Abstract
Background: The fecal microbiota has been characterized in some adult populations, but little is known about its community structure during lactation.
Objectives: We characterized the maternal fecal microbiome during lactation and explored possible mediating factors such as nutrition.
Methods: Fecal samples were collected from 20 lactating women from 2 d to 6 mo postpartum, and bacterial taxa were characterized with the use of high-throughput sequencing. Bacterial community structure (at each taxonomic level) and relations between bacterial taxa and environmental and dietary variables were visualized and analyzed with the use of stacked bar charts, principal component analysis, and multivariate analyses such as nonmetric multidimensional scaling and canonical correlation analysis.
Results: Complex bacterial community structure was somewhat similar to those previously published for other adult populations (although there were some notable differences), and there were no clear associations with time postpartum or anthropometric or environmental variables. However, Spearman rank correlations suggested that increased intake of pantothenic acid, riboflavin, vitamin B-6, and vitamin B-12 were related to increased relative abundance of Prevotella (r = 0.45, 0.39, 0.34, and 0.24, respectively; P ≤ 0.01) and decreased relative abundance of Bacteroides (r = −0.55, −0.46, −0.32, and −0.35, respectively; P ≤ 0.01). Intakes of copper, magnesium, manganese, and molybdenum were positively associated with Firmicutes (r = 0.33, 0.38, 0.44, and 0.51, respectively; P ≤ 0.01) and negatively associated with Bacteroidetes (r = −0.38, −0.44, −0.48, and −0.53, respectively; P ≤ 0.01). Overall, data consistently suggest that increased consumption of a more nutrient- and calorie-rich diet was positively associated with relative abundance of Firmicutes.
Conclusions: The fecal microbiome of lactating women is relatively stable in the postpartum period and somewhat similar to that of other adult populations. Variation in dietary constituents may be related to that of relative abundance of individual bacterial taxa. Controlled dietary intervention studies will be required to determine whether these associations are causal in nature.
Keywords: feces, gastrointestinal, lactation, maternal, microbiome, microbiota, nutrients
Introduction
Although experts have long known that the gastrointestinal tract contains myriad bacterial taxa, recent technologic advances have led to a heightened interest in and understanding of how variation in this ecosystem influences and may be influenced by health. For example, twin studies coupled with experimental animal data suggest that a malnourished child’s risk of developing kwashiorkor might be causally related to gastrointestinal microbial community structure (1). The fecal microbiota is also quite different among vegans, vegetarians, and omnivores (2), and variation has been linked to irritable bowel syndrome (3), Crohn disease (4), and type 2 diabetes (5, 6). As such, understanding what is normal in terms of gastrointestinal microbial communities, what constitutes a dysbiosis or disease state, and factors related to variation in community structure of gastrointestinal microbes may afford important information related to optimal health (7).
One area of particular interest is understanding the origin of gastrointestinal microbes (8). Although researchers have historically thought that the infant’s first bacterial inoculation occurs during or soon after birth, newborns are actually first exposed to bacteria in utero (9, 10); these bacteria originate at least in part from the mother’s gastrointestinal tract (11). Whereas an abundance of literature supports the importance of the vaginal microbiota, delivery mode, and antibiotics on the infant’s early gastrointestinal microbiota (12), human milk also delivers a rich diversity of bacteria to the infant’s gastrointestinal tract (13–18). As with the bacteria in the mother’s uterus during pregnancy, these milk-borne bacteria likely originate at least in part from the mother’s gastrointestinal tract (19). Hence, characterizing and understanding factors that shape a woman’s gastrointestinal microbiome during her reproductive years may lend insight as to not only how they influence her own health, but also that of her offspring.
Several research groups have begun to document the maternal gastrointestinal microbiota during pregnancy and lactation (18, 20). One factor that might impact a woman’s fecal microbiome is diet. In relation to energy balance, Ley et al. (21) and others (22) have provided evidence that gastrointestinal bacterial communities of obese individuals have relatively higher concentrations of bacteria in the Firmicutes phylum. Conversely, Collado et al. (23) reported no difference in Clostridium histolyticum group bacteria (members of the Firmicutes phylum) between normal-weight and obese pregnant women, and increased concentrations of Bacteroides–Prevotella group bacteria (members of the Bacteroidetes phylum) in obese compared with normal-weight pregnant women. Data from the Human Microbiome Project (24), however, do not suggest a strong relation between BMI and microbial community structure. Nonetheless, there is reason to believe that acute or long-term nutritional status may impact gastrointestinal microbial ecology, at least in some situations. For instance, there may be an inverse correlation between relative abundance of Firmicutes and fat intake and a positive correlation with fiber intake (25). However, there remains much to learn concerning the nature of the relation between dietary choices and the gastrointestinal microbiome. Moreover, to our knowledge, this relation has not been explored during lactation.
The primary objectives of this study were 2-fold: to characterize the gastrointestinal microbiome of healthy lactating US women and to investigate possible relations between nutritional status (as assessed with the use of BMI and dietary intake variables) and variation in the gastrointestinal microbiome in this population. In general, we hypothesized that the gastrointestinal microbiota of healthy breastfeeding women would be similar to those of other previously characterized healthy adult populations. We also tested several hypotheses based on previously published studies investigating nutrient–microbe associations in nonlactating subjects. For instance, we hypothesized that BMI would be positively associated with the relative abundance of Firmicutes and negatively associated with that of Bacteroidetes. In addition, we hypothesized that carbohydrate intake would be positively associated with relative abundance of Prevotella and inversely associated with that of Bacteroides, dietary fat intake would be positively associated with relative abundance of Bacteroides and negatively associated with that of Prevotella, and saturated fat intake would be positively associated with relative abundance of Bacteroides and Parabacteroides. We also conducted extensive hypothesis-generating, exploratory analyses to appraise other possible relations between dietary intake variables and gastrointestinal microbial populations of healthy lactating women.
Methods
Subjects and study design.
This was a prospective, longitudinal investigation of 20 self-reported healthy breastfeeding women who, in their third trimester of pregnancy, were recruited from the Pullman, Washington, and Moscow, Idaho area. Written informed consent was obtained in accordance with procedures approved by the Washington State University and the University of Idaho institutional review boards. Samples and data were collected on 2, 5, and 10 d (± 1 d), and 1, 2, 3, 4, 5, and 6 mo (± 1 d) postpartum. BMI was considered in 2 ways: prepregnancy BMI (as reported by each subject at enrollment) and current BMI (as measured at each sampling period). Each woman was classified as either normal weight (<25 kg/m2) or overweight/obese (≥25 kg/m2).
Sample collection.
Maternal fecal samples were collected at each time point either at the subject’s home or at Washington State University or the University of Idaho. A tissue wipe was placed by the subject into a sterile plastic bag and stored immediately at –80°C (when collected at a university site) or in a home freezer until able to be transferred to a −80°C freezer (when collected at home).
Maternal dietary records.
With the assistance of trained study personnel, a comprehensive, quantitative 24 h dietary recall was completed for each subject at each time point. All foods and beverages (but not dietary supplements) were recorded and included in the analysis. Diet records were entered into and energy, macronutrient intake, and selected micronutrient intake estimated with the use of Genesis R&D version 7.6 software (ESHA Research).
Extraction and amplification of bacterial DNA.
DNA was extracted from ∼0.05 g feces with the use of a QIAamp DNA Stool Mini Kit (QIAGEN Catalog no. 51504), following the manufacturer’s protocol. A solution of TE 50 (10 mM Tris-HCl and 50 mM EDTA, pH 8; 500 μL) was used as a negative control. Extracted DNA was eluted in AE buffer and stored at −80°C until further analysis. A dual-barcoded 2-step PCR was conducted to amplify the V1–V3 hypervariable region of the bacterial 16S rRNA gene; a 7-fold degenerate forward primer targeting position 27F and a reverse primer targeting position 534R were used. Primer sequences are provided in Supplemental Table 1.
DNA was amplified in a dedicated PCR hood, and the PCR mixture (50 μL) contained the following: 0.05 μM primers (Integrated DNA Technologies), 1 μL DNA extract, and a PCR master mix containing a final concentration of 1× PCR buffer (Life Technologies), 3.12 mM MgCl2 (Life Technologies), 0.24 g/L BSA (Sigma), 0.2 mM deoxyribonucleotide triphosphate (Life Technologies), and 25 U/mL AmpliTaq DNA 360 polymerase (Life Technologies). PCRs were conducted with the use of an Applied Biosystems 2720, Veriti, or ProFlex model thermocycler under the following conditions: 95°C for 2 min, then 95°C for 1 min, 51°C for 1 min, and 72°C for 1 min for 30 cycles, and then a final extension step of 72°C for 10 min. Samples were held at 4°C in the thermocycler until being stored at −20°C.
Products from the first PCR were electrophoresed on 1% agarose gels made with tris-acetate-ethylenediamine tetraacetic acid (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) buffer and containing ethidium bromide (0.0007 g/L). Gels were allowed to run for 30 min at 80 V, and bands were viewed with the use of a BioRad UltraCam Digital Imaging System. Samples with high-quality amplicons (a relatively bright band of interest at ∼534 bp), low primer-dimers, and absence of unwanted bands or smears were deemed acceptable for the second PCR reaction. PCR products were diluted (1:14) with nuclease-free water and subjected to a second round of PCR in a reaction mix containing 0.75 μL primers with dual-index barcodes and Illumina sequencing adapters (University of Idaho Institute for Bioinformatics and Evolutionary Studies Genomics Resources Core Facility). The quality of the second PCR amplicons was evaluated with the use of a QIAxcel DNA screening cartridge (QIAGEN), and DNA quantified with the use of PicoGreen (Life Technologies).
An appropriate volume of each amplicon (containing 50 ng DNA) was pooled to create a composite sample for high-throughput sequencing. Amplicon pools were size-selected with the use of AMPure beads. The cleaned amplicon pool was quantified with the use of a KAPA Illumina library quantification kit (KAPA Biosciences) and the Applied Biosystems StepOne Plus real-time PCR system. Sequences were obtained with the use of an Illumina MiSeq v3 paired-end 300 bp protocol for 600 cycles.
Sequence analysis.
Raw DNA sequence reads were processed with the use of the python application dbcAmplicons (26). The dbcAmplicons application was designed to process Illumina double-barcoded amplicons generated in the manner described above. Specifically, the application performs processing of raw reads (preprocessing), joining of overlapping paired reads into longer single reads, classification of reads to the genus level, and generation of abundance tables. During preprocessing, barcodes were allowed to have ≤1 mismatch (Hamming distance), and primers were allowed to have ≤4 mismatches (Levenshtein distance), as long as the final 4 bases of the primer perfectly matched the target sequence. Reads identified as not having a corresponding barcode and primer sequence were discarded. dbcAmplicons uses a modified version of FLASH (27) to join overlapping reads and the Ribosomal Database Project Bayesian classifier (28) to assign sequences to phylotypes. Reads were assigned to the first Ribosomal Database Project taxonomic level with a bootstrap value ≥50%.
Characterization of bacterial community composition and diversity.
Sequence counts were standardized and converted to relative abundance values with the use of the decostand function in R (vegan package, version 0.98.945). Alpha and β diversity indexes, including richness, Shannon diversity index, inverse Simpson index, Shannon evenness, Simpson evenness, and Pielou evenness, were calculated in R (29).
Spearman rank correlation analysis.
To visualize and characterize simple associations present between bacterial taxa and nutritional status (BMI or nutrient intake as continuous variables), heat maps of Spearman rank correlation coefficients were constructed with the use of the vegan and gplots packages in R. To avoid possible influences of both the birthing process and the introduction of supplementary foods to the infant, only data collected from 1 to 3 mo postpartum were included in this portion of the analysis. To help control for multiple comparisons, associations were deemed significant in this discovery phase of the analysis at P ≤ 0.01; however, because of the exploratory nature of this analysis, we also denote weaker trends at P ≤ 0.05.
Exploratory multivariate analysis.
All further exploratory and statistical analyses were performed in SAS version 9.3. Principal component analysis (PCA)9 and nonmetric multidimensional scaling (NMDS) analysis were carried out to examine patterns and similarity among complex bacterial community structures. Variables including relative abundance of top bacteria and diversity indexes were analyzed with the use of the PRINCOMP. Relative bacterial abundance was double–square-root transformed if needed before NMDS was performed. Selected metadata (e.g., birth mode and dietary variables) were used to enhance both the resulting PCA and NMDS analysis plots in order to visually assess potential associations and groupings.
Inferential multivariate statistics.
To examine potential relations between relative bacterial abundance and dietary (quartiles of intake) with anthropometric measures, canonical correlation analyses were performed with the use of the SAS CANCORR procedure. As part of this analysis, standardized canonical coefficients of relevant axes were visualized over each of the 9 time points to examine potential changes in the contribution of response to the canonical components. To address unequal and low sample numbers at each time point, as well as increase the statistical power, bacterial abundance data were grouped together over time into 3 possible scenarios. These included the following: 1) days 2–5, day 10 and month 1, months 2–3, and months 4–6; 2) days 2–10, months 1–3, and months 4–6; and 3) days 2–10 and months 1–9. The groups in each scenario were subsequently subjected to multivariate analysis of variance (MANOVA) (SAS GLM) to examine effects and complex associated interactions of time, bacterial community structure, and nutrient intake data.
Additional analyses to relate selected metadata (e.g., birth mode and dietary variables) to variation in complex microbial community structure between 1 and 3 mo postpartum were assessed with the use of ANOVA and MANOVA as appropriate. The dependent variables included in these models were the proportions of the top 4 most abundant phyla (Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria) and/or diversity indexes discussed previously; independent variables were categorical metadata such as subject, parity, birth mode, and classified nutritional data.
Results
Subject description and final sample deposition.
Basic anthropometric and reproductive history information of all 20 women at enrollment is provided in Table 1, and subject weights at each time point are provided in Table 2. In general, women were ∼31 y old, weighed 64 kg, and delivered vaginally in a hospital. All infants were exclusively breastfed for at least 4 mo. Although fecal samples were collected from most women at each time point, several fecal samples were not used in the subsequent analyses. For instance, 20 samples had been collected from women who reported antibiotic use in the time elapsed since the previous sample was collected; these samples were not used in the final analysis because we wished to characterize fecal bacterial communities of “healthy” women. Other samples (n = 16) were excluded because of an insufficient electrophoresis band after DNA amplification. In the end, a total of 120 fecal samples were used for this study: 10, 15, 15, 15, 16, 17, 12, 11, and 9 samples at days 2, 5, and 10 and months 1, 2, 3, 4, 5, and 6, respectively.
TABLE 1.
Anthropometric and descriptive variables of the lactating women participating in this study1
| Variable | Value |
| Age, y | 31 ± 3 (25–37) |
| Height, cm | 168 ± 12 (125–181) |
| Prepregnancy weight, kg | 64 ± 7 (53–77) |
| Prepregnancy BMI, kg/m2 | 23.5 ± 3.3 (19.3–34.5) |
| Birth method | |
| Vaginal | 16 |
| Cesarean | 4 |
| Birth location | |
| Hospital | 18 |
| Home | 2 |
| Parity | |
| 1 | 7 |
| 2 | 10 |
| 3 | 2 |
| 4 | 1 |
Values are means ± SDs (ranges) or n; total n = 20 women.
TABLE 2.
Body weights, energy intake, macronutrient intakes, and selected micronutrient intakes from 2 d to 6 mo postpartum in healthy lactating women1
| Variable | Day 2 (n = 13) | Day 5 (n = 18) | Day 10 (n = 17) | 1 mo (n = 17) | 2 mo (n = 18) | 3 mo (n = 18) | 4 mo (n = 15) | 5 mo (n = 14) | 6 mo (n = 13) | Effect of time (P) |
| Body weight, kg | 73 ± 12 (57–99) | 72 ± 9 (58–92) | 69 ± 7 (57–81) | 72 ± 9 (54–90) | 70 ± 10 (51–92) | 68 ± 9 (50–90) | 69 ± 11 (48–89) | 66 ± 11 (48–88) | 66 ± 11 (47–89) | 0.34 |
| Energy, kcal/d | 2680 ± 790a,b (1400–4170) | 2870 ± 630a (2170–4200) | 2490 ± 760a,b (920–4370) | 2180 ± 710b,c (1190–4000) | 2180 ± 750b,c (1010–3460) | 2240 ± 630b,c (1097–3341) | 2160 ± 636b,c (1395–3185) | 1852 ± 406c (1355–2473) | 1946 ± 325b,c (1504–2478) | 0.008 |
| Energy from fat, kcal/d | 843 ± 488 (273–2100) | 893 ± 242 (526–1260) | 777 ± 275 (203–1200) | 750 ± 242 (285–2040) | 768 ± 345 (147–1550) | 8253 ± 335 (242–1592) | 773 ± 310 (441–1242) | 581 ± 200 (310–951) | 726 ± 281 (420–1155) | 0.56 |
| Energy from carbohydrates, kcal/d | 1183 ± 409 (694–1895) | 1579 ± 515 (972–2670) | 1385 ± 531 (624–2890) | 1112 ± 279 (607–1447) | 1088 ± 417 (306–1870) | 1045 ± 336 (506–2080) | 1099 ± 317 (677–1710) | 974 ± 267 (641–1497) | 916 ± 159 (709–1102) | 0.24 |
| Energy from protein, kcal/d | 393 ± 135 (214–621) | 433 ± 87 (305–584) | 394 ± 116 (124–569) | 349 ± 133 (212–634) | 381 ± 196 (124–880) | 431 ± 187 (149–925) | 324 ± 112 (161–513) | 327 ± 91 (195–533) | 316 ± 103 (190–514) | 0.21 |
| Protein, g/d | 99 ± 34 (53–155) | 108 ± 22 (76–146) | 99 ± 29 (31–142) | 87 ± 33 (53–159) | 95 ± 49 (31–220) | 108 ± 47 (37–231) | 81 ± 28 (40–128) | 82 ± 23 (49–133) | 79 ± 26 (47–128) | 0.21 |
| Carbohydrates, g/d | 296 ± 102b,c (173–474) | 395 ± 129a (243–667) | 346 ± 133a,b (156–722) | 278 ± 70b,c (152–362) | 272 ± 104c (76–468) | 261 ± 84c (126–520) | 275 ± 79b,c (169–427) | 243 ± 67c (160–374) | 229 ± 40c (177–276) | 0.0004 |
| Fiber, g/d | 25 ± 11 (10–43) | 33 ± 15 (16–60) | 27 ± 12 (9–56) | 27 ± 10 (6–48) | 24 ± 14 (9–54) | 25 ± 15 (10–59) | 27 ± 11 (11–53) | 25 ± 7 (13–34) | 23 ± 9 (9–39) | 0.71 |
| Insoluble fiber, g/d | 5 ± 4 (0–13) | 7 ± 6 (1–20) | 9 ± 6 (0–23) | 9 ± 7 (0–20) | 5 ± 3 (0–12) | 6 ± 6 (0–21) | 9 ± 7 (0–27) | 6 ± 4 (0–15) | 4 ± 5 (0–15) | 0.30 |
| Soluble fiber, g/d | 3 ± 4 (0–12) | 3 ± 2 (0–7) | 4 ± 3 (0–11) | 3 ± 3 (0–7) | 2 ± 1 (0–5) | 3 ± 4 (0–14) | 3 ± 2 (0–6) | 3 ± 4 (0–13) | 2 ± 2 (0–5) | 0.55 |
| Fat, g/d | 94 ± 54 (30–233) | 99 ± 27 (58–140) | 86 ± 31 (23–133) | 83 ± 47 (32–227) | 85 ± 38 (16–172) | 92 ± 37 (27–177) | 86 ± 34 (46–138) | 65 ± 22 (35–106) | 81 ± 31 (47–128) | 0.56 |
| Saturated fat, g/d | 24 ± 22 (11–88) | 31 ± 13 (21–68) | 29 ± 11 (8–55) | 28 ± 16 (14–71) | 29 ± 17 (2–55) | 31 ± 16 (4–75) | 29 ± 19 (7–61) | 20 ± 7 (7–28) | 24 ± 10 (10–44) | 0.31 |
| Polyunsaturated fat, g/d | 16 ± 7 (2–21) | 13 ± 7 (4–23) | 11 ± 7 (1–27) | 11 ± 15 (0–63) | 8 ± 4 (2–16) | 12 ± 12 (2–54) | 12 ± 5 (6–22) | 10 ± 9 (1–29) | 11 ± 7 (3–23) | 0.96 |
| Monounsaturated fat, g/d | 34 ± 22 (11–88) | 36 ± 14 (16–68) | 29 ± 11 (8–55) | 28 ± 16 (14–71) | 29 ± 17 (2–55) | 31 ± 16 (4–75) | 29 ± 19 (7–61) | 20 ± 7 (7–28) | 24 ± 10 (10–44) | 0.76 |
| Calcium, g/d | 1.06 ± 0.04 (0.36–1.76) | 1.35 ± 0.61 (0.43–2.88) | 1.28 ± 0.53 (0.43–2.15) | 1.21 ± 0.54 (0.42–2.19) | 0.83 ± 0.51 (0.24–1.90) | 1176 ± 611 (272–2686) | 1120 ± 584 (403–2244) | 1028 ± 491 (370–2091) | 790 ± 299 (317–1132) | 0.12 |
| Iron, mg/d | 16.5 ± 6.4 (7.7–24.8) | 21.5 ± 10.4 (12.8–52.1) | 35.6 ± 57.4 (8.2–238.7) | 17.2 ± 10.1 (3.1–41.0) | 15.8 ± 7.2 (7.1–32.6) | 19.6–14.3 (8.1–58.9) | 19.1 ± 11.0 (9.2–46.6) | 25.4 ± 26.1 (6.6–78.0) | 12.1 ± 3.2 (7.9–17.2) | 0.35 |
| Vitamin B-12, μg/d | 4.7 ± 3.1 (1.3–13.0) | 4.6 ± 2.6 (0.2–9.0) | 5.0 ± 2.7 (0.5–10.4) | 4.2 ± 3.7 (0.4–13.6) | 3.4 ± 3.3 (0.1–13.3) | 5.5 ± 4.8 (0.0–18.4) | 4.9 ± 3.6 (0.3–12.1) | 7.9 ± 8.7 (0.3–29.4) | 3.6 ± 3.5 (0.1–12.2) | 0.34 |
Values are means ± SEMs (ranges); n = 20 healthy lactating women at each time point. Labeled means in a row without a common letter differ, P < 0.05.
Fecal bacterial community structure and diversity (all time points).
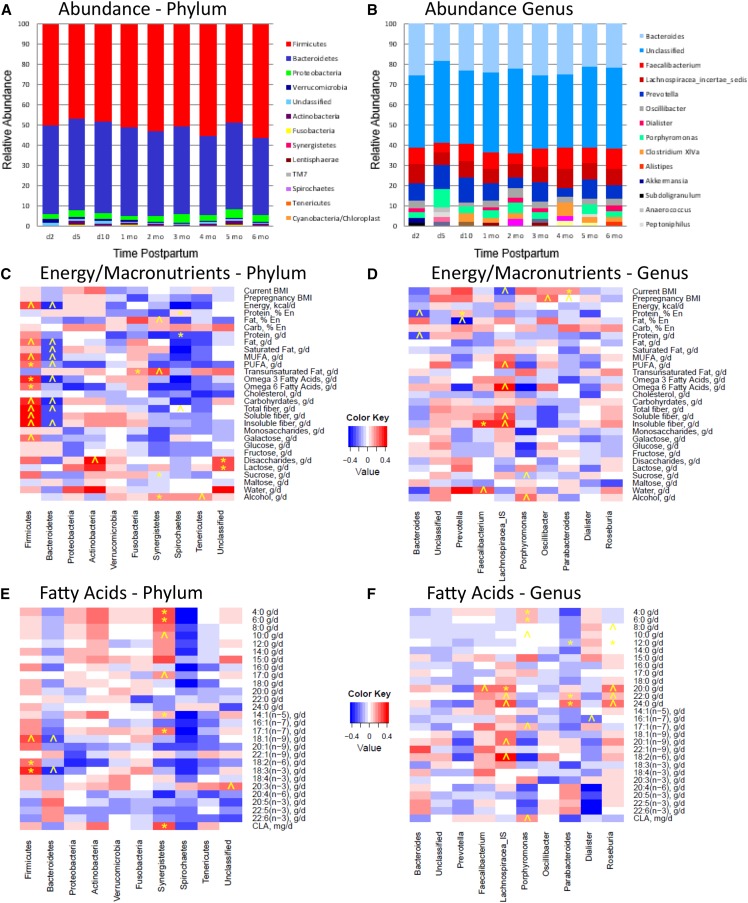
Stacked bar charts illustrating the relative distributions of the 10 most abundant bacterial groups at each time point for phylum and genus levels are shown in Figure 1 (additional charts illustrating the 10 most abundant members of the other taxonomic levels are found in Supplemental Figures 1–3). In general, members of the Firmicutes and Bacteroidetes phyla were most abundant, followed by lesser amounts of Proteobacteria, Verrucomicrobia, unclassified bacteria, and Actinobacteria. At the genus level, bacterial communities were dominated by unclassified bacteria followed by Bacteroides, Faecalibacterium, Lachnospiracea incertae sedis (denoted as Lachnospiracea unclassified in some figures), and Prevotella. Diversity indexes including richness, Shannon diversity index, inverse Simpson index, Simpson diversity number, Pielou evenness, Simpson evenness, and Shannon evenness are shown in Table 3. Neither canonical correlation analysis nor MANOVA revealed an effect of time on bacterial membership abundance or any of the diversity indexes considered. In other words, number (diversity indexes) and relative distribution (evenness) of the complex bacterial communities present did not change over time.
FIGURE 1.
Top 10 most abundant bacterial phyla (A) and genera (B) in fecal samples collected from 20 healthy lactating women from 2 d to 6 mo postpartum (n = 10, 15, 15, 15, 16, 17, 12, 11, and 9 samples at days 2, 5, and 10, and months 1, 2, 3, 4, 5, and 6, respectively) and Spearman rank correlations between BMI, energy intake, and nutrient intakes and relative abundance of the 10 most abundant bacterial phyla (C and E) and genera (D and F) in fecal samples collected from healthy lactating women from 1–3 mo postpartum. ˄Trend (P ≤ 0.05); *Significant (P ≤ 0.01). A total of 120 fecal samples from 20 women were used in this analysis. % En, percentage of energy.
TABLE 3.
Mean richness, Shannon diversity index, inverse Simpson index, Shannon evenness, Simpson evenness, and Pielou evenness at each taxonomic level of the fecal microbiome of healthy lactating women1
| Phylum | Class | Order | Family | Genus | |
| Richness | 8 ± 2 (4–12) | 16 ± 3 (6–21) | 21 ± 4 (5–33) | 37 ± 9 (9–59) | 94 ± 23 (14–162) |
| Shannon diversity index | 2.4 ± 0.5 (1.2–4.0) | 3.0 ± 0.7 (1.8–5.1) | 3.0 ± 0.7 (1.8–5.3) | 6.4 ± 2.2 (2.8–13.5) | 10.5 ± 3.7 (5.1–23.3) |
| Inverse Simpson index | 2.1 ± 0.4 (1.0–3.4) | 2.4 ± 0.5 (1.4–4.2) | 2.4 ± 0.5 (1.4–4.3) | 4.5 ± 1.7 (1.8–10.6) | 6.1 ± 2.3 (2.4–14.3) |
| Shannon evenness | 0.3 ± 0.1 (0.1–0.6) | 0.2 ± 0.1 (0.1–0.5) | 0.1 ± 0.1 (0.1–0.5) | 0.2 ± 0.1 (0.1–0.7) | 0.1 ± 0.1 (0.1–0.6) |
| Simpson evenness | 0.26 ± 0.01 (0.12–0.54) | 0.16 ± 0.05 (0.08–0.39) | 0.12 ± 0.05 (0.06–0.47) | 0.13 ± 0.08 (0.04–0.58) | 0.07 ± 0.05 (0.02–0.38) |
| Pielou evenness | 0.4 ± 0.1 (0.1–0.7) | 0.4 ± 0.1 (0.2–0.6) | 0.4 ± 0.1 (0.2–0.6) | 0.5 ± 0.1 (0.3–0.9) | 0.5 ± 0.1 (0.4–0.8) |
Values are means ± SDs (ranges); n = 120 fecal samples from 20 healthy lactating women.
Dietary intake (all time points).
Mean dietary energy and macronutrient and selected micronutrient intake at each time point are provided in Table 2. In general, women consumed energy and nutrients at levels that would be expected for well-nourished lactating women (30). ANOVA results revealed an effect of time (P < 0.05) on carbohydrate (grams per day) and energy (kilocalories per day) intake such that these dietary components generally decreased as time postpartum advanced; there was no effect of time on intake of other nutrients.
Associations between diet and relative abundance of single bacterial taxa (1–3 mo).
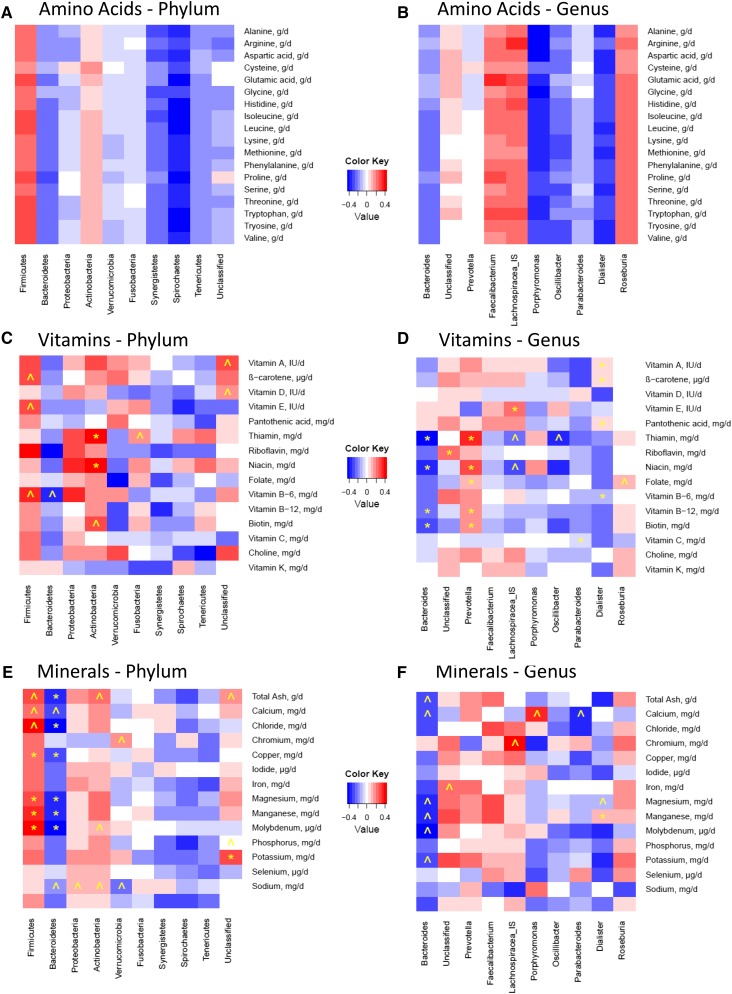
Heat maps illustrating correlations between dietary variables (BMI, energy, and nutrient intake values) and relative abundance of the top 10 bacterial phyla and genera are provided in Figure 1 and Figure 2 (additional heat maps constructed for the other taxonomic levels are provided in Supplemental Figures 4–18). Significant correlations (P ≤ 0.01), as well as trends (P ≤ 0.05), between several dietary intake values and relative abundance of particular bacterial groups were found. Some of these associations are highlighted and summarized in Table 4. In general, overall consumption of a more nutrient-rich diet appeared to be related to higher relative abundance of Firmicutes and lower relative abundance of Bacteroidetes members.
FIGURE 2.
Spearman rank correlations between BMI, energy intake, and nutrient intakes and relative abundance of the 10 most abundant bacterial phyla (A, C, and E) and genera (B, D, and F) in fecal samples collected from healthy lactating women from 1–3 mo postpartum. ˄Trend (P ≤ 0.05); *Significant (P ≤ 0.01). A total of 120 fecal samples from 20 women were used in this analysis.
TABLE 4.
Selected associations between nutrient intakes and relative bacterial abundance at the phylum and genus levels in healthy lactating women determined by Spearman rank correlations
| Nutrient group | Phylum level (n = 120) | Genus level (n = 120) |
| Macronutrients and energy | •↑Carbohydrate, protein → ↑Firmicutes (r = 0.37 and 0.18, respectively; P ≤ 0.01) | •↑Current BMI → ↑Parabacteroides (r = 0.19; P ≤ 0.01) |
| •↑Carbohydrate, fat, protein → ↓Bacteroidetes (r = −0.38, −0.32, and −0.21, respectively; P ≤ 0.01) | •↑Insoluble fiber → ↑Faecalibacterium (r = 0.36; P ≤ 0.01) | |
| • ↑Protein→ ↓Spirochaetes (r = −0.29; P ≤ 0.01) | ||
| FAs | • ↑18:2(n–6) → ↑Firmicutes (r = 0.24; P ≤ 0.01) | •↑4:0, 6:0 → ↑Porphyromonas (r = 0.14 and 0.12, respectively; P ≤ 0.01) |
| •↑4:0, 6:0, 14:1(n–5), 17:1(n–7), CLA → ↑Synergistetes (r = 0.32, 0.29, 0.14, 0.24, and 0.34, respectively; P ≤ 0.01) | ||
| •↑n–3, n–6 FAs → ↑Firmicutes (r = 0.35 and 0.42, respectively; P ≤ 0.01) | ||
| Amino acids1 | • ↑Essential amino acids → ↑Firmicutes and Actinobacteria | •↑Essential amino acids → ↑Faecalibacterium, Lachnospiracea, and Roseburia |
| •↑Essential amino acids → ↓Bacteroidetes, Synergistetes, Spirochaetes, and Tenericutes | •↑Essential amino acids → ↓ Bacteroides, Porphyromonas, Oscillibacter, Parabacteroides, and Dialister | |
| Vitamins | •↑Folate → ↑Firmicutes (r = 0.32) and ↓Bacteroidetes (r = −0.34; P ≤ 0.05) | •↑Pantothenic acid, riboflavin, vitamin B-6, vitamin B-12 → ↑Prevotella (r = 0.45, 0.39, 0.34, and 0.24, respectively; P ≤ 0.01) and ↓Bacteroides (r = −0.55, −0.46, −0.32, and −0.35, respectively; P ≤ 0.01) |
| • ↑Vitamin E → ↑Firmicutes (r = 0.26; P ≤ 0.05) | •↑Vitamin A, β-carotene, vitamin D → ↑Dialister (r = 0.10, 0.10, and 0.06, respectively; P ≤ 0.01) | |
| • ↑β-carotene → ↑Firmicutes (r = 0.29; P ≤ 0.05) | •↑Folate → ↓Dialister (r = −0.18; P ≤ 0.01) | |
| • ↑Pantothenic acid → ↑Actinobacteria (r = 0.39; P ≤ 0.01) | ||
| • ↑Riboflavin → ↑Actinobacteria (r = 0.40; P ≤ 0.01) | ||
| Minerals | •↑Copper, magnesium, manganese, and molybdenum → ↑Firmicutes (r = 0.33, 0.38, 0.44, and 0.51, respectively; P ≤ 0.01) and ↓Bacteroidetes (r = −0.38, −0.44, −0.48, and −0.53, respectively; P ≤ 0.01) | •↑Total ash, calcium, magnesium, manganese, molybdenum, and potassium → ↓Bacteroides (r = −0.10, −0.31, −0.10, −0.11, and −0.27, respectively; P ≤ 0.05) |
| •↑Total ash, chloride → ↓Bacteroidetes (r = −0.46 and −0.56, respectively; P ≤ 0.01) |
Although none of these relations were significant at either the P ≤ 0.05 or P ≤ 0.01 level, the relations described were notably consistent in terms of whether they were positive or negative (see Figure 2A and B for visualizations of this observation). Ranges for P values of correlations between each essential amino acid and taxonomic group at the phylum level were 0.16–0.82, 0.19–0.69, 0.55–0.93, 0.11–0.38, 0.23–0.60, and 0.34–0.64 for Firmicutes, Bacteroidetes, Actinobacteria, Synergistetes, Spirochaetes, and Tenericutes, respectively. Ranges for P values of correlations between each essential amino acid and taxonomic group at the genus level were 0.21–0.88, 0.20–0.57, 0.06–0.36, 0.06–0.33, 0.13–0.59, 0.47–0.82, 0.36–0.59, 0.26–0.66, and 0.28–0.61 for Bacteroides, Prevotella, Faecalibacterium, Lachnospiracea, Porphyromonas, Oscillibacter, Parabacteroides, Dialister, and Roseburia, respectively.
Univariate and multivariate analysis of bacterial community structure (1–3 mo).
PCA of bacterial abundance in feces collected from 1–3 mo suggested that variation in the 4 most abundant phyla explained 70% of the total community variation. At lower taxonomic levels, a large number of bacterial taxa (>8) were necessary to explain at least 70% of the variation; this number represented too many variables to include in these types of statistical analyses. Consequently, only 4 bacterial phyla (Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria) were used for further multivariate analysis (PCA, NMDS, and MANOVA).
Visual examination of PCA plots (plots not shown) suggested no clustering patterns by subject, time postpartum, delivery mode, place of birth (home vs. hospital), prepregnancy BMI classification, current BMI classification, or parity. NMDS plots (plots not shown) also showed little to no clustering by subject, delivery mode, place of birth (home vs. hospital), prepregnancy BMI classification, or current BMI classification. There was no obvious clustering of data by quartiles of energy, total carbohydrates, protein, fat, saturated fat, polyunsaturated fat, fiber, vitamin B-12, calcium, or iron intake (plots not shown). When relative abundance of each of the phyla were considered along with the diversity indexes as a complex multivariate dependent outcome variable, results garnered from MANOVA also suggested little to no effect of subject, birth location, delivery mode, current or prepregnancy BMI category, energy intake quartile, macronutrient intake quartiles, or micronutrient intake quartiles on complex bacterial community structure.
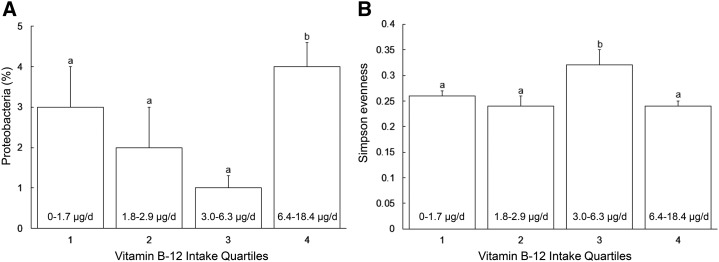
Results from univariate ANOVA for the effects of selected anthropometric, dietary, and environmental variables on the relative abundance of individual bacterial groups and diversity indexes also showed no significance for the specified effects, with the exception of prepregnancy BMI, which was related to relative abundance of Firmicutes (46.5% and 56.7% in normal and overweight/obese women, respectively; P < 0.05), and vitamin B-12 intake, which was related to Simpson evenness and relative abundance of Proteobacteria (P < 0.05) (Figure 3). Compared with all other quartiles, relative abundance of Proteobacteria was higher in individuals consuming the highest quartile of vitamin B-12 (P < 0.05). Evenness was higher in women grouped in the third quartile of vitamin B-12 intake (P < 0.05) compared with all other nutrient intake quartiles.
FIGURE 3.
Relative abundance of Proteobacteria (A) and Simpson evenness score (B) as they relate to vitamin B-12 intake quartiles in 20 healthy lactating women (n = 5 per quartile) between 1 and 3 mo postpartum. Values are means ± SEMs; values in the lower portion of each bar represent ranges of vitamin B-12 intake in the quartile. Bars within a panel without a common letter differ, P ≤ 0.05. Data related to relative abundance of Proteobacteria were transformed for the statistical analysis, although data presented here have been back-transformed.
Discussion
Data gleaned from this largely exploratory, hypothesis-generating prospective study suggest that the fecal microbiome of healthy lactating women is generally similar to that of other adult human populations—at least in terms of the most abundant bacterial taxa. Similar to reports for other adult populations, the primary phylum and genus detected in feces of lactating women were Firmicutes and Bacteroides, respectively. Members of the Bacteroides and Prevotella genera were among the most abundant in both subjects studied in the Human Microbiome Project (nonlactating healthy adults) and the women participating in our study (24). However, unlike in the Human Microbiome Project, we did not find Haemophilus, Veillonella, Lactobacillus, Propionibacterium, and Streptococcus in our most abundant taxa (although they were detected as less abundant genera). Similar to our findings, Jost et al. (18) reported that one of the most abundant taxa in the fecal microbiome of lactating women included Bacteroides. However, their findings suggest that Bifidobacterium, Blauta, and Streptococcus were more dominant than in our population. Whether these differences are genuine—for example, because of population differences—or a result of methodologic dissimilarity (sequencing platform and/or primers) is not known.
Supporting previously published data concerning the maternal fecal microbiota during late pregnancy and early lactation (20), microbial community structure appears to be quite stable throughout the first 6 mo postpartum. However, our analyses suggest that many factors that we had hypothesized might be related to fecal microbial variability (e.g., delivery mode, parity, and time postpartum) did not explain much of the observed community structure variation, either at the complex community level or at the individual phylum level. As such, our findings concur with conclusions put forth previously by the Human Microbiome Project Consortium (24) that “the human microbiome is not well explained by these phenotypic metadata, and other important factors such as short- and long-term diet, daily cycles, founder effects such as mode of delivery (of host), and host genetics should be considered in future analyses.”
Although our data supported few of our a priori hypotheses, they revealed a host of previously undescribed but potentially interesting relations between individual taxa and nutrient intake levels. To our knowledge, these findings represent only the second evaluation to date of potential associations between consumption of essential nutrients (rather than dietary patterns such as veganism) and fecal microbiome in any population. In the other report by Wu et al. (25) 98 men and women between the ages of 18 and 40 y and who had not used antibiotics for at least 6 mo were studied. Both 24 h recalls and FFQs were used to assess acute and chronic nutrient intake, respectively. In general, and unlike what our data suggest, this research team reported few associations between fecal bacteria and acute nutrient intake. Moreover, when chronic nutrient intake ascertained from FFQs were considered, Wu et al. described many relations that were seemingly at odds with what we report here. For instance, they found that relative abundance of Bacteroides was positively associated with fat and protein consumption; and higher carbohydrate intake was related to increased abundance of Prevotella. In contrast, our results suggest an inverse trend between protein intake and relative abundance of Bacteroides (P = 0.02; r = −0.29), and no relation between members of this genus and fat intake (P = 0.73; r = −0.006). Furthermore, our data suggest that relative abundance of Prevotella members is not associated with carbohydrate consumption (P = 0.37; r = 0.05). And, whereas Wu et al. reported that saturated fat intake was positively associated with relative abundance of Bacteroides and Parabacteroides, we did not (P = 0.37; r = −0.03 and P = 0.17; r = −0.13, respectively).
It is unclear what factors are driving these disparate results, although it is possible that there are significant sex and life-stage differences that need to be more carefully and systematically explored. It is also possible that differences in DNA extraction, amplification, and sequencing methods and technologies might also be involved. For instance, whereas Wu et al. (25) used the Mo Bio PowerSoil kit for DNA extraction and primers for the V5–V6 hypervariable region, we used the QIAamp DNA Stool Mini Kit and primers for the V1–V3 hypervariable region. It is possible that these differences resulted in disparate relative abundance values for the bacterial genera, leading to the characterization of differential relations with dietary variables. Alternatively, differences in estimation of dietary intake data (FFQ vs. 24 h recalls) might be partially responsible. To help address these potential confounded results, we recommend that researchers in the future simultaneously study nutrient–microbiome relations in men and women (at least, when not considering the impact of reproductive stage).
Our results somewhat support results from Ley et al. (21) and Turnbaugh et al. (22), such that multivariate analyses suggest that higher prepregnancy BMI was related to increased relative abundance of Firmicutes. However, this was not found for current BMI, and simple associations (shown in the heat maps) suggest that these relations are weak at best. It is possible that our relatively homogeneous population did not allow robust detection of this relation. It is also conceivable that BMI did not adequately reflect adiposity status in our lactating women. However, it is noteworthy that others (3, 24) have also reported no relation between BMI and fecal bacterial community structures. Why these discrepancies exist is not known and deserves more focused investigation with the use of more sensitive measures of body composition, such as DXA.
It is noteworthy that there are several key limitations to this study that should be considered carefully when interpreting its results. For instance, our sample size was relatively small and our subjects homogeneous in terms of age and overall nutritional status. These factors may have limited our ability to detect associations between metadata and fecal bacterial community structure. In addition, several fecal samples could not be used in our analyses because of antibiotic use or inability to amplify sufficient amounts of microbial DNA, thus possibly introducing some bias and reducing our power to detect potentially important relations. Furthermore, we used 24 h recalls to estimate dietary intake rather than more comprehensive food duplicate analyses, FFQs, or prospective 3 d diet records. We also did not include information concerning dietary supplements and prebiotic/probiotic foods/supplements in our analyses. Clearly, our population of lactating women likely consumed prenatal vitamins and other supplements in response to general recommendations for breastfeeding women; intake from supplements was not included in our dietary assessment analyses. As such, our data may be more qualitative than quantitative in terms of actual nutrient intake (31), and do not account for consumption of foods and supplements intentionally containing live bacteria (probiotics) or compounds that support them (prebiotics). Total nutrient intake (from diet and supplements) and factors known to influence the gastrointestinal microbiota (e.g., pre- and probiotics) should be addressed in future studies examining dietary influences on the microbiome of lactating women and other populations. In addition, and as mentioned previously, BMI was used rather than more accurate methods such as DXA to estimate body fatness (32). Nonetheless, we believe that the longitudinal nature of this study, coupled with the careful collection and analysis of a range of potentially important metadata (including nutrient intake) in a previously understudied population (lactating women), support its importance in the current available literature concerning the potential relation between diet and the gastrointestinal microbiome.
Of particular interest, we uncovered a multitude of previously undescribed associations and patterns that might be biologically relevant in terms of identifying dietary factors that could drive microbial ecology in the gastrointestinal tract. For instance, in general, increased consumption of energy-yielding nutrients was related to higher relative abundance of Firmicutes and lower relative abundance of Bacteroidetes. Similarly, consumption of a more nutrient-rich diet was associated with increased relative abundance of Firmicutes and an inverse association with Bacteroidetes. This finding appeared to be relatively consistent across all nutrient classes, from macronutrients (including FAs and amino acids) to micronutrients. Noteworthy was the somewhat consistent relation between intake of the various B vitamins (particularly vitamin B-12) and bacterial diversity. Indeed, although almost nothing is known about the prebiotic effects of vitamin intake on bacterial microbial assembly and maintenance, others have posited that this may be an underappreciated link between nutrition and the gastrointestinal microbiome (33). Of course, because of the observational nature of this study and the compositional nature of how the data are reported (as relative abundance rather than absolute amount), it is impossible to determine if these nutrient–microbe relations are causal and which bacterial taxa are actually being influenced. Additional controlled dietary intervention studies with the use of more quantitative measures of bacterial abundance will be required to answer these questions.
In summary, our largely exploratory data analyses suggest that the fecal microbiome of breastfeeding women is relatively similar to those of other adult populations (although differences are noted), is not substantially related to maternal adiposity or a variety of reproductive factors, and is quite stable. Although we were unsuccessful in relating current dietary intake variables to variation in complex global fecal communities, many univariate relations were unveiled when individual members of each taxonomic level were considered independently of each other. Whether these correlations are causal in nature and biologically important is yet to be determined. Nonetheless, we anticipate that the descriptive data garnered from this study will be useful in future research designed to examine connections between the maternal diet, maternal fecal microbiome, milk microbiome, and fecal microbiome of breastfeeding infants. These studies should be designed to assess nutrient intake from both diet and supplements and also should aim to evaluate intake of pre- and probiotic foods. Gaining a more comprehensive understanding of these relations is fundamental to understanding how early maternal and infant environments (including nutrition and microbial exposure) influence lifelong health and disease.
Acknowledgments
We thank Dan New at the University of Idaho Institute for Bioinformatics and Evolutionary Studies core for his expert guidance and patient work in sequencing and data analysis. MAM and MKM equally designed the research and provided general oversight for its conduct, laboratory and statistical analyses, and interpretation; MAY, SLB, and JEW collected the samples; JMC, SLB, KAL, and JEW carried out the laboratory analyses with the assistance of MLS; JMC, JEW, BS, and WJP conducted the statistical analyses; all authors collectively wrote the manuscript; and MKM had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: MANOVA, multivariate analysis of variance; NMDS, nonmetric multidimensional scaling; PCA, principal component analysis.
References
- 1.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013;339:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmer J, Lange B, Frick JS, Sauer H, Zimmermann K, Schwiertz A, Rusch K, Klosterhalfen S, Enck P. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr 2012;66:53–60. [DOI] [PubMed] [Google Scholar]
- 3.Mättö J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome–a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol 2005;43:213–22. [DOI] [PubMed] [Google Scholar]
- 4.Hall LJ, Walshaw J, Watson AJ. Gut microbiome in new-onset Crohn’s disease. Gastroenterology 2014;147:932–4. [DOI] [PubMed] [Google Scholar]
- 5.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085. [DOI] [PMC free article] [PubMed]
- 6.Burcelin R, Serino M, Chabo C, Blasco-Baque V, Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetol 2011;48:257–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado MC, Rautava S, Isolauri E, Salminen S. Gut microbiota: source of novel tools to reduce the risk of human disease? Pediatr Res 2015;77:182–8. [DOI] [PubMed] [Google Scholar]
- 8.Munyaka PM, Khafipour E, Ghia JE. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr 2014;2:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiulio DB. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med 2012;17:2–11. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, Narbad A, Olivares M, Xaus J, Rodríguez JM. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol 2005;51:270–4. [DOI] [PubMed] [Google Scholar]
- 11.Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, Fernández L, Rodríguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol 2008;159:187–93. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010;107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín R, Heilig HG, Zoetendal EG, Jiménez E, Fernández L, Smidt H, Rodríguez JM. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol 2007;158:31–7. [DOI] [PubMed] [Google Scholar]
- 14.Collado MC, Delgado S, Maldonado A, Rodríguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol 2009;48:523–8. [DOI] [PubMed] [Google Scholar]
- 15.Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 2011;6:e21313. [DOI] [PMC free article] [PubMed]
- 16.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 2012;96:544–51. [DOI] [PubMed] [Google Scholar]
- 17.McGuire MK, McGuire MA. Human milk: Mother Nature’s prototypical probiotic food? Adv Nutr 2015;6:112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 2014;16:2891–904. [DOI] [PubMed] [Google Scholar]
- 19.Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013;69:1–10. [DOI] [PubMed] [Google Scholar]
- 20.Jost T, Lacroix C, Braegger C, Chassard C. Stability of the maternal gut microbiota during late pregnancy and early lactation. Curr Microbiol 2014;68:419–27. [DOI] [PubMed] [Google Scholar]
- 21.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr 2008;88:894–9. [DOI] [PubMed] [Google Scholar]
- 24.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settles M. GitHub. Analysis of double barcoded Illumina amplicon data [cited 2015 Mar 9]. Available from: https://github.com/msettles/dbcAmplicons.
- 27.Magoč T, Salzberg A. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole RJ. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson G, Solymos P, Henry H, Stevens H, et al. vegan: Community ecology package. R package version 2.0–10. 2013 [cited 2015 Mar 9]. Available from: http://CRAN.R-project.org/package=vegan.
- 30.Institute of Medicine. Dietary reference intakes. The essential guide to nutrient requirements. The National Academies Press, Washington (DC); 2006.
- 31.Shim JS, Oh K, Kim HC. Dietary assessment methods in epidemiologic studies. Epidemiol Health 2014;36:e2014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domer MC, Beerman KA, Ahmadzadeh A, Dasgupta N, Williams JE, McGuire MA, McGuire MK. Loss of body fat and associated decrease in leptin in early lactation are related to shorter duration of postpartum anovulation in healthy U.S. women. J Hum Lact 2015;31:282–93. [DOI] [PubMed] [Google Scholar]
- 33.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab 2014;20:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]