Abstract
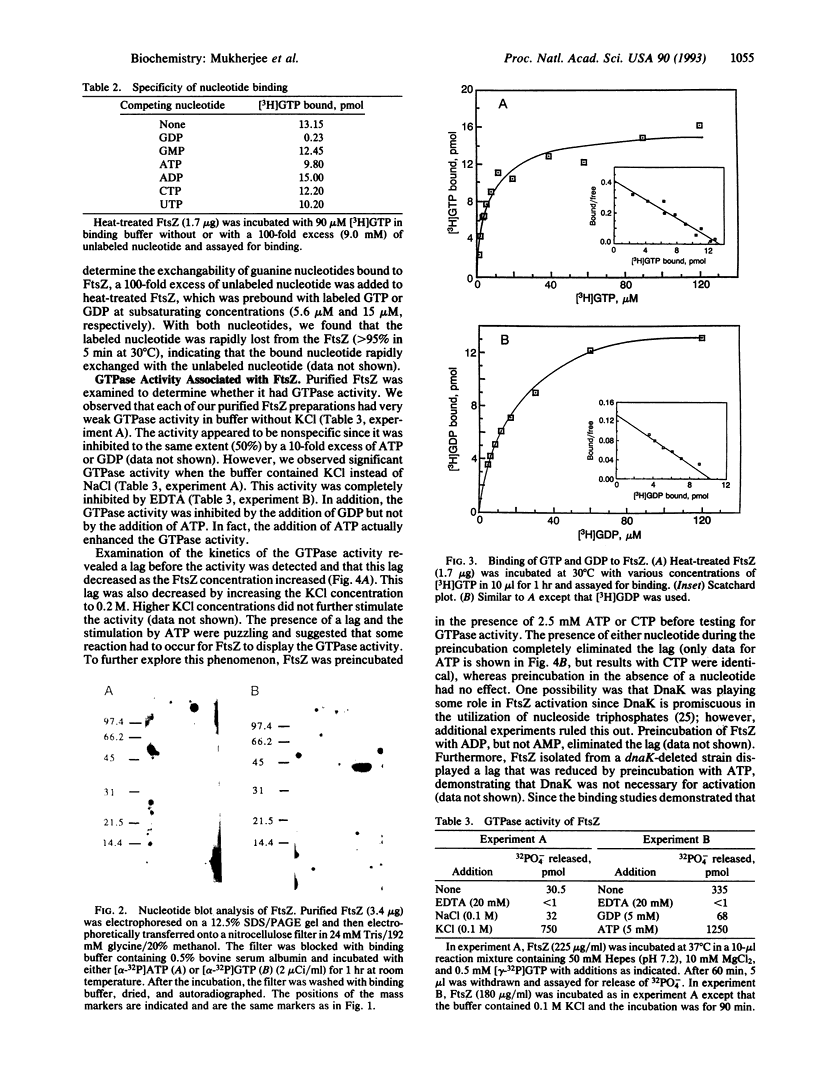
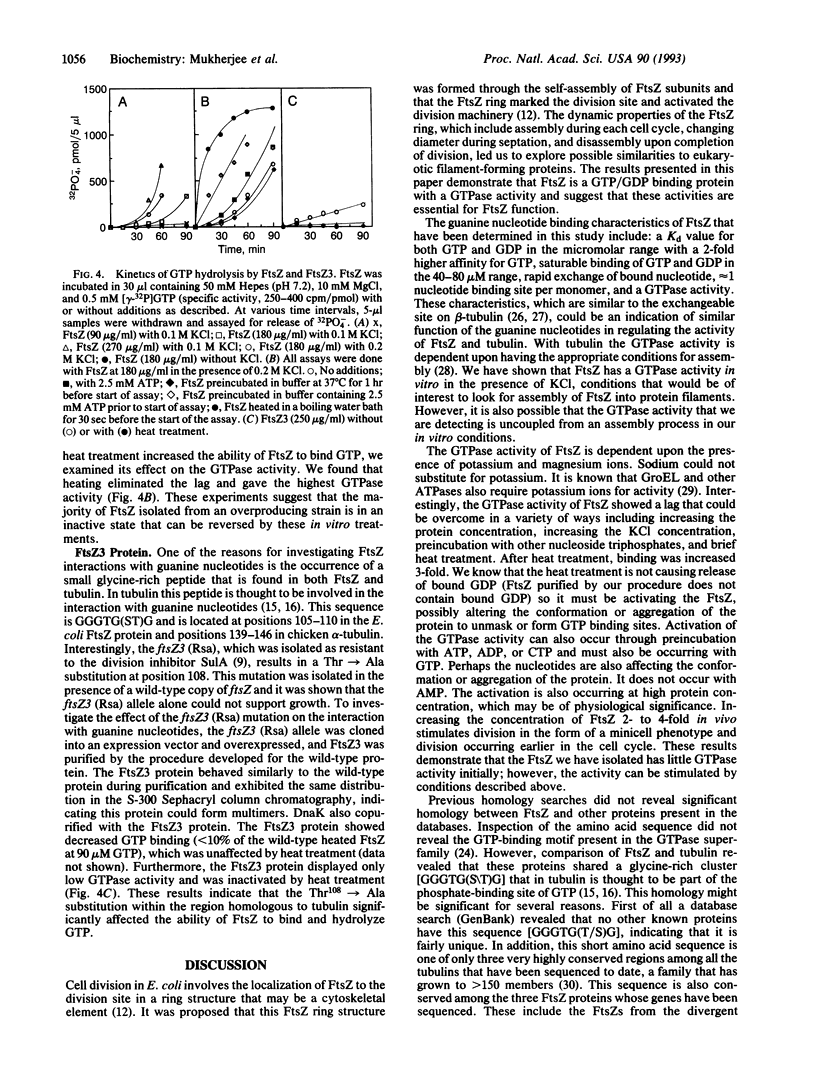
FtsZ is an essential cell division protein in Escherichia coli that forms a ring structure at the division site under cell cycle control. The dynamic nature of the FtsZ ring suggests possible similarities to eukaryotic filament forming proteins such as tubulin. In this study we have determined that FtsZ is a GTP/GDP binding protein with GTPase activity. A short segment of FtsZ is homologous to a segment in tubulin believed to be involved in the interaction between tubulin and guanine nucleotides. A lethal ftsZ mutation, ftsZ3 (Rsa), that leads to an amino acid alteration in this homologous segment decreased GTP binding and hydrolysis, suggesting that interaction with GTP is essential for ftsZ function.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beall B., Lowe M., Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988 Oct;170(10):4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991 Nov 14;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J Bacteriol. 1990 Oct;172(10):5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990 Oct;172(10):5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol. 1992 Aug;174(16):5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989 May;171(5):2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns R. G. Alpha-, beta-, and gamma-tubulins: sequence comparisons and structural constraints. Cell Motil Cytoskeleton. 1991;20(3):181–189. doi: 10.1002/cm.970200302. [DOI] [PubMed] [Google Scholar]
- Carlier M. F. Guanosine-5'-triphosphate hydrolysis and tubulin polymerization. Review article. Mol Cell Biochem. 1982 Sep 3;47(2):97–113. doi: 10.1007/BF00234410. [DOI] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991 Jun;173(11):3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Geahlen R. L., Haley B. E. Use of a GTP photoaffinity probe to resolve aspects of the mechanism of tubulin polymerization. J Biol Chem. 1979 Dec 10;254(23):11982–11987. [PubMed] [Google Scholar]
- Hesse J., Thierauf M., Ponstingl H. Tubulin sequence region beta 155-174 is involved in binding exchangeable guanosine triphosphate. J Biol Chem. 1987 Nov 15;262(32):15472–15475. [PubMed] [Google Scholar]
- Jacobs M., Smith H., Taylor E. W. Tublin: nucleotide binding and enzymic activity. J Mol Biol. 1974 Nov 5;89(3):455–468. doi: 10.1016/0022-2836(74)90475-6. [DOI] [PubMed] [Google Scholar]
- Korn E. D., Carlier M. F., Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987 Oct 30;238(4827):638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Linse K., Mandelkow E. M. The GTP-binding peptide of beta-tubulin. Localization by direct photoaffinity labeling and comparison with nucleotide-binding proteins. J Biol Chem. 1988 Oct 15;263(29):15205–15210. [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V., Bekesi E., Kung H. F. Ha-ras proteins exhibit GTPase activity: point mutations that activate Ha-ras gene products result in decreased GTPase activity. Proc Natl Acad Sci U S A. 1985 Jan;82(2):376–380. doi: 10.1073/pnas.82.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V., Yamazaki S., Kung H. F. Guanosine nucleotide binding by highly purified Ha-ras-encoded p21 protein produced in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6953–6957. doi: 10.1073/pnas.81.22.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W., Corbo J. C., Long S. R. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991 Sep;173(18):5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla J., Sánchez M., Palacios P., Vicente M., Aldea M. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol Microbiol. 1991 Jul;5(7):1681–1686. doi: 10.1111/j.1365-2958.1991.tb01915.x. [DOI] [PubMed] [Google Scholar]
- Schmitt H. D., Wagner P., Pfaff E., Gallwitz D. The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell. 1986 Nov 7;47(3):401–412. doi: 10.1016/0092-8674(86)90597-0. [DOI] [PubMed] [Google Scholar]
- Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991 Oct 18;67(2):239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Siekierka J., Manne V., Mauser L., Ochoa S. Polypeptide chain initiation in eukaryotes: reversibility of the ternary complex-forming reaction. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1232–1235. doi: 10.1073/pnas.80.5.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht H., Yaffe M. B., Farr G. W. A model of the nucleotide-binding site in tubulin. FEBS Lett. 1987 Apr 20;214(2):226–235. doi: 10.1016/0014-5793(87)80061-3. [DOI] [PubMed] [Google Scholar]
- Taschner P. E., Huls P. G., Pas E., Woldringh C. L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988 Apr;170(4):1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétart F., Bouché J. P. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol. 1992 Mar;6(5):615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Lubben T. H., Reed J., Goloubinoff P., O'Keefe D. P., Lorimer G. H. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990 Jun 19;29(24):5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- Wang X. D., de Boer P. A., Rothfield L. I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991 Nov;10(11):3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg R. C. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972 Sep 22;177(4054):1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36(3):241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- Zylicz M., LeBowitz J. H., McMacken R., Georgopoulos C. The dnaK protein of Escherichia coli possesses an ATPase and autophosphorylating activity and is essential in an in vitro DNA replication system. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6431–6435. doi: 10.1073/pnas.80.21.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]





