively) (Fig. 4B). LAIV-vaccinated animals had mean titers of MN/10(β) virus in the trachea on dpi 5 similar to those measured from NV/CH pigs (Fig. 4B). Additionally, mean viral titers in the lung lavage fluid of all immunization groups challenged with MN/10(β) did not differ from the levels seen with NV/CH pigs (Fig. 4C). Pigs exposed to WT NY/09 or LAIV and challenged with IL/10(γ) did not have any virus recovered from trachea wash or lung lavage fluid (Fig. 4E and F) on dpi 5. Ad5-HA-vaccinated animals also exhibited increased cross-protection against IL/10(γ) (see Fig. S2E and F in the supplemental material).
Measures of IAV-specific cross-reactive immunity.
Since both live-virus exposure and LAIV vaccination provided increased cross-protection against the γ-cluster H1 variant and less cross-protection against the β-cluster H1 variant, we evaluated whether levels of IAV-specific IgA in a live-animal sample source were predictive of cross-protection, given that the standard HI assay was unable to measure cross-reactivity to either challenge virus (Table 3). Using NW and OF samples collected from all pigs at 42 dpv, the cross-reactivity of IgA to eight H1 viruses from different phylogenetic clusters (listed in Table 2) and one H3 virus (Fig. 5) was determined. Mucosal IAV-specific IgA reactive to homologous, H1N1pdm09 lineage viruses as well as IgA cross-reactive to representative viruses from each H1 cluster and an H3 virus was detected in NW from WT NY/09 virus-immunized animals (P < 0.05), while LAIV-immunized animals did not have detectable IgA reactive to all representative clusters (Fig. 5A). OF samples from WT NY/09- and LAIV-immunized animals had significantly higher levels of IAV-specific IgA than those from the NV controls (P < 0.0001 for all clusters and vaccine groups) (Fig. 5B). IgA in NW from LAIV-vaccinated animals had significant reductions in OD values for γ-cluster viruses (P = 0.003), δ-2 cluster viruses (P = 0.0006), and H3 viruses (P = 0.01) compared to the OD values for homologous NY/09 antigen. However, IgA in OF from LAIV-vaccinated animals had lower OD values across all heterologous viruses compared to those seen with viruses of the pandemic cluster (P < 0.001).
FIG 5.
LAIV vaccination elicits mucosal IgA cross-reactive to heterologous H1 and H3 IAV. Nasal wash and oral fluid samples were collected at 42 dpv and used to measure levels of cross-reactive IAV-specific IgA by whole-virus ELISA. Samples were tested against representative viruses from each H1 cluster and an H3 virus. (A and B) Cross-reactive IgA data expressed as means ± SEM at an optical density (OD) of 405 nm in nasal wash (A) and oral (B) fluids. ns, no significant difference from NV control. (C and D) Reciprocal endpoint titers of IAV-specific IgA averaged by cluster in nasal wash samples pooled by respective group (C) and oral-fluid group (D). Each dot represents a viral antigen in the indicated cluster with the mean endpoint dilution for the cluster indicated (see Materials and Methods).
Since reductions in OD values compared to those of homologous IAV antigen were similar across tested heterologous H1 variants, average endpoint titers of IgA specific to each H1 cluster were determined to further elucidate differences in cross-reactive IgA. IAV-specific IgA endpoint titers in NW and OF against heterologous H1 cluster viruses were reduced compared to the homologous H1 endpoint titers (Fig. 5C and D). Differences between the OF IgA endpoint titer against the NY/09 vaccine virus and mean IgA endpoint titers in OF from WT NY/09-exposed pigs were an average of 1.8-fold for similar H1N1pdm09 lineage viruses, 4-fold for γ-cluster viruses, 4.8-fold for β-cluster viruses, 6.7-fold for δ-cluster viruses, and 8.4-fold for H3 virus (Fig. 5D). IgA endpoint titers from OF collected from LAIV-vaccinated animals were reduced by an average of 1.6-fold compared to those seen with similar H1N1pdm09 lineage viruses, 2.5-fold compared to those seen with γ-cluster viruses, 2-fold compared to those seen with β-cluster viruses, 3.8-fold compared to those seen with δ-cluster viruses, and 2.1-fold compared to those seen with H3 viruses.
DISCUSSION
In this study, we evaluated the ability of H1N1pdm09 cluster H1 LAIV and a replication-defective viral vector encoding IAV H1N1pdm09 HA to provide cross-protection against representative heterologous γ- and β-cluster H1 IAV infections compared to live virus infections and investigated predictive immune measures of cross-protection in samples collected from a live animal. In evaluations of viral loads, vaccination with LAIV or exposure to wild-type virus resulted in significant cross-protection against the γ-cluster challenge IL/10 IAV; however, protection against β-cluster MN/10 was more limited. The H1N1pdm09 cluster and γ-cluster viruses are more closely phylogenetically and antigenically related than the β-cluster and H1N1pdm09 cluster viruses (17, 18); thus, the increased protection against IL/10(γ) compared to MN/10(β) was somewhat anticipated. Together, these data suggest that a multivalent next-generation swine IAV vaccine may be necessary to protect against the diverse H1 viruses cocirculating in U.S. swine.
Although cold-adapted, temperature-sensitive LAIV has been approved for use in humans (51) and in horses (52), LAIV vaccines have not been approved for use in pigs. Original work on the LAIV vaccine focused on H3N2 surface genes (25, 34, 35). Here we describe the first use of an H1N1 NS1-truncated virus as an LAIV vaccine and compared the levels of immunogenicity and efficacy following exposure to WT NY/09 virus, which shares the same HA and NA as the LAIV. Similarly to previous reports of H3 NS1-truncated LAIV vaccination in pigs (35) and H1 NS1-truncated LAIV vaccination in mice (33), the H1 NS1-truncated LAIV in pigs was attenuated, with significantly less virus detected in the nose, trachea, and lungs as well as reduced levels of macroscopic pneumonia (Fig. 1). A previous study used a lower dose of challenge WT virus (3 Log10 TCID50 less than was used in the current study) (27), and it was as pathogenic as the typical dose of WT virus used in the current study. This suggests that, regardless of dose, WT IAV induces significant pathology, and although LAIV was administered at a level 1.8 Log10 TCID50 lower than that of the WT virus, the LAIV was attenuated. In addition, there are significant reports that truncating the NS1 gene of IAV leads to attenuation (33, 34, 53, 54) Despite the dose of LAIV vaccine administered and the minimal evidence of LAIV replication in vivo, the LAIV elicited a measureable IAV-specific antibody response, similarly to previous studies (33, 35). Administering a higher dose of LAIV may increase the degree of cross-protection, similarly to the cross-protection observed with live virus; however, this may not translate into a substantial increase in immune responses. In a follow-up study using a 1.5-Log10-higher dose of LAIV administered i.n. to 21-day-old pigs, minimal LAIV was recovered from NS and IAV-specific mucosal IgA endpoint titers were not increased over the titers observed in the current study (data not shown).
Direct and indirect transmission of IAV is a major animal health as well as public health concern. Direct pig-to-pig transmission has been shown with the 2009 pandemic H1N1 virus (55), as well as indirect aerosol transmission with emerging H3N2 variant viruses (38). Agricultural fairs present a unique interface for interspecies transmission of IAV between pigs and people (6, 7, 12). WIV vaccines for IAV have been shown to limit disease but do not always limit virus shedding (21, 38). An IAV vaccine that limits nasal shedding of virus would likely have the ability to limit further transmission; therefore, we evaluated nasal shedding as a measure of cross-protection following challenge. All animals challenged with MN/10(β) had detectable virus in NS as early as 1 dpi. All immunized animals had reduced viral shedding of MN/10(β) in nasal swabs compared to NV/CH controls at 1 dpi, though all animals had similar levels of shedding by the end of the study (5 dpi). In contrast, LAIV-vaccinated pigs challenged with IL/10(γ) demonstrated reduced virus replication in the nose that was cleared by 4 dpi. These reductions in nasal shedding in vaccinated, challenged animals suggests that LAIV vaccination could limit the transmission of heterologous virus to naive or vaccinated contact animals (38, 56, 57), thus slowing or breaking the transmission cycle. Studies are under way to investigate this hypothesis.
A common parameter used to evaluate IAV vaccine efficacy is the prevention or significant reduction of lung lesions following challenge compared to nonvaccinated challenged animal results (45). LAIV-vaccinated animals challenged with IL/10(γ) had pneumonia scores that were not significantly different from the scores calculated for NV/CH controls. However, pneumonia scores following challenge were relatively low across all treatment groups, with NV/CH pigs averaging 3.5%, making significant reductions in pneumonia difficult to assess. Our observation of low pneumonia scores following challenge is consistent with previous studies of H1 IAV intranasal challenge in pigs and is not uncommon in 10-week-old pigs (24, 58). Despite the low pneumonia scores, virus was recovered from the lungs of all NV/CH pigs, and IL/10(γ) was not recovered from lung lavage fluid or trachea washes of LAIV-vaccinated pigs, indicating protection against virus replication. The observation of lung pathology in the absence of virus in the respiratory tract is consistent with previous work evaluating LAIV or Ad5-HA efficacy in pigs (24, 29, 53). Whereas Ad5-HA-vaccinated animals had increased macroscopic lung lesions following MN/10(β) challenge compared to the NV/CH controls (see Fig. S2B in the supplemental material), the microscopic lesion characteristics were not consistent with previous descriptions of lesions associated with vaccine-associated enhanced respiratory disease (47). Collectively, these observations suggest that intranasal LAIV and/or Ad5-HA vaccination primes a mucosal cell-mediated immune response that is rapidly activated upon heterologous challenge and that this activation may control viral burden but result in immunopathology (59). Future work evaluating the mucosal IAV-specific cell-mediated immune response following intranasal IAV vaccination may provide insight into the mechanism associated with pneumonia following heterologous challenge; however, such an evaluation is unlikely to provide a useful live-animal sample that could be readily used for evaluating LAIV immunogenicity and cross-protection. While lung lesions are often used to evaluate IAV vaccine efficacy, together, these data indicate that some vaccines may significantly limit virus replication but that lung lesions may not be reduced.
The HI assay is commonly used to evaluate WIV vaccine immunogenicity and efficacy, and hyperimmune antisera (generated through WIV vaccination) are often used to evaluate antigenic relatedness between swine IAV strains (17, 60). Previous studies have shown that LAIV or Ad5-HA vaccination results in lower serum HI titers to homologous IAV and undetectable cross-reactive HI titers to IAV (24–26, 38). Consistent with previous reports, LAIV and Ad5-HA vaccination resulted in low HI titers to homologous IAV and did not elicit cross-reactive HI titers to heterologous IAV in the current study. These data reemphasize that neither LAIV vaccination nor Ad5-HA vaccination induces a robust peripheral HI antibody response and that the commonly used serum HI assay may not be suitable for predicting LAIV or Ad5-HA vaccine efficacy. In contrast, serum virus neutralization antibody was detected following LAIV vaccination and neutralization titers may be a useful measure of LAIV immunogenicity and efficacy. The greater sensitivity of the neutralization assay than the HI assay has made it an important tool in the development of IAV vaccines that are poorly immunogenic (37, 61, 62). Serum from LAIV vaccinates neutralized homologous H1N1pdm09 virus and IL/10(γ) virus equally. Pigs exposed to WT NY/09 or LAIV had significant reductions in serum neutralization against MN/10(β) compared to homologous NY/09 virus neutralization. Higher serum neutralization titers measured against IL/10(γ) virus than against MN/10(β) virus were associated with control of virus replication in the lungs and trachea. These data suggest that the serum neutralization assay may be an effective tool to predict cross-protection of LAIV vaccines in pigs, though further work is warranted to evaluate cross-reactive titers against the panel of H1 viruses, particularly as it relates to cross-protection.
While serum can be an easy sample source for evaluating vaccine immunogenicity, compartmentalization of immune responses may limit the utility of peripheral blood and antibody assays to predict immunogenicity and efficacy associated with intranasally delivered vaccines. Previous studies have shown that IAV antigen-specific antibody can be detected by ELISA in NW and OF samples collected following vaccination or infection (24, 38, 39, 63). Experimental IAV infection elicits an IAV-specific antibody response detectable in OF (63), and OF have become a rapidly evolving diagnostic sample source for several human and animal pathogens (63, 64). We detected IAV-specific antibody in both NW and OF following WT NY/09 exposure and LAIV immunization using whole-virus ELISA. OF samples from immunized animals had increased IAV-specific antibody compared to NW samples as determined by a standard OD reading performed with a single dilution of sample or by sample endpoint titration. Previous studies have shown that intranasal LAIV vaccination induces a local IAV-specific IgA response measureable by ELISA and that the response was associated with protection from homologous challenge in humans (65) as well as in pigs (21, 38). Although we did not evaluate protection from homologous challenge, the higher antibody response observed in OF than in NW following vaccination would suggest that OF could serve as a sample source to evaluate LAIV immunogenicity and homologous protective efficacy as well as provide a measure of herd immune status (63).
Cross-reactive IAV-specific IgA in NW following LAIV vaccination has been shown to correlate with improved heterologous cross-protection in humans and in animal models of human IAV infection (26, 65, 66). Given the reductions in heterologous virus nasal shedding and in measureable IAV-specific IgA levels in mucosal samples prechallenge (42 dpv), we evaluated the association of cross-reactive mucosal IgA in immunized pigs with the level of cross-protection observed. Both NW and OF had measurable cross-reactive IgA; however, using ELISA OD levels as an association value, a broadly cross-reactive response to all H1 cluster viruses as well to as an H3 virus was observed, with OD values nearly the same across heterologous phylogenetic clusters. Using this association alone, we would have predicted complete protection against both challenge viruses, since mucosal IAV-specific IgA was cross-reactive with both MN/10(β) and IL/10(γ) antigens at similar OD values; however, this was not the case. Endpoint titrations of IAV-specific IgA in NW and OF from WT NY/09-exposed pigs separated the data showing cross-reactivity between virus clusters, and a general trend for decreasing IgA endpoint titers was evident, as phylogenetically distant viruses were used as the test antigen (pdm>γ-cluster>β-cluster>δ-cluster) (19), and these levels were associated with cross-protection. Due to the reduced immunogenicity of the LAIV vaccine, there was no clear association between IgA endpoint titers and cross-protection. Performing this analysis with OF samples from WT NY/09-inoculated animals, which had higher IAV-specific IgA titers, did show an association with antigenic relatedness, cross-reactive IgA titers, and cross-protection (Fig. 4 and 5B). Collectively, these data suggest that IAV-specific IgA in OF could be used to assess antigenic relatedness and cross-protection of circulating and emerging IAVs but that it may require that pigs be inoculated with wild-type viruses to elicit a response such that sufficient levels of mucosal IAV-specific IgA are induced. Vaccine or surveillance researchers would be able to curate a collection of OF samples from IAV-infected pigs and measure the antigenic relatedness of the IAV immunogen to currently circulating IAV strains and update and modify vaccine strains as necessary. This approach is somewhat similar to that of human seasonal IAV vaccine selection, where ferrets are infected with live IAV to elicit a high serum HI titer in order to assess the antigenic relatedness of circulating IAV isolates (67, 68).
ACKNOWLEDGMENTS
We thank Zahra Olson, Lilia Walther, and Steven Kellner for technical assistance and Jason Huegel, Tyler Standley, and Jason Crabtree for assistance with animal work.
Mention of trade names or products is solely for the purpose of providing specific information and does not imply endorsement by the USDA. USDA is an equal opportunity provider and employer.
The findings and conclusions in the manuscript are ours and do not necessarily represent the views of the USDA.
Funding for this work is provided in part by The National Pork Board (no. 13-122) and Boehringer-Ingelheim Vetmedica, Inc.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00358-15.
REFERENCES
- 1.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. 2011. Immune responses to influenza virus infection. Virus Res 162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A. 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 3.Ito T. 2000. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol Immunol 44:423–430. doi: 10.1111/j.1348-0421.2000.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 4.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 5.Bowman AS, Nelson SW, Page SL, Nolting JM, Killian ML, Sreevatsan S, Slemons RD. 2014. Swine-to-human transmission of influenza A(H3N2) virus at agricultural fairs, Ohio, USA, 2012. Emerg Infect Dis 20:1472–1480. doi: 10.3201/eid2009.131082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Z, Gomez J, Bowman AS, Ye J, Long LP, Nelson SW, Yang J, Martin B, Jia K, Nolting JM, Cunningham F, Cardona C, Zhang J, Yoon KJ, Slemons RD, Wan XF. 2013. Antigenic characterization of H3N2 influenza A viruses from Ohio agricultural fairs. J Virol 87:7655–7667. doi: 10.1128/JVI.00804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent AL, Swenson SL, Lager KM, Gauger PC, Loiacono C, Zhang Y. 2009. Characterization of an influenza A virus isolated from pigs during an outbreak of respiratory disease in swine and people during a county fair in the United States. Vet Microbiol 137:51–59. doi: 10.1016/j.vetmic.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Altmüller A, Kunerl M, Muller K, Hinshaw VS, Fitch WM, Scholtissek C. 1992. Genetic relatedness of the nucleoprotein (NP) of recent swine, turkey, and human influenza A virus (H1N1) isolates. Virus Res 22:79–87. doi: 10.1016/0168-1702(92)90091-M. [DOI] [PubMed] [Google Scholar]
- 9.Choi YK, Lee JH, Erickson G, Goyal SM, Joo HS, Webster RG, Webby RJ. 2004. H3N2 influenza virus transmission from swine to turkeys, United States. Emerg Infect Dis 10:2156–2160. doi: 10.3201/eid1012.040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, Buranathal C, Chaisingh A, Auewarakul P, Hanh NT, Ma SK, Hui PY, Guan Y, Peiris JS, Webster RG. 2005. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol 79:10821–10825. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wideman G. 2013. Impact of influenza A in pork production, p 191–193. Proceedings of the London Swine Conference, London, Ontario, Canada. [Google Scholar]
- 12.Bowman AS, Nolting JM, Nelson SW, Slemons RD. 2012. Subclinical influenza virus A infections in pigs exhibited at agricultural fairs, Ohio, USA, 2009–2011. Emerg Infect Dis 18:1945–1950. doi: 10.3201/eid1812.121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. 2008. Swine influenza viruses: a North American perspective. Adv Virus Res 72:127–154. doi: 10.1016/S0065-3527(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 14.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39:176–185. doi: 10.1007/s11262-009-0386-6. [DOI] [PubMed] [Google Scholar]
- 16.Ma W, Vincent AL, Lager KM, Janke BH, Henry SC, Rowland RR, Hesse RA, Richt JA. 2010. Identification and characterization of a highly virulent triple reassortant H1N1 swine influenza virus in the United States. Virus Genes 40:28–36. doi: 10.1007/s11262-009-0413-7. [DOI] [PubMed] [Google Scholar]
- 17.Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, Lewis NS. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92:919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nfon CK, Berhane Y, Hisanaga T, Zhang S, Handel K, Kehler H, Labrecque O, Lewis NS, Vincent AL, Copps J, Alexandersen S, Pasick J. 2011. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. J Virol 85:8667–8679. doi: 10.1128/JVI.00801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL. 2015. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res 201:24–31. doi: 10.1016/j.virusres.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Gauger PC, Loving CL, Vincent AL. 2014. Enzyme-linked immunosorbent assay for detection of serum or mucosal isotype-specific IgG and IgA whole-virus antibody to influenza A virus in swine. Methods Mol Biol 1161:303–312. doi: 10.1007/978-1-4939-0758-8_25. [DOI] [PubMed] [Google Scholar]
- 21.Heinen PP, van Nieuwstadt AP, de Boer-Luijtze EA, Bianchi AT. 2001. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet Immunol Immunopathol 82:39–56. doi: 10.1016/S0165-2427(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 22.Vincent AL, Ciacci-Zanella JR, Lorusso A, Gauger PC, Zanella EL, Kehrli ME Jr, Janke BH, Lager KM. 2010. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine 28:2782–2787. doi: 10.1016/j.vaccine.2010.01.049. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Madson D, Miller CL, Harris DL. 2012. Vaccine development for protecting swine against influenza virus. Anim Health Res Rev 13:181–195. doi: 10.1017/S1466252312000175. [DOI] [PubMed] [Google Scholar]
- 24.Braucher DR, Henningson JN, Loving CL, Vincent AL, Kim E, Steitz J, Gambotto AA, Kehrli ME Jr. 2012. Intranasal vaccination with replication-defective adenovirus type 5 encoding influenza virus hemagglutinin elicits protective immunity to homologous challenge and partial protection to heterologous challenge in pigs. Clin Vaccine Immunol 19:1722–1729. doi: 10.1128/CVI.00315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent AL, Ma W, Lager KM, Janke BH, Webby RJ, Garcia-Sastre A, Richt JA. 2007. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25:7999–8009. doi: 10.1016/j.vaccine.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustin KM, Maines TR, Belser JA, van Hoeven N, Lu X, Dong L, Isakova-Sivak I, Chen LM, Voeten JT, Heldens JG, van den Bosch H, Cox NJ, Tumpey TM, Klimov AI, Rudenko L, Donis RO, Katz JM. 2011. Comparative immunogenicity and cross-clade protective efficacy of mammalian cell-grown inactivated and live attenuated H5N1 reassortant vaccines in ferrets. J Infect Dis 204:1491–1499. doi: 10.1093/infdis/jir596. [DOI] [PubMed] [Google Scholar]
- 27.Gauger PC, Loving CL, Khurana S, Lorusso A, Perez DR, Kehrli ME Jr, Roth JA, Golding H, Vincent AL. 2014. Live attenuated influenza A virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology 471–473:93–104. [DOI] [PubMed] [Google Scholar]
- 28.Masic A, Booth JS, Mutwiri GK, Babiuk LA, Zhou Y. 2009. Elastase-dependent live attenuated swine influenza A viruses are immunogenic and confer protection against swine influenza A virus infection in pigs. J Virol 83:10198–10210. doi: 10.1128/JVI.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandbulte MR, Platt R, Roth JA, Henningson JN, Gibson KA, Rajao DS, Loving CL, Vincent AL. 2014. Divergent immune responses and disease outcomes in piglets immunized with inactivated and attenuated H3N2 swine influenza vaccines in the presence of maternally-derived antibodies. Virology 464–465:45–54. [DOI] [PubMed] [Google Scholar]
- 30.Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, Gauger PC, Loving CL, Webby RJ, Garcia-Sastre A. 2012. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol 86:10597–10605. doi: 10.1128/JVI.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wesley RD, Lager KM. 2006. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet Microbiol 118:67–75. doi: 10.1016/j.vetmic.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. 2000. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol 74:11566–11573. doi: 10.1128/JVI.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci U S A 97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solórzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol 79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A, Garcia-Sastre A. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol 80:11009–11018. doi: 10.1128/JVI.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wesley RD, Tang M, Lager KM. 2004. Protection of weaned pigs by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of H3N2 swine influenza virus. Vaccine 22:3427–3434. doi: 10.1016/j.vaccine.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 37.Katz JM, Hancock K, Xu X. 2011. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther 9:669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 38.Loving CL, Lager KM, Vincent AL, Brockmeier SL, Gauger PC, Anderson TK, Kitikoon P, Perez DR, Kehrli ME Jr. 2013. Efficacy in pigs of inactivated and live attenuated influenza virus vaccines against infection and transmission of an emerging H3N2 similar to the 2011–2012 H3N2v. J Virol 87:9895–9903. doi: 10.1128/JVI.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clements ML, Murphy BR. 1986. Development and persistence of local and systemic antibody responses in adults given live attenuated or inactivated influenza A virus vaccine. J Clin Microbiol 23:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pena L, Vincent AL, Ye J, Ciacci-Zanella JR, Angel M, Lorusso A, Gauger PC, Janke BH, Loving CL, Perez DR. 2011. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 85:456–469. doi: 10.1128/JVI.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loving CL, Kehrli ME Jr, Brockmeier SL, Bayles DO, Michael DD, Schlink SN, Lager KM. 2013. Porcine granulocyte-colony stimulating factor (G-CSF) delivered via replication-defective adenovirus induces a sustained increase in circulating peripheral blood neutrophils. Biologicals 41:368–376. doi: 10.1016/j.biologicals.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Loving CL, Brockmeier SL, Vincent AL, Palmer MV, Sacco RE, Nicholson TL. 2010. Influenza virus coinfection with Bordetella bronchiseptica enhances bacterial colonization and host responses exacerbating pulmonary lesions. Microb Pathog 49:237–245. doi: 10.1016/j.micpath.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Ramirez A, Wang C, Prickett JR, Pogranichniy R, Yoon KJ, Main R, Johnson JK, Rademacher C, Hoogland M, Hoffmann P, Kurtz A, Kurtz E, Zimmerman J. 2012. Efficient surveillance of pig populations using oral fluids. Prev Vet Med 104:292–300. doi: 10.1016/j.prevetmed.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol 41:3198–3205. doi: 10.1128/JCM.41.7.3198-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. 1995. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 47.Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME Jr, Roth JA. 2012. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 48.Vincent AL, Lager KM, Ma W, Lekcharoensuk P, Gramer MR, Loiacono C, Richt JA. 2006. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol 118:212–222. doi: 10.1016/j.vetmic.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 49.Kitikoon P, Nilubol D, Erickson BJ, Janke BH, Hoover TC, Sornsen SA, Thacker EL. 2006. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol 112:117–128. doi: 10.1016/j.vetimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Reed LJ, Muench H. 1938. A simple method for estimating fifty percent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 51.Belshe RB. 2004. Current status of live attenuated influenza virus vaccine in the US. Virus Res 103:177–185. doi: 10.1016/j.virusres.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 52.Paillot R, Hannant D, Kydd JH, Daly JM. 2006. Vaccination against equine influenza: quid novi? Vaccine 24:4047–4061. doi: 10.1016/j.vaccine.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 53.Kappes MA, Sandbulte MR, Platt R, Wang C, Lager KM, Henningson JN, Lorusso A, Vincent AL, Loving CL, Roth JA, Kehrli ME Jr. 2012. Vaccination with NS1-truncated H3N2 swine influenza virus primes T cells and confers cross-protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 30:280–288. doi: 10.1016/j.vaccine.2011.10.098. [DOI] [PubMed] [Google Scholar]
- 54.Wacheck V, Egorov A, Groiss F, Pfeiffer A, Fuereder T, Hoeflmayer D, Kundi M, Popow-Kraupp T, Redlberger-Fritz M, Mueller CA, Cinatl J, Michaelis M, Geiler J, Bergmann M, Romanova J, Roethl E, Morokutti A, Wolschek M, Ferko B, Seipelt J, Dick-Gudenus R, Muster T. 2010. A novel type of influenza vaccine: safety and immunogenicity of replication-deficient influenza virus created by deletion of the interferon antagonist NS1. J Infect Dis 201:354–362. doi: 10.1086/649428. [DOI] [PubMed] [Google Scholar]
- 55.Lange E, Kalthoff D, Blohm U, Teifke JP, Breithaupt A, Maresch C, Starick E, Fereidouni S, Hoffmann B, Mettenleiter TC, Beer M, Vahlenkamp TW. 2009. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J Gen Virol 90:2119–2123. doi: 10.1099/vir.0.014480-0. [DOI] [PubMed] [Google Scholar]
- 56.Price GE, Lo CY, Misplon JA, Epstein SL. 2014. Mucosal immunization with a candidate universal influenza vaccine reduces virus transmission in a mouse model. J Virol 88:6019–6030. doi: 10.1128/JVI.03101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houser KV, Pearce MB, Katz JM, Tumpey TM. 2013. Impact of prior seasonal H3N2 influenza vaccination or infection on protection and transmission of emerging variants of influenza A(H3N2)v virus in ferrets. J Virol 87:13480–13489. doi: 10.1128/JVI.02434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent AL, Lager KM, Janke BH, Gramer MR, Richt JA. 2008. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet Microbiol 126:310–323. doi: 10.1016/j.vetmic.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 59.La Gruta NL, Kedzierska K, Stambas J, Doherty PC. 2007. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol 85:85–92. doi: 10.1038/sj.icb.7100026. [DOI] [PubMed] [Google Scholar]
- 60.Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. 2014. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 88:4752–4763. doi: 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Hoschler K, Saville M, Vogel FR, Barclay W, Donatelli I, Zambon M, Wood J, Haaheim LR. 2009. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 27:1889–1897. doi: 10.1016/j.vaccine.2009.01.116. [DOI] [PubMed] [Google Scholar]
- 62.Keitel WA, Atmar RL. 2009. Vaccines for pandemic influenza: summary of recent clinical trials. Curr Top Microbiol Immunol 333:431–451. [DOI] [PubMed] [Google Scholar]
- 63.Panyasing Y, Goodell CK, Gimenez-Lirola L, Kittawornrat A, Wang C, Schwartz KJ, Zimmerman JJ. 2013. Kinetics of influenza A virus nucleoprotein antibody (IgM, IgA, and IgG) in serum and oral fluid specimens from pigs infected under experimental conditions. Vaccine 31:6210–6215. doi: 10.1016/j.vaccine.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 64.Nokes DJ, Enquselassie F, Nigatu W, Vyse AJ, Cohen BJ, Brown DW, Cutts FT. 2001. Has oral fluid the potential to replace serum for the evaluation of population immunity levels? A study of measles, rubella and hepatitis B in rural Ethiopia. Bull World Health Organ 79:588–595. [PMC free article] [PubMed] [Google Scholar]
- 65.Clements ML, Betts RF, Tierney EL, Murphy BR. 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol 24:157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asahi-Ozaki Y, Yoshikawa T, Iwakura Y, Suzuki Y, Tamura S, Kurata T, Sata T. 2004. Secretory IgA antibodies provide cross-protection against infection with different strains of influenza B virus. J Med Virol 74:328–335. doi: 10.1002/jmv.20173. [DOI] [PubMed] [Google Scholar]
- 67.Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, Gust ID, Hampson AW, Hay AJ, Hurt AC, de Jong JC, Kelso A, Klimov AI, Kageyama T, Komadina N, Lapedes AS, Lin YP, Mosterin A, Obuchi M, Odagiri T, Osterhaus AD, Rimmelzwaan GF, Shaw MW, Skepner E, Stohr K, Tashiro M, Fouchier RA, Smith DJ. 2008. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 26(Suppl 4):D31–D34. doi: 10.1016/j.vaccine.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 68.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]