Abstract
Immunization with the pneumococcal proteins pneumolysin (Ply), choline binding protein A (CbpA), or pneumococcal surface protein A (PspA) elicits protective responses against invasive pneumococcal disease in animal models. In this study, we used different mouse models to test the efficacy of a variety of multivalent protein-based vaccines that comprised various combinations of full-length or peptide regions of the immunogens Ply, CbpA, or PspA: Ply toxoid with the L460D substitution (referred to herein as L460D); L460D fused with protective peptide epitopes from CbpA (YPT-L460D-NEEK [YLN]); L460D fused with the CD2 peptide containing the proline-rich region (PRR) of PspA (CD2-L460D); a combination of L460D and H70 (L460D+H70), a slightly larger PspA-derived peptide containing the PRR and the SM1 region; H70+YLN; and other combinations. Each mouse was immunized either intraperitoneally (i.p.) or subcutaneously (s.c.) with three doses (at 2-week intervals) of the various antigen combinations in alum adjuvant and then challenged in mouse models featuring different infection routes with multiple Streptococcus pneumoniae strains. In the i.p. infection sepsis model, H70+YLN consistently provided significant protection against three different challenge strains (serotypes 1, 2, and 6A); the CD2+YLN and H70+L460D combinations also elicited significant protection. Protection against intravenous (i.v.) sepsis (type 3 and 6A challenge strains) was largely dependent on PspA-derived antigen components, and the most protection was elicited by H70 with or without L460D or YLN. In a type 4 intratracheal (i.t.) challenge model that results in progression to meningitis, antigen combinations that contained YLN elicited the strongest protection. Thus, the trivalent antigen combination of H70+YLN elicited the strongest and broadest protection in diverse pneumococcal challenge models.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is responsible for almost 1 million deaths worldwide in children <5 years of age each year (1) and is a leading cause of both invasive pneumococcal diseases (IPD) (e.g., bacteremia, bacteremic pneumonia, and meningitis) and noninvasive diseases (e.g., nonbacteremic pneumonia, otitis media, and sinusitis) (2). Collectively, these conditions account for greater global morbidity and mortality than diseases caused by any other pathogen. Antipneumococcal vaccination strategies currently target the capsular polysaccharides, which are serologically varied, with >93 distinct serotypes identified to date (3). A 23-valent pneumococcal polysaccharide vaccine (PPV23) has been used in many countries for several decades; it provides strictly serotype-dependent protection but does not elicit immunological memory and shows poor immunogenicity in children <2 years, the age group with the highest incidence of IPD (4).
A 7-valent pneumococcal conjugate vaccine (PCV7) was licensed in 2000 to overcome the abovementioned shortcomings of PPV23. Unlike PPV23, PCV7 was highly protective in young children against IPD caused by the included serotypes. There was also a marked herd immunity effect due to its capacity to reduce nasopharyngeal carriage of vaccine-type pneumococci, thereby reducing the transmission of these serotypes to nonvaccinated individuals within the community (5). However, the declines in the incidences of IPD and nasopharyngeal colonization by vaccine-type pneumococci have been offset to various extents by increases in both carriage and disease due to nonvaccine serotypes (5–7). This serotype replacement can result from two scenarios: first, unmasking of nonvaccine types already present in the nasopharynx in low numbers or elsewhere in the community, and second, by acquisition of a nonvaccine serotype capsule biosynthesis locus by a vaccine type strain through natural genetic transformation (3, 8). This second scenario may be of greater potential significance, as it enables highly transmissible, virulent, and often antibiotic-resistant clones to escape the PCV. More recently licensed PCVs have increased serotype coverage (10- and 13-valent) (9, 10), but this is at best only a stop-gap measure. The emergence of a high proportion of virulent strains outside the 13-valent vaccine coverage has recently been documented in the high-risk sickle cell disease population (11).
Pneumococcal protein-based vaccine formulations comprising combinations of conserved virulence proteins that function at different stages of pathogenesis offer an attractive alternative to PCVs. Three proteins stand out as the best candidate vaccine antigens, namely, pneumolysin (Ply), pneumococcal surface protein A (PspA), and choline binding protein A (CbpA) (also known as PspC). These proteins elicit some of the highest antibody titers in sera from humans exposed to pneumococci (12–15), and their functions in pathogenesis are well characterized, involving processes in all body compartments (16–22).
Ply is a cholesterol-binding toxin produced by virtually all strains of S. pneumoniae. It was one of the first virulence-related proteins identified in S. pneumoniae, and its cytotoxicity emanates from its capacity to insert into eukaryotic cell membranes and oligomerize to form large transmembrane pores, resulting in cell lysis (17). A number of immunogenic pneumolysoids generated by point mutations in the ply gene have reduced cytotoxicity and provide significant protection against different serotypes of S. pneumoniae at multiple stages of infection in various murine models. One such toxoid, PdB, has a single substitution (W433F) that reduces hemolytic activity by >99% (23), and it is one of the most well-studied antigens. It protects mice from diverse S. pneumoniae strains when used as either a single immunogen or in combination with other proteins, particularly PspA and CbpA (24–28). More recently, a triple mutant toxoid, PlyD1 (comprising T65C, G293C, and C428A mutations), with only 0.001% residual hemolytic activity, was also shown to elicit protection against pneumococcal infection and lung injury (29). A novel Ply toxoid devoid of detectable cytotoxicity contains an L460D substitution, which disrupts a threonine-leucine pair that comprises the cholesterol recognition motif (CRM) and is critical for cholesterol binding on target cells by several cholesterol-dependent cytolysins (CDCs) (30). In GenBank, this CRM region is strictly conserved in approximately 50 different bacterial species that express a CDC. Due to its undetectable cytotoxicity, Ply L460D (referred to as L460D herein) is potentially a more suitable vaccine candidate than PdB.
PspA is one of the best-characterized members of the choline binding protein family. It is found on the surfaces of all pneumococci and has important roles in the pathogenesis of disease (31, 32). During invasive disease, it interferes with complement-dependent host defense and thereby reduces the deposition of complement on the cell surface; concomitantly, it impairs complement receptor-mediated clearance. PspA also blocks killing by host cationic bactericidal peptides (33) and interferes with phagocytosis by a complement-independent mechanism (34). The PspA protein consists of three main regions: the N-terminal alpha-helical domain, a proline-rich region (PRR), and a choline binding domain. Previous studies have focused on the N-terminal alpha-helical domain of PspA, which elicits protection against invasive disease (28, 35–37). However, the alpha-helical domain of PspA is variable in terms of its amino acid sequence and antigenic epitopes (38–41). Attention has recently shifted to the PRR, as this region is much more conserved and contains epitopes on the pneumococcal surface that are antibody accessible (42). The PRR has also been shown to elicit broader cross-reactive antibodies and cross-protection against pneumococcal infection (42, 43).
Another well-characterized pneumococcal choline binding protein is CbpA, which participates in pneumococcal adherence to host cells and helps pneumococci evade complement attack and opsonophagocytosis. CbpA plays important roles in many stages of disease and elicits protection against pneumococcal infection in different animal models (20, 26, 27, 44–46). The N-terminal domain of CbpA binds to the secretory component of secretory immunoglobulin A (sIgA), C3, factor H, and the laminin receptor (47–52). This domain consists of two nearly identical repeat regions that each assemble into antiparallel helices (not-coiled coils), and the turns between the helices show extremely high conservation across most pneumococcal strains (49, 53). An RRNYPT sequence in the first turn binds to the epithelial polymeric immunoglobulin receptor (pIgR), and the EPRNEEK sequence in the second turn binds to the laminin receptor on the blood-brain barrier endothelium (52). Recently, a recombinant fusion protein consisting of these two turns fused to either terminus of L460D has been shown to elicit broad protection against multiple stages of pneumococcal infection (54).
The aim of the present study was to develop a multivalent and broadly efficacious pneumococcal protein vaccine via a thorough examination of the protective immunogenicity of the proteins L460D, CbpA, and PspA. Vaccine candidates consisted of either full-length or peptide derivatives of these proteins, recombinant fusion proteins comprising immunogenic regions from these proteins fused to the full length or portions of one of the other proteins, or these in various combinations. The resultant multivalent protein candidates were tested for protective efficacy in different animal models against a wide range of pneumococcal challenge strains to establish the optimal formulation of a multivalent vaccine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. pneumoniae serotype 4 bioluminescent strain TIGR4X was grown in chemically defined medium (C+Y) without aeration at 37°C in 5% CO2 to an optical density at 620 nm (OD620) of 0.4 to 0.5 (54). S. pneumoniae strains D39, 1861, and P9 were grown in serum broth to an OD600 of 0.18, as previously described (55). S. pneumoniae strains DBL6A, A66.1, and EF3030 were grown in Todd-Hewitt broth to an OD600 of 0.4, as previously described (42).
TABLE 1.
S. pneumoniae challenge strains used in this study
Construction and production of recombinant proteins.
The features of the recombinant proteins and expression constructs used in this study are presented in Table 2. The PCR primers used for subcloning are listed in Table 3. All expression constructs were sequenced at the St. Jude Children's Research Hospital (SJCRH) Hartwell Center. All proteins were purified by the SJCRH Protein Production Facility and stored in 10 mM histidine buffer (pH 6.0) with 15% trehalose.
TABLE 2.
Features of recombinant proteins and expression constructs
| Construct | Protein(s) expressed (aa) | Source strain(s) | Template for cloning | 5′ primer | 3′ primer |
|---|---|---|---|---|---|
| L460D | Pneumolysoid | D39 | pQE30-L460D | PLYNDE | PLYSAC |
| CD2 | PspA proline-rich region | AC094 | gDNA | PI5-up-LIC | PI5-down-LIC |
| H70 | PspA proline-rich region and SM1 | Rx1 | pIDTSMART-H70a | H70NDE | H70SAC |
| pUAB055 | PspA (1–302) | Rx1 | gDNA | pspA1-F | pspA6-R |
| rCbpA | CbpA (175–443) | TIGR4 | gDNA | A175NDE | A443SAC |
| YPT-L460D-NEEK (YLN) | Pneumolysoid and CbpA peptides | TIGR4 and D39 | pET28-L460D | YPTNDE | NEEKSAC |
| CD2-L460D | PspA peptide and pneumolysoid | AC094 and D39 | pCD2, pET28-L460D | CD2NDE | PLYSAC |
Initial construct made by Integrated DNA Technologies.
TABLE 3.
PCR primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| PLYNDE | CGCGCGCGCATATGGCAAATAAAGCAGTAAATGAC |
| PLYSAC | GCGCGCGAGCTCTTACTAGTCATTTTCTACCTTATCC |
| YPTNDE | CGCGCGCGCATATGGCTTGTAAAAAAGCCGAGGATCAAAAAGAAGAAGATCGCCGTAACTACCCAACCAATACTTACAAAACGCTTGAACTTGAATGTGCTGAGGGTGGTGCAAATAAAGCAGTAAATG |
| NEEKSAC | CGCGCGGAGCTCCTATTTACATTGCTTAACTTTTTCCTCGTTTCGAGGTTCCTTAGCACACTCTTTGTCATTTTCTACCTTATCCTC |
| A175NDE | GCGCGCCATATGCCAGGAGAAAAGGTAGCAG |
| A443SAC | GCGCGCGAGCTCTTATGGTTTTTCTTTAACTTTATC |
| CD2NDE | GCGCGCCATATGGCCATGGCTGACCTTAAG |
| CD2F2 | CGGTATGTGGATGGCAAATAAAGCAGTAAATG |
| CD2R1 | CTGCTTTATTTGCCATCCACATACCGTTTTCTTG |
| H70NDE | GCGCGCCATATGTATTTTAAAGAAGGGTTAG |
| H70SAC | GCGCGAGCTCCTAACCATTTTCTTGTTTCCACCCAG |
| PI5-up-LIC | GACGACGACAAGATGGCTGACCTTAAGAAAGC |
| PI5-down-LIC | GAGGAGAAGCCCGGTTTACCACATACCGTTTTCTTG |
| pspA1-F | GCCATGGAAGAATCTCCCGTAGCC |
| pspA6-R | CTCGAGTTCTGGGGCTGGAGTTTC |
| EKLIC-BglII | GACGACGACAAGAGATCTTACTTTAAAGAAGGTTTAGAG |
| EKLIC_SM1-ProR | GAGGAGAAGCCCGGTCTAGGATCCCATACCGTTTTCTTGTTTCC |
Pneumolysoid.
The ply gene was first PCR amplified from strain D39 and then cloned into vector pQE30. The L460D substitution was then introduced by overlapping PCR, as previously described (56). The resulting plasmid, pQE30-L460D, was used as the template to obtain the L460D gene using primers PLYNDE and PLYSAC, and the L460D gene was subsequently cloned into vector pET28a (Novagen) (54). The recombinant plasmid pET28a-L460D was then transformed into Escherichia coli strain BL21(DE3) cells for protein production.
CbpA derivatives and fusion proteins.
CbpA derivatives YPT and NEEK from TIGR4 were constructed as previously described (54). Briefly, YPT-L460D-NEEK (YLN) and L460D-NEEK were both constructed by incorporating the constrained CbpA sequences of YPT and NEEK into the primers used to amplify L460D. The construction of the YLN fusion protein was previously described (54). Each fragment was cloned into the vector pET28a (Novagen), and the desired constructs were verified by DNA sequencing.
PspA derivatives and fusion proteins.
The construction of pUAB055, carrying genes encoding a truncated version of PspA containing the N-terminal region (amino acids [aa]1 to 302), was described previously (39). Briefly, a pUAB055 fragment was amplified by PCR from S. pneumoniae strain Rx1 using primers pspA1-F and pspA6-R and was then cloned into vector pCRII (Invitrogen) to form plasmid pCRII-pUAB055. The plasmid was digested with NcoI and XhoI to obtain the 918-bp pUAB055 fragment that was then cloned into the NcoI/XhoI sites of vector pET20b. The resulting plasmid was then transformed into the E. coli strain RosettaBlue (DE3) pLysS for protein production.
The construction of peptide CD2 was previously described (42). It contains the proline-rich region (PRR) of the PspA from S. pneumoniae strain AC94. Briefly, the PRR (315 bp) was PCR amplified using the primers PI5-up-LIC and PI5-down-LIC and cloned into vector pET-46Ek/LIC. This recombinant plasmid was then transformed into E. coli BL21 Star (DE3) cells for protein production.
Peptide H70 is slightly larger than CD2, comprising the PRR and an SM1 peptide, which is naturally located upstream of the PRR in strain Rx1. The region containing the SM1 and PRR was PCR amplified from strain Rx1 using the primers EKLIC-BglII and EKLIC_SM1-ProR. The resulting PCR fragment was then cloned into vector pET46-Ek/LIC and verified by DNA sequence to confirm its insertion of the expected 393-bp fragment. This recombinant plasmid was then used as the template to obtain an H70 fragment using primers H70NDE and H70SAC, and it was subsequently cloned into vector pET28a (Novagen). The recombinant plasmid pET28a-H70 was then transformed into BL21(DE3) cells for protein production.
Intraperitoneal sepsis model.
All animal experiments were carried out in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (57) and the South Australian Animal Welfare Act 1985 (58) and were approved by the animal ethics committee of the University of Adelaide, South Australia, Australia. Outbred 4- to 6-week-old female CD1 (Swiss) mice were used in all intraperitoneal (i.p.) sepsis experiments. Each mouse was immunized i.p. three times with 100 μl of 0.5% saline containing 10 μg of each protein and 200 μg of alum adjuvant (Alhydrogel; InvivoGen) at 14-day intervals. Serum samples were collected from the mice by submandibular bleeding 1 week after the third immunization. A week later, the mice were challenged i.p. with strains D39 (2.5 × 104 CFU, serotype 2), P9 (5 × 104 CFU, serotype 6A), and 1861 (5 × 102 CFU, serotype 1). The infected mice were observed for up to 21 days to determine the number of days until they were moribund.
Meningitis/sepsis model.
The experiments were approved by the St. Jude Institutional Animal Care and Use Committee and were carried out essentially as previously described (54). Briefly, 6- to 7-week-old female BALB/c mice were immunized i.p. with 10 μg of protein containing 130 μg of alum (Brenntag), and subsequent boosts were given at weeks 2 and 4. Serum samples were collected from the mice at week 5 for functional antibody analysis. The mice were challenged intratracheally (i.t.) with 1 × 107 CFU of TIGR4X (serotype 4) at week 7, and survival was monitored for 14 days. Meningitis was observed by physical signs and confirmed through Xenogen imaging and plating of cerebrospinal fluid samples. Nasal washes of the survivors were collected and plated on day 14 postchallenge.
Intravenous sepsis model and i.t. focal pneumonia models.
These experiments were approved by the University of Alabama at Birmingham (UAB) Animal Care Committee. This intravenous (i.v.) sepsis model was carried out as previously described (42). Briefly, CBA/CaHNVrkxid/J (CBA/N) mice were immunized subcutaneously (s.c.) three times at 2-week intervals with 100 μl containing 5 μg of protein adjuvanted with alum. Three weeks after the last immunization, blood samples were taken from the mice for enzyme-linked immunosorbent assay (ELISA). The following day, the mice were infected i.v. with 4 × 103 CFU of strain A66.1 (serotype 3) or DBL6A (serotype 6B) diluted from a frozen infectious stock in lactated Ringer's solution containing 7.8% glycerol. The infected mice were observed for up to 21 days to determine the number of days until they were moribund.
In the i.t. focal pneumonia model, CBA/N mice were immunized as described above, except for the challenge, for which mice were first anesthetized lightly with isoflurane and then challenged i.t. with 5 × 106 CFU of S. pneumoniae strain EF3030 (serotype 19F), as described previously (28). On day 5 postinfection, the mice were euthanized, and blood and lungs were harvested and plated for CFU recovery.
Measurement of antibody responses.
Antigen-specific antibody responses were determined by ELISA using serum samples collected from immunized mice, as previously described (42, 59).
Hemolysis and anti-hemolytic activity assays.
The hemolytic activity of pneumolysin and the capacity of murine serum to neutralize the hemolytic activity of wild-type Ply were assayed, as previously described (60).
Statistical analysis.
Statistical analysis was carried out using GraphPad Prism 6. The Mann-Whitney U test (two-tailed) was used for all statistical comparisons carried out in this study (median survival, CFU recovery, and antibody levels). P values of <0.05 were considered statistically significant.
RESULTS
Construction of multivalent protein vaccine candidates.
L460D, CbpA, PspA, and their various derivatives (Fig. 1) were tested to determine the optimum formulation of a multivalent vaccine. The immunogens were formulated as mixtures of one, two, or three proteins/peptides. Formulations that included L460D consisted of full-length L460D alone or as a fusion construct with the PspA peptide CD2 (CD2-L460D) or with either one or two constrained CbpA peptides (L460D-NEEK or YPT-L460D-NEEK [YLN] [54]). Derivatives of PspA were also mixed as separate proteins with L460D or YLN.
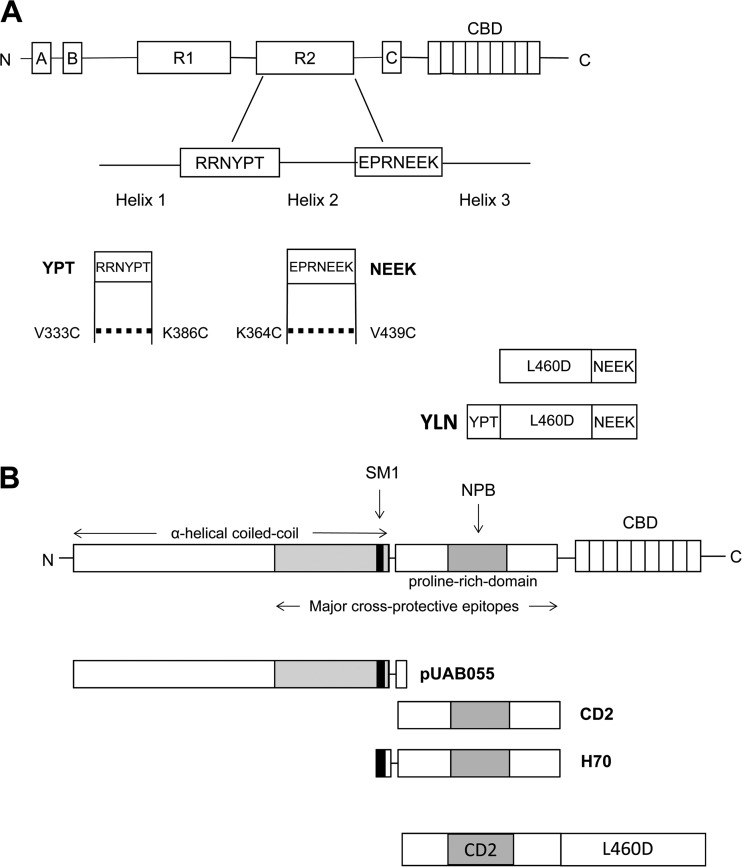
FIG 1.
Schematic representation of CbpA and PspA derivatives used in vaccine constructs. (A) Full-length CbpA (top) and regions used to make each construct, YPT and NEEK (bottom). The dotted lines indicate disulfide bridges formed by dual substitutions made in YPT (V333C and K386C) and NEEK (K364C and V439C) to engender a constrained structure of native CbpA. Fusion constructs of peptides with L460D are shown. (B) Full-length PspA (top) and regions used to make each vaccine construct: pUAB055, CD2, H70, and CD2-L460D (bottom).
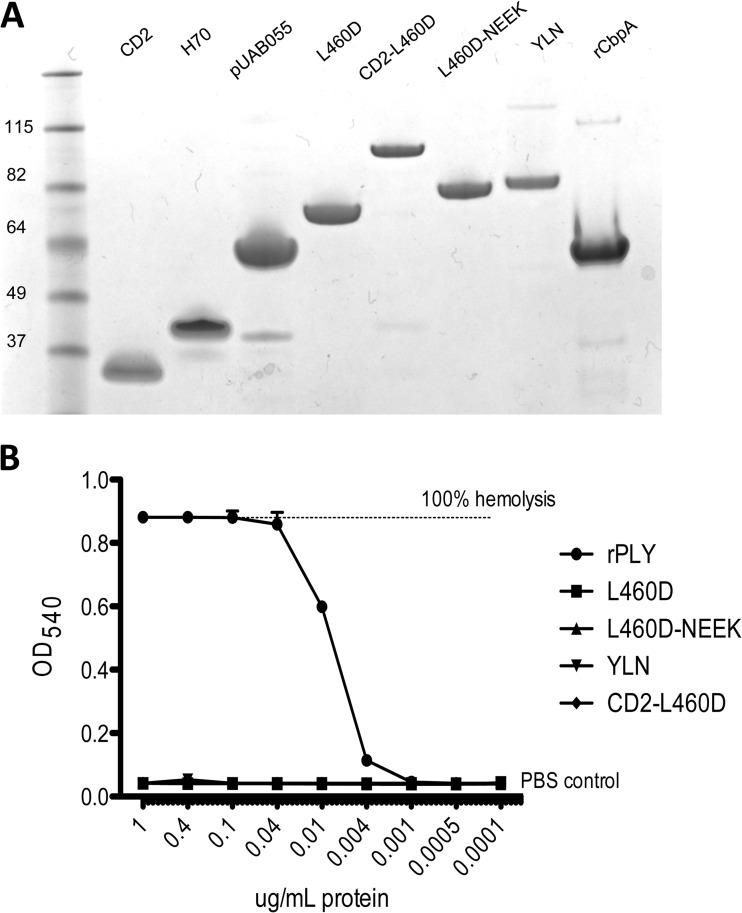
All of the constructs were purified at the St. Jude Protein Production Facility and then tested for purity, size, concentration, and contamination with endotoxin. Analysis by SDS-PAGE (Fig. 2A) showed that all proteins migrated according to their respective molecular sizes, except for CD2 and H70, which migrated more slowly than predicted: CD2 is 13 kDa but migrated at 25 kDa, and H70 is 15 kDa but migrated at 33 kDa. This anomalous electrophoretic mobility is likely due to their high proline content. The complete absence of hemolytic activity of purified L460D and its fusion derivatives was confirmed by hemolysis assay (Fig. 2B).
FIG 2.
Properties of vaccine constructs. (A) SDS-PAGE analysis of purified recombinant proteins stained with Coomassie blue. A molecular size marker ladder is also shown. (B) In vitro hemolytic activity of L460D, L460D fusions, and wild-type Ply. The indicated dilutions of Ply and L460D constructs were incubated with 1% washed human erythrocytes for 30 min at 37°C. After centrifugation, hemoglobin release was quantitated by absorbance at 540 nm. Triton X-100- and PBS-treated erythrocytes were used as controls for 100% and 0% lysis, respectively.
Immunogenicity of the multivalent protein vaccine candidates.
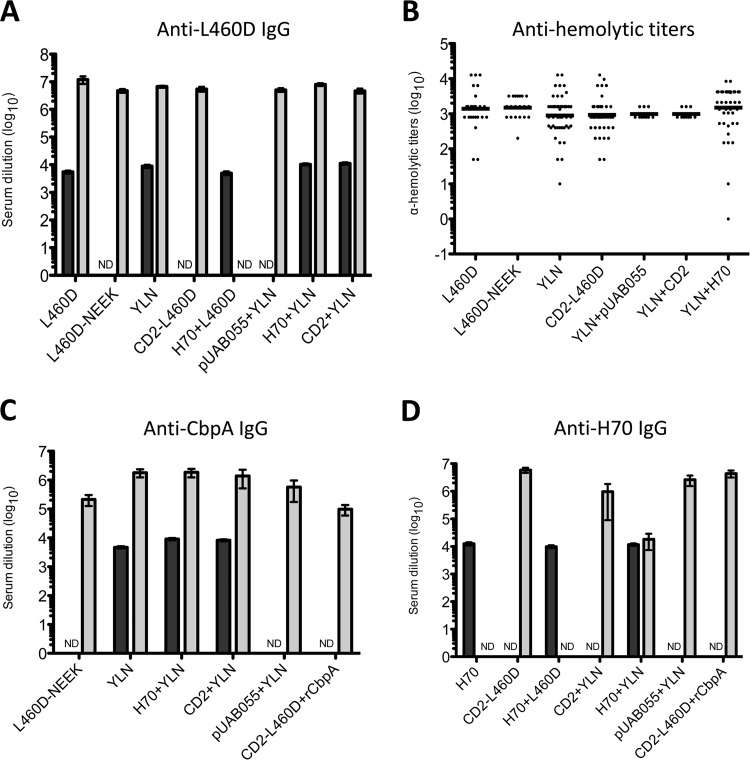
Groups of 12 to 15 mice (BALB/c or CD1) were immunized intraperitoneally (i.p.) or subcutaneously (s.c.) with each of the various antigens, singly or in combination, using alum adjuvant. Mice were injected three times at 2-week intervals (see Materials and Methods). A serum sample was then collected from each mouse 1 week after the final immunization for analysis of antibody responses. Antigen-specific IgG responses were measured by ELISA, using L460D, recombinant CbpA (rCbpA), or H70 as a coating antigen (Fig. 3). All vaccine formulations elicited strong antibody responses to L460D, with geometric mean (GM) anti-L460D IgG titers of >4,000 for all groups, compared to <200 for the alum placebo group (Fig. 3A).
FIG 3.
Immune responses in mice. (A, C, and D) Serum IgG responses to the various antigens were measured by ELISA using L460D (A), rCbpA (C), or H70 (D) as a coating antigen. BALB-C mice, gray bars; CD1 mice, black bars. The data are shown as the geometric mean (GM) titer, with the error bars denoting the 95% confidence intervals (CI). ND, not determined. (B) Functional activity of L460D antibodies elicited by various vaccine constructs was measured by the ability to neutralize 50% of the hemolytic activity of a standard dose of wild-type Ply. The data are shown as the GM antihemolytic titers. In this assay, the average hemolytic titer of the alum control group was 16.
Pneumolysin antibodies are expected to neutralize the wild-type toxin. Accordingly, the capacity of sera from mice immunized with the various L460D-containing formulations to neutralize the hemolytic activity of wild-type pneumolysin was also assessed (Fig. 3B). All antigens elicited significant toxin-neutralizing responses (GM antihemolytic titers, ∼300, compared with approximately 20 for the alum placebo group). Analysis of CbpA-specific antibody levels also showed strong responses (GM ELISA titers, >3,000) for all groups immunized with antigens containing YLN relative to the responses for the alum placebo group and antigen formulations lacking the CbpA-derived peptide epitopes (GM ELISA titers, <200) (Fig. 3C). However, in CD1 mice, both the H70-plus-YLN combination (H70+YLN) and CD2+YLN elicited significantly higher anti-CbpA levels than YLN alone (P < 0.0001 for both groups), although the biological significance of this numerically modest difference is uncertain. In CD1 mice, the three formulations containing H70 (H70, H70+L460D, and H70+YLN) elicited similarly strong IgG responses (GM ELISA titers, >10,000) compared with the response in the alum placebo group (GM titer, <200) (Fig. 3D). However, in BALB/c mice, anti-H70 responses to H70+YLN were lower than those elicited by the formulations containing either CD2 or pUAB055. Nevertheless, even for H70+YLN, the GM titer was >10,000, compared with <200 for the alum placebo.
Protective efficacy in sepsis models.
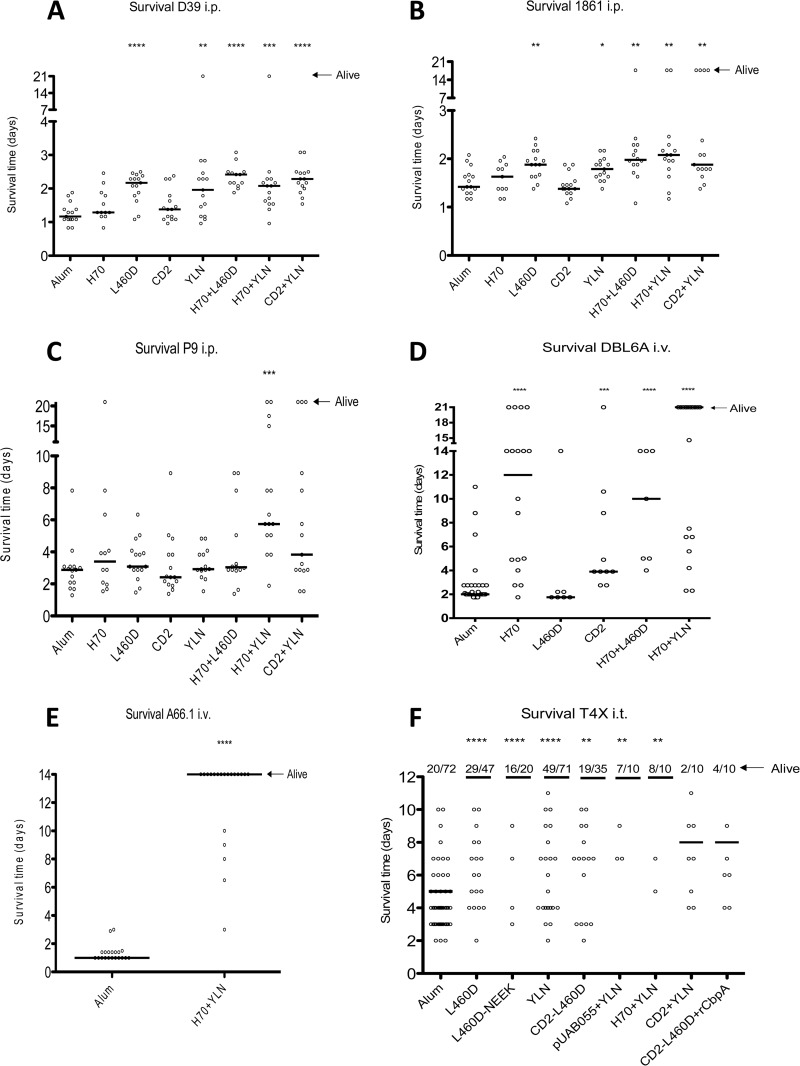
The efficacy of the vaccine formulations was determined using two sepsis models: i.p. challenge with one of three strains, D39 (serotype 2), P9 (serotype 6A), or 1861 (serotype 1), or i.v. challenge with strain DBL6A or A66.1 (serotypes 6B and 3, respectively). Immunized mice were challenged 2 weeks after the final immunization, and animals were monitored for survival for up to 21 days. In the i.p. challenge model, the H70+YLN, CD2+YLN, H70+L460D, YLN, and L460D vaccine groups all exhibited significant protection against two challenge strains, D39 and 1861 (Fig. 4A and B). However, only the trivalent H70+YLN group exhibited significant protection against strain P9, a clinical isolate that causes meningitis in approximately 50% of unprotected mice (Fig. 4C). In this experiment, any mice infected with P9 that displayed clinical signs of meningitis were promptly euthanized. The difference in protection seen between the trivalent H70+YLN and all the other groups, including the placebo control, was largely attributable to a lower rate of progression to meningitis, with only approximately 20% of mice immunized with H70+YLN showing symptoms, compared to approximately 50% for all other groups (results not shown). Peptides H70 and CD2 were not protective when given alone, but when each was combined with YLN, the level of protection increased significantly relative to that with YLN alone, indicating that the strongest protection is achieved by the combination of pneumolysoid and immunogenic portions of PspA and CbpA.
FIG 4.
Survival time of vaccinated mice in three sepsis models. (A to C) Intraperitoneal sepsis model. Mice were challenged with strain D39 (A), 1861 (B), or P9 (C). The solid lines denote the median survival time for each group, and mice that survived to 21 days postinfection were plotted as alive. Significant protection compared to that with the alum control group is indicated in each panel by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (D and E) Intravenous sepsis model. Mice were challenged with strain DBL6A (D) or A66.1 (E). The solid lines denote the median survival time for each group, and mice that survived to 21 days postinfection were plotted as alive. (F) Intratracheal sepsis/meningitis model. Mice were challenged with strain TIGR4X. The solid lines denote the median survival time for each group. The number of mice in each group and the number of mice alive at day 14 are depicted at the top of the panel.
Mice immunized with either H70 or CD2 exhibited significant protection against challenge with DBL6A in the i.v. sepsis model compared to that in the alum control group (Fig. 4D). H70 immunization elicited slightly superior protection to that with CD2, but this was not statistically significant. When protection following immunization with either L460D or H70 was compared with that for the H70+L460D group, significant protection relative to that in the alum placebo was seen for both H70 (P < 0.0001) and H70+L460D (P < 0.0001) but not for L460D alone (Fig. 4D). However, the combination of H70+YLN elicited the strongest protection against strain DBL6A (P < 0.0001) (Fig. 4D) and very strong protection against A66.1 (P < 0.0001) (Fig. 4E).
Multivalent protein vaccine candidates confer protection in a meningitis/sepsis infection model.
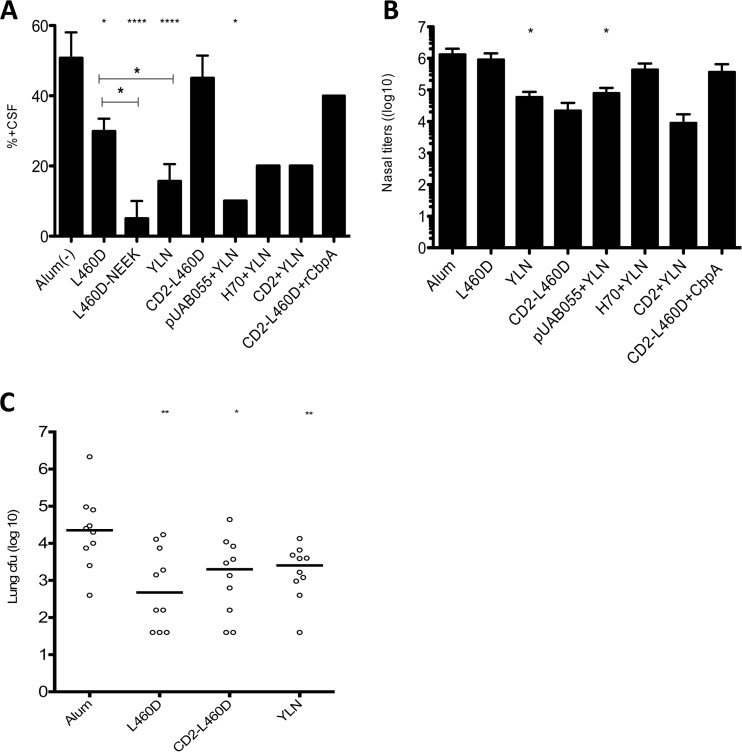
When mice are infected i.t. with strain TIGR4X (serotype 4), disease rapidly progresses from infection of the upper and lower respiratory tract to sepsis and meningitis. The survival time was the longest in mice immunized with combinations of all three antigens, followed by L460D or YLN alone (P < 0.0001) (Fig. 4F). Other groups, such as CD2-L460D, YLN+H70, and YLN+pUAB055, elicited modestly higher protection than that in the alum group. The proportion of mice developing meningitis, indicated by the degree of CFU recovery in cerebrospinal fluid, was significantly lower in mice immunized with L460D alone than in the alum group (P < 0.05). Further significant reductions were seen in groups immunized with L460D-NEEK and YLN (P < 0.05 in both cases), and similar low levels of meningitis were observed in any of the other groups immunized with antigen combinations including YLN (Fig. 5A). The numbers of bacteria in the nasopharynx of survivors at day 14 indicated that only animals immunized with YLN or pUAB055+YLN showed a significant decrease in bacterial load (Fig. 5B).
FIG 5.
Organ-specific analysis of vaccine protection. (A) Percentage of mice infected with strain TIGR4X that developed meningitis in each vaccine group, as determined by positive cerebrospinal fluid (CSF) culture (mean ± standard deviation). (B) Bacterial density in the nasopharynx of mice surviving to day 14 postinfection with TIGR4X (mean ± standard deviation). (C) Focal pneumonia: Mice were challenged with EF3030, and lungs were harvested at day 5 for CFU recovery. The solid lines denote the group medians. For all graphs, significant differences relative to the alum control group are indicated by asterisks (*, P < 0.05; **, P < 0.01; ****, P < 0.0001).
Single-valent and multivalent protein vaccine candidates confer protection against pneumonia.
In a focal pneumonia model (without subsequent progression to sepsis), mice were challenged i.t. with type 19F strain EF3030. After 5 days, mice were euthanized, and blood and lungs were harvested for analysis of CFU recovery. No CFU were recovered from blood of any group. Analysis of CFU recovery from the lungs revealed that immunization with L460D alone (P = 0.0031) or in combination in either the trivalent YLN (P = 0.007) or the divalent CD2-L460D (P = 0.023) constructs afforded significant protection compared to that in the alum control group (Fig. 5C).
DISCUSSION
This, to our knowledge, is one of the most comprehensive studies conducted to identify the best potential protein-based alternative to the current polysaccharide-based pneumococcal vaccines. Here, we describe the testing of new vaccine formulations comprising multiple protein antigens in different challenge models. The protein antigens L460D, PspA, and CbpA used in this study are the best characterized pneumococcal proteins to date in terms of their roles in pathogenicity and protective efficacy. Furthermore, several derivatives of PspA have recently been shown to potentially confer protection with fewer safety concerns (42). It is therefore reasonable to test them in various combinations in order to identify the best formulation of combinations of these protein antigens. The protein antigens L460D, pUAB055, H70, CD2, and YLN were tested singly or in various combinations in models of sepsis, pneumonia, and meningitis using three different mouse backgrounds and six different challenge strains. All combinations were highly immunogenic, with no indication of interference. Fusion constructs elicited titers to individual component antigens that were equal or higher than those of single proteins. Antihemolytic titers against wild-type pneumolysin were equivalent, regardless of construct. Intraperitoneal and subcutaneous routes of immunization were also equivalent.
Pneumolysin has long been recognized as a critical virulence factor in pneumococci, capable of lysing cells by forming membrane pores and interacting with the immune system by activating complement and inducing inflammation (17, 61–63). More recently, pneumolysin has been shown to be involved in evading recognition by pattern recognition receptors in the innate immune system (64, 65). Thus, a pneumolysin toxoid, such as L460D, is a highly desirable vaccine component. To limit the number of separate proteins in the multicomponent product, L460D was used as a backbone for several fusion proteins. The addition of CbpA and PspA derivatives (YPT-L460D-NEEK [YLN] and CD2-L460D) expanded the anti-Ply-mediated protection to the pathogenic processes of adherence, penetration of the blood-brain barrier, and opsonization. In the case of pneumonia, PspA and L460D derivatives were important for protection, while CbpA components played a strong protective role against meningitis. The trivalent combinations CD2+YLN or H70+YLN elicited protection against all six serotypes tested in the challenge models.
A previous study characterized YLN in the i.t. meningitis model and found that immunization with this fusion protected against sepsis, meningitis, pneumonia, and otitis media (54). Passive protection was also examined; mice immunized with anti-YLN rabbit serum were also protected against these diseases. YLN was further shown to provide cross-protection against meningococcal and Haemophilus infection, as the sequence EPRNEEK in the NEEK peptide is able to bind to the same domain of the laminin receptor of the blood-brain barrier as Haemophilus influenzae and Neisseria meningitidis (52).
The PspA peptide CD2 has been examined in the i.v. sepsis model in a prior study (42). As mentioned above, CD2 contains the highly conserved proline-rich region of PspA present in all PspAs and almost all CbpAs. CD2 is advantageous over the N-terminal region of PspA for inclusion in a vaccine, as the CD2 region varies only slightly between PspA and CbpA and elicits cross-reactive antibodies to both PspA and CbpA. Immunization with CD2 or passive immunization with CD2 monoclonal antibodies elicited protection of mice against three challenge strains. Also, upon exposure of children to pneumococci, cross-reactive CD2 antibodies were detected, regardless of the PspA family of the infecting strains (43). In this study, we have shown that in a comparison of H70 and CD2, H70 appeared to more consistently elicit stronger protection than CD2, either singly or in combination with YLN. CD2 and H70 both contain the proline-rich region of PspA, while H70 has the additional SM1 peptide. This SM1 peptide is located just upstream of the proline-rich region of PspA in strain Rx1. Human lactoferrin is known to kill many species of bacteria, including pneumococci, and PspA has been observed to specifically inhibit in vitro killing of pneumococci by apolactoferrin (the iron-free form of lactoferrin); this action is believed to be due to the SM1 peptide that blocks the binding of lactoferrin to PspA (66). Thus, the addition of the SM1 peptide in H70 is likely to have provided enhanced protection over CD2.
Taken together, these results clearly indicate that the trivalent H70+YLN combination is the optimal protein vaccine candidate tested here. It consistently demonstrated significantly broad and strong protection against multiple diseases, such as sepsis, pneumonia, and meningitis, caused by different strains/serotypes in different animal models. We also show that the ability of the L460D antibody to neutralize the cytolytic activity of the wild-type toxin Ply was not compromised by creating fusion constructs that minimize vaccine components.
ACKNOWLEDGMENTS
This work was supported by grants from PATH (to J.C.P., E.I.T., and D.E.B.). E.I.T. was also supported by NIAID R0127913, the Schafer Family Gift, and the American Lebanese Syrian Associated Charities. J.C.P. is a National Health and Medical Research Council of Australia (NHMRC) Senior Principal Research Fellow. D.E.B. and S.K.H. were also supported by NIH grant R01-AI021458. R.K.T. is supported by grant 1R01 AI037657 from the NIH National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2008. 23-valent pneumococcal polysaccharide vaccine. WHO position paper. Wkly Epidemiol Rec 83:373–384. [PubMed] [Google Scholar]
- 3.Weinberger DM, Trzcinski K, Lu YJ, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog 5:e1000476. doi: 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman C, Anderson R. 2014. Review: current and new generation pneumococcal vaccines. J Infect 69:309–325. doi: 10.1016/j.jinf.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Miller E, Andrews NJ, Waight PA, Slack MP, George RC. 2011. Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: an observational cohort study. Lancet Infect Dis 11:760–768. doi: 10.1016/S1473-3099(11)70090-1. [DOI] [PubMed] [Google Scholar]
- 6.Pai R, Moore MR, Pilishvili T, Gertz RE, Whitney CG, Beall B, Active Bacterial Core Surveillance Team. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J Infect Dis 192:1988–1995. doi: 10.1086/498043. [DOI] [PubMed] [Google Scholar]
- 7.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger DM, Malley R, Lipsitch M. 2011. Serotype replacement in disease after pneumococcal vaccination. Lancet 378:1962–1973. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingues CM, Verani JR, Montenegro Renoiner EI, de Cunto Brandileone MC, Flannery B, de Oliveira LH, Santos JB, de Moraes JC, Brazilian Pneumococcal Conjugate Vaccine Effectiveness Study Group. 2014. Effectiveness of ten-valent pneumococcal conjugate vaccine against invasive pneumococcal disease in Brazil: a matched case-control study. Lancet Respir Med 2:464–471. doi: 10.1016/S2213-2600(14)70060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore CE, Paul J, Foster D, Mahar SA, Griffiths D, Knox K, Peto TE, Walker AS, Crook DW, Oxford Invasive Pneumococcal Surveillance Group. 2014. Reduction of invasive pneumococcal disease 3 years after the introduction of the 13-valent conjugate vaccine in the Oxfordshire region of England. J Infect Dis 210:1001–1011. doi: 10.1093/infdis/jiu213. [DOI] [PubMed] [Google Scholar]
- 11.Carter R, Wolf J, van Opijnen T, Muller M, Obert C, Burnham C, Mann B, Li Y, Hayden RT, Pestina T, Persons D, Camilli A, Flynn PM, Tuomanen EI, Rosch JW. 2014. Genomic analyses of pneumococci from children with sickle cell disease expose host-specific bacterial adaptations and deficits in current interventions. Cell Host Microbe 15:587–599. doi: 10.1016/j.chom.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCool TL, Cate TR, Tuomanen EI, Adrian P, Mitchell TJ, Weiser JN. 2003. Serum immunoglobulin G response to candidate vaccine antigens during experimental human pneumococcal colonization. Infect Immun 71:5724–5732. doi: 10.1128/IAI.71.10.5724-5732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rapola S, Jäntti V, Haikala R, Syrjänen R, Carlone GM, Sampson JS, Briles DE, Paton JC, Takala AK, Kilpi TM, Käyhty H. 2000. Natural development of antibodies to pneumococcal surface protein A, pneumococcal surface adhesin A, and pneumolysin in relation to pneumococcal carriage and acute otitis media. J Infect Dis 182:1146–1152. doi: 10.1086/315822. [DOI] [PubMed] [Google Scholar]
- 14.Simell B, Korkeila M, Pursiainen H, Kilpi TM, Käyhty H. 2001. Pneumococcal carriage and otitis media induce salivary antibodies to pneumococcal surface adhesin A, pneumolysin, and pneumococcal surface protein A in children. J Infect Dis 183:887–896. doi: 10.1086/319246. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Choo S, Finn A. 2002. Immune responses to novel pneumococcal proteins pneumolysin, PspA, PsaA, and CbpA in adenoidal B cells from children. Infect Immun 70:5363–5369. doi: 10.1128/IAI.70.10.5363-5369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briles DE, Paton JC, Swiatlo E, Hollingshead S, Crain M. 2006. Pneumococcal vaccines, p 289–298. In Frischetti V, Novick R, Ferretti J, Portnoy D, Rood J (ed), Gram-positive pathogens, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 17.Paton JC. 1996. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol 4:103–106. doi: 10.1016/0966-842X(96)81526-5. [DOI] [PubMed] [Google Scholar]
- 18.Paton JC. 1998. Novel pneumococcal surface proteins: role in virulence and vaccine potential. Trends Microbiol 6:85–87. doi: 10.1016/S0966-842X(98)01220-7. [DOI] [PubMed] [Google Scholar]
- 19.Paton JC. 2004. New pneumococcal vaccines: basic science developments, p 382–402. In Tuomanen EI, Mitchell TJ, Morrison DA, Spratt BG (ed), The pneumococcus. ASM Press, Washington, DC. [Google Scholar]
- 20.Rosenow C, Ryan P, Weiser JN, Johnson S, Fontan P, Ortqvist A, Masure HR. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol 25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 21.Watson DA, Musher DM, Verhoef J. 1995. Pneumococcal virulence factors and host immune responses to them. Eur J Clin Microbiol Infect Dis 14:479–490. doi: 10.1007/BF02113425. [DOI] [PubMed] [Google Scholar]
- 22.Mukerji R, Mirza S, Roche AM, Widener RW, Croney CM, Rhee DK, Weiser JN, Szalai AJ, Briles DE. 2012. Pneumococcal surface protein A inhibits complement deposition on the pneumococcal surface by competing with the binding of C-reactive protein to cell-surface phosphocholine. J Immunol 189:5327–5335. doi: 10.4049/jimmunol.1201967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton JC, Lock RA, Lee CJ, Li JP, Berry AM, Mitchell TJ, Andrew PW, Hansman D, Boulnois GJ. 1991. Purification and immunogenicity of genetically obtained pneumolysin toxoids and their conjugation to Streptococcus pneumoniae type 19F polysaccharide. Infect Immun 59:2297–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander JE, Lock RA, Peeters CC, Poolman JT, Andrew PW, Mitchell TJ, Hansman D, Paton JC. 1994. Immunization of mice with pneumolysin toxoid confers a significant degree of protection against at least nine serotypes of Streptococcus pneumoniae. Infect Immun 62:5683–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogunniyi AD, Folland RL, Briles DE, Hollingshead SK, Paton JC. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect Immun 68:3028–3033. doi: 10.1128/IAI.68.5.3028-3033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton JC. 2007. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect Immun 75:350–357. doi: 10.1128/IAI.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogunniyi AD, Woodrow MC, Poolman JT, Paton JC. 2001. Protection against Streptococcus pneumoniae elicited by immunization with pneumolysin and CbpA. Infect Immun 69:5997–6003. doi: 10.1128/IAI.69.10.5997-6003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis 188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 29.Salha D, Szeto J, Myers L, Claus C, Sheung A, Tang M, Ljutic B, Hanwell D, Ogilvie K, Ming M, Messham B, van den Dobbelsteen G, Hopfer R, Ochs MM, Gallichan S. 2012. Neutralizing antibodies elicited by a novel detoxified pneumolysin derivative, PlyD1, provide protection against both pneumococcal infection and lung injury. Infect Immun 80:2212–2220. doi: 10.1128/IAI.06348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. 2010. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc Natl Acad Sci U S A 107:4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel LS, Yother J, Vijayakumar M, McGarry L, Guild WR, Briles DE. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J Exp Med 165:381–394. doi: 10.1084/jem.165.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu AH, Fulgham RL, McCrory MA, Briles DE, Szalai AJ. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun 67:4720–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaper M, Hollingshead SK, Benjamin WH Jr, Briles DE. 2004. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect Immun 72:5031–5040. doi: 10.1128/IAI.72.9.5031-5040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genschmer KR, Accavitti-Loper MA, Briles DE. 2013. A modified surface killing assay (MSKA) as a functional in vitro assay for identifying protective antibodies against pneumococcal surface protein A (PspA). Vaccine 32:39–47. doi: 10.1016/j.vaccine.2013.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briles DE, King JD, Gray MA, McDaniel LS, Swiatlo E, Benton KA. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858–867. doi: 10.1016/0264-410X(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel LS, Ralph BA, McDaniel DO, Briles DE. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb Pathog 17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 37.McDaniel LS, Sheffield JS, Delucchi P, Briles DE. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun 59:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks-Walter A, Briles DE, Hollingshead SK. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun 67:6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollingshead SK, Becker R, Briles DE. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun 68:5889–5900. doi: 10.1128/IAI.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iannelli F, Oggioni MR, Pozzi G. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63–71. doi: 10.1016/S0378-1119(01)00896-4. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel LS, Sheffield JS, Swiatlo E, Yother J, Crain MJ, Briles DE. 1992. Molecular localization of variable and conserved regions of pspA and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb Pathog 13:261–269. doi: 10.1016/0882-4010(92)90036-N. [DOI] [PubMed] [Google Scholar]
- 42.Daniels CC, Coan P, King J, Hale J, Benton KA, Briles DE, Hollingshead SK. 2010. The proline-rich region of pneumococcal surface proteins A and C contains surface-accessible epitopes common to all pneumococci and elicits antibody-mediated protection against sepsis. Infect Immun 78:2163–2172. doi: 10.1128/IAI.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melin M, Coan P, Hollingshead S. 2012. Development of cross-reactive antibodies to the proline-rich region of pneumococcal surface protein A in children. Vaccine 30:7157–7160. doi: 10.1016/j.vaccine.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Balachandran P, Brooks-Walter A, Virolainen-Julkunen A, Hollingshead SK, Briles DE. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect Immun 70:2526–2534. doi: 10.1128/IAI.70.5.2526-2534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis 190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Masi AW, Barniak V, Mountzouros K, Hostetter MK, Green BA. 2001. Recombinant PhpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infect Immun 69:3827–3836. doi: 10.1128/IAI.69.6.3827-3836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Q, Finkel D, Hostetter MK. 2000. Novel purification scheme and functions for a C3-binding protein from Streptococcus pneumoniae. Biochemistry 39:5450–5457. doi: 10.1021/bi992157d. [DOI] [PubMed] [Google Scholar]
- 48.Dave S, Brooks-Walter A, Pangburn MK, McDaniel LS. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect Immun 69:3435–3437. doi: 10.1128/IAI.69.5.3435-3437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammerschmidt S, Talay SR, Brandtzaeg P, Chhatwal GS. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol 25:1113–1124. doi: 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 50.Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem 275:37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 51.Jarva H, Janulczyk R, Hellwage J, Zipfel PF, Björck L, Meri S. 2002. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8–11 of factor H. J Immunol 168:1886–1894. doi: 10.4049/jimmunol.168.4.1886. [DOI] [PubMed] [Google Scholar]
- 52.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala'Aldeen DA, Tuomanen EI. 2009. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest 119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo R, Mann B, Lewis WS, Rowe A, Heath R, Stewart ML, Hamburger AE, Sivakolundu S, Lacy ER, Bjorkman PJ, Tuomanen E, Kriwacki RW. 2005. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J 24:34–43. doi: 10.1038/sj.emboj.7600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mann B, Thornton J, Heath R, Wade KR, Tweten RK, Gao G, El Kasmi K, Jordan JB, Mitrea DM, Kriwacki R, Maisonneuve J, Alderson M, Tuomanen EI. 2014. Broadly protective protein-based pneumococcal vaccine composed of pneumolysin toxoid-CbpA peptide recombinant fusion protein. J Infect Dis 209:1116–1125. doi: 10.1093/infdis/jit502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harvey RM, Ogunniyi AD, Chen AY, Paton JC. 2011. Pneumolysin with low hemolytic activity confers an early growth advantage to Streptococcus pneumoniae in the blood. Infect Immun 79:4122–4130. doi: 10.1128/IAI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harvey RM, Hughes CE, Paton AW, Trappetti C, Tweten RK, Paton JC. 2014. The impact of pneumolysin on the macrophage response to Streptococcus pneumoniae is strain-dependent. PLoS One 9:e103625. doi: 10.1371/journal.pone.0103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Health and Medical Research Council. 2004. Australian code of practice for the care and use of animals for scientific purposes, 7th ed Australian Government, National Health and Medical Research Council, Canberra, Australian Capital Territory, Australia: https://www.nhmrc.gov.au/_files_nhmrc/publications/attachments/ea16_animal_code.pdf. [Google Scholar]
- 58.Parliament of South Australia. 1985. Animal Welfare Act 1985. Parliament of South Australia, Adelaide, South Australia, Australia. [Google Scholar]
- 59.Jalonen E, Paton JC, Koskela M, Kerttula Y, Leinonen M. 1989. Measurement of antibody responses to pneumolysin–a promising method for the presumptive aetiological diagnosis of pneumococcal pneumonia. J Infect 19:127–134. doi: 10.1016/S0163-4453(89)91864-1. [DOI] [PubMed] [Google Scholar]
- 60.Paton JC, Lock RA, Hansman DJ. 1983. Effect of immunization with pneumolysin on survival time of mice challenged with Streptococcus pneumoniae. Infect Immun 40:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Braun JS, Novak R, Gao G, Murray PJ, Shenep JL. 1999. Pneumolysin, a protein toxin of Streptococcus pneumoniae, induces nitric oxide production from macrophages. Infect Immun 67:3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cockeran R, Theron AJ, Steel HC, Matlola NM, Mitchell TJ, Feldman C, Anderson R. 2001. Proinflammatory interactions of pneumolysin with human neutrophils. J Infect Dis 183:604–611. doi: 10.1086/318536. [DOI] [PubMed] [Google Scholar]
- 63.Kadioglu A, Gingles NA, Grattan K, Kerr A, Mitchell TJ, Andrew PW. 2000. Host cellular immune response to pneumococcal lung infection in mice. Infect Immun 68:492–501. doi: 10.1128/IAI.68.2.492-501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marriott HM, Mitchell TJ, Dockrell DH. 2008. Pneumolysin: a double-edged sword during the host-pathogen interaction. Curr Mol Med 8:497–509. doi: 10.2174/156652408785747924. [DOI] [PubMed] [Google Scholar]
- 65.Witzenrath M, Pache F, Lorenz D, Koppe U, Gutbier B, Tabeling C, Reppe K, Meixenberger K, Dorhoi A, Ma J, Holmes A, Trendelenburg G, Heimesaat MM, Bereswill S, van der Linden M, Tschopp J, Mitchell TJ, Suttorp N, Opitz B. 2011. The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. J Immunol 187:434–440. doi: 10.4049/jimmunol.1003143. [DOI] [PubMed] [Google Scholar]
- 66.Mirza S, Wilson L, Benjamin WH Jr, Novak J, Barnes S, Hollingshead SK, Briles DE. 2011. Serine protease PrtA from Streptococcus pneumoniae plays a role in the killing of S. pneumoniae by apolactoferrin. Infect Immun 79:2440–2450. doi: 10.1128/IAI.00489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]