Abstract
Biological systems use complex ‘information-processing cores’ composed of molecular networks to coordinate their external environment and internal states. An example of this is the acquired, or adaptive, immune system (AIS), which is composed of both humoral and cell-mediated components. Here we report the step-by-step construction of a prototype mimic of the AIS that we call an adaptive immune response simulator (AIRS). DNA and enzymes are used as simple artificial analogues of the components of the AIS to create a system that responds to specific molecular stimuli in vitro. We show that this network of reactions can function in a manner that is superficially similar to the most basic responses of the vertebrate AIS, including reaction sequences that mimic both humoral and cellular responses. As such, AIRS provides guidelines for the design and engineering of artificial reaction networks and molecular devices.
Supplementary information
The online version of this article (doi:10.1038/nchem.2325) contains supplementary material, which is available to authorized users.
Subject terms: DNA computing, DNA, Networks and systems biology
A cascade reaction network has been created that can function in a manner that is superficially similar to the most basic steps of the vertebrate adaptive immune response. This reaction network uses DNA and enzymes as simple artificial analogues of the components of the acquired immune system.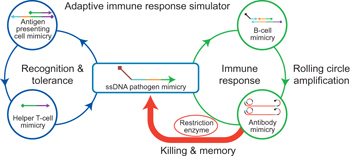
Supplementary information
The online version of this article (doi:10.1038/nchem.2325) contains supplementary material, which is available to authorized users.
Main
All organisms, including plants, have developed mechanisms of defence against pathogenic and other challenges, and thus solve a key biological problem: survival. A stunning example of this in humans is the development of learned, or adaptive, immunity, which is composed of both humoral and cell-mediated components. Adaptive immunity involves the production of antibodies and immunological memory, but also cell-mediated responses, including the key steps of antigen presentation, antigen binding, co-stimulation and destruction of the foreign body (phagocytosis). More broadly, specialized cells, tissues and organs combine together as a network to recognize, respond and remember specific antigens or pathogens. Functionally, each step in the sequence of humoral and cellular immune response involves the activation and deployment of B and T lymphocytes via highly complex signalling pathways. A fully implemented artificial adaptive immune system (AIS) has not been well studied, possibly as a result of the difficulty in controlling programmability and transferring the high-level biological codes into simple artificial modules and physical implementations. In addition, the formation of robust molecular structures and the precise control of their temporal dynamics may pose other difficulties. However, in the present work, we hypothesize the feasibility of designing a network of reactions able to function in a manner similar to the vertebrate AIS, using DNA and enzymes as simplified artificial analogues to mimic the activation and deployment of an adaptive immune response at its most fundamental level.
This study reports the step-by-step construction of such a prototype, termed hereinafter as an adaptive immune response simulator, or AIRS. We first asked what building materials would best replicate the intricate networks of chemical reactions, signalling pathways and cell–cell communication that control host immunity1. As a carrier of genetic information with well-regulated and predictable structures, DNA offers an excellent platform for the design of reaction networks with arbitrary topologies and high programmability2,3. Building on the richness of DNA computing4,5 and DNA–enzyme manipulation6,7, synthetic DNA-based systems have been used to explore the possibilities of mimicking both simple and complex systems. For example, small-scale in vitro circuits that encode elementary functions, such as cascades8, bistable memory9,10 or oscillations11, have been engineered successfully. In addition, larger networks, including those that mimic neural networks12 or predator–prey ecosystems13,14, confirm the value of DNA chemistry for implementing the type of system proposed in this study.
Pathogenic challenge results in the production of antibodies that lock onto a part of the foreign body known as an antigen. As this event triggers a complex cascade of cell-mediated reactions, we next asked which bioengineering techniques would best replicate the intricate networks of chemical reactions, signalling pathways and cell–cell communication that control host immunity. To address both questions, in a stepwise manner we report the construction of a chemical reaction network, AIRS, that couples DNA–enzyme cascade interactions, as described above, with DNA-strand-displacement cascades in which a new product can be activated by the presence of specific initiators15,16, and thus allows many such reactions to be linked into a cascade and even to form a complex network, in particular one such as adaptive immunity in which biological events follow each other sequentially in a rolling fashion.
Results and discussion
System construction
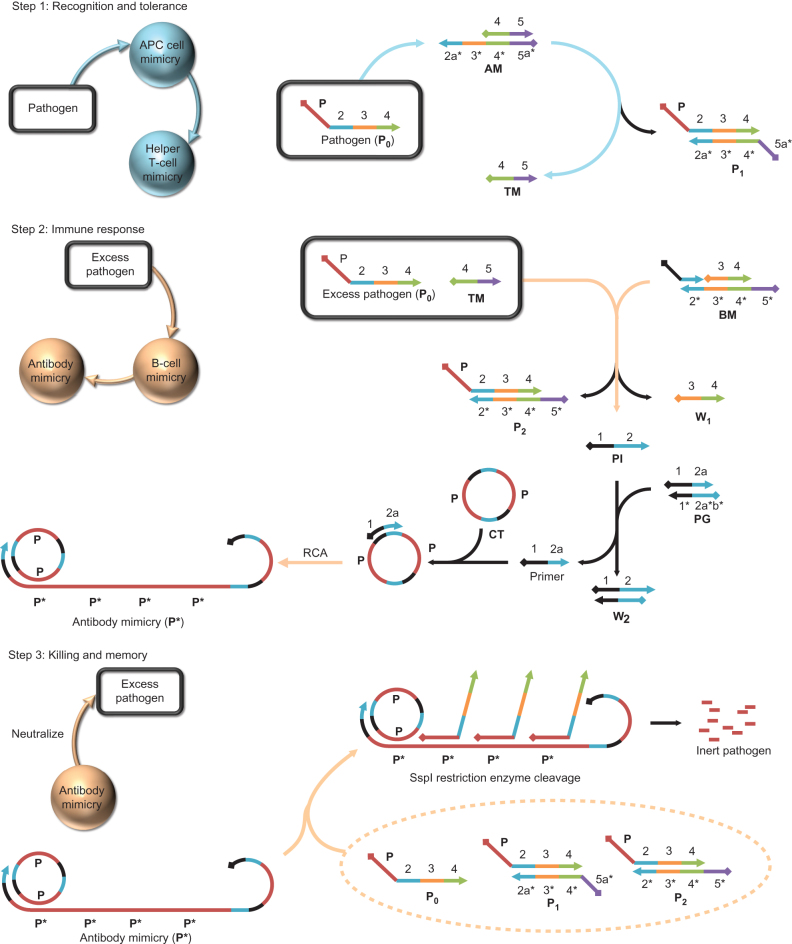
Once a foreign target has crossed the threshold of immune tolerance, the humoral and cellular components of the vertebrate immune system are called to action. A key function is antigen presentation, which provides the mechanism for recognition and response. B cells secrete antibodies, which bind to and tag an antigen, whereas T cells perform the job of attacking target cells. Both T and B cells also have roles in forming memory cells. As shown in Fig. 1, we divided this series of events into three steps (recognition and tolerance, immune response, and killing and memory) under the control of four functional DNA components, including the DNA duplexes AM (antigen-presenting cell (APC) mimicry), BM (B-cell mimicry), PG (primer generator, which generates primers for the analogue antibody) and single-stranded circular DNA CT (circular template, which controls the sequence of the analogue antibody), as well as two enzymes (Phi29 DNA polymerase and SspI restriction enzyme) able to autonomously and programmably respond to an incoming piece of single-stranded DNA (ssDNA) pathogen input (P0) taken from bacterial genome. When no P0 is present, the system is maintained in a steady, balanced state by the effective blocking of the functional domains in each component. However, when challenged by P0, these functional domains are activated in a series of steps designed to mimic the three steps given above.
Figure 1. Working principle of the AIRS.
AIRS consists of three steps: (1) recognition and tolerance, (2) immune response and (3) killing and memory. These functions are blocked in the absence of ssDNA pathogen input (P0), but once P0 is introduced to the AIRS system it is driven by a series of DNA toehold-mediated strand displacement and DNA–enzyme reactions. Coloured bars indicate DNA strands with different domains. All x domains are complementary to x*; P0 is the pathogen sequence that possesses infection ability in the ssDNA form. AM (antigen-presenting cell mimicry), BM (B-cell mimicry) and PG (primer generator, which generates primers for the analogue antibody) are initially present as duplex components, along with CT (circular template, which controls the sequence of the analogue antibody) and two functional enzymes, Phi29 polymerase and SspI restriction enzyme. TM (T Cell Mimicry); RCA (rolling cycle amplification). P1 and P2 are displacement reaction products.
In step 1, in the absence of P0, the functional components of the system remain in stasis. Mimicking immune tolerance, the unresponsive state of immune defence, the reaction priority of P0 to duplex DNA AM, as well as to BM, is controlled through the lengths of their corresponding toeholds, which are short ssDNA overhang segments in duplexes. Accordingly, we designed a ten nucleotide (nt) toehold in AM with a displacement reaction rate k of 106 M−1 s−1 and a 0 nt toehold in BM with a k value of 103 M−1 s−1 for the initial P0 hybridization17,18. AM can display P0, which has two ssDNA domains, one having a sequence taken from the genome of Bacillus anthracis, a Gram-positive, rod-shaped bacterium (P), and the other designed (ssDNA domain 2-3-4) to control downstream reactions through a DNA-displacement reaction. However, at the recognition and tolerance phase, our system can only proceed to step 2, immune response, when the amount of pathogen accumulated exceeds the threshold of AM.
Once the threshold of immune tolerance is reached in the system, step 2, the analogue humoral immune response, is activated. As previously noted, during the depletion of AM by the displacement reaction, one of the displaced products, ssDNA TM (T-cell mimicry), accumulates and can be used as a catalyst to increase the reaction rate between P0 and BM, and thus release a primer (ssDNA domain 12a) for the DNA polymerase-catalysed rolling-circle amplification (RCA), which this system will employ to produce DNA ‘antibody’ mimicry (P*) to the antigen B. anthracis (P). Thus, to replicate the host-defence immune response, we incorporated two B. anthracis genomic sequence fragments (P) in the CT, which resulted in a rapid generation of the complementary sequences P*, essentially by enzymatic amplification, using Phi29 DNA polymerase, the replicative polymerase from the B. subtilis phage phi29, and deoxyribonucleotide triphosphate (dNTP), to mimic the fast and antibody-specific generation naturally produced by B cells.
In step 3, killing and memory, of the proposed system it was necessary to mimic the process by which cell-mediated immune response eliminates pathogens after the formation of an antibody–antigen complex from the body, which, as noted above, results from activation of the humoral immune response by B lymphocytes. To accomplish this, three infectious states were assumed: original pathogen strand P0 and the displacement reaction products (with AM and BM) P1 and P2. Together, these all contain an infectious pathogen domain (P). Then, to form the antibody–antigen complex in the system, the P domain in P0, P1 and P2 hybridizes with a large amount of P* and forms stable duplex regions PP*. However, although the vertebrate host immune system uses phagocytosis as a key strategy in removing pathogens, we designed a purely biomechanical method to mimic the activation of the humoral immune response, that is the binding between the generated P* and the P domain from the pathogen, followed by a restriction enzyme-based step that mimics cell-mediated killing by specifically cutting the ‘antibody–antigen’ duplex. More specifically, this latter step is accomplished when the hybridization of the P domain with P* creates an active recognition site for the restriction enzyme SspI, which is extracted from an E. coli strain that carries the cloned and modified (Y98F) sspi gene from the restriction site of Sphaerotilus species. In this way, the P domain in P0, P1 and P2 can be specifically cut into two fragments with the nucleotide numbers 56 and 15 (ref. 19), thus losing infectivity.
Next, in the adaptive immune system, some T and B cells will become memory cells able to ‘remember’ specific pathogens. These memory cells will trigger a much faster immune response on subsequent exposure to the same pathogen. Therefore, to create a mechanism to mimic the memory function of B cells, we turned to ssDNA TM, which is produced by the displacement reaction between P0 and AM and remains in the system from the first exposure to P0. We also describe its catalysis of P0 and BM displacement to release primer quickly, and thus trigger a faster RCA reaction that creates antibody for the next exposure to the same pathogen sequence. This reaction is defined as entropy driven because it is one that spontaneously evolves towards thermodynamic equilibrium (Supplementary Fig. 7)18. Without TM, the displacement rate between P0 and BM would be slowed by the effective blocking of active domains on BM. However, in the presence of TM, BM can hybridize with TM through its 4 nt toehold (domain 5*) and then release the ssDNA primer initiator (PI) and the side product W1, which results in a new toehold binding region (domain 3*) for the hybridization between P0 and BM, which is driven forwards thermodynamically by the entropic gain of the liberated molecules. Thus, the effect of memory is produced by speeding up the reaction rate between P0 and BM using the catalytic effect of the remaining TM produced by the initial recognition. Thus, by leaving TM as a ‘catalytic fuel’ in the system, a memory effect for a specific pathogen input is achieved.
Verification of signal transduction
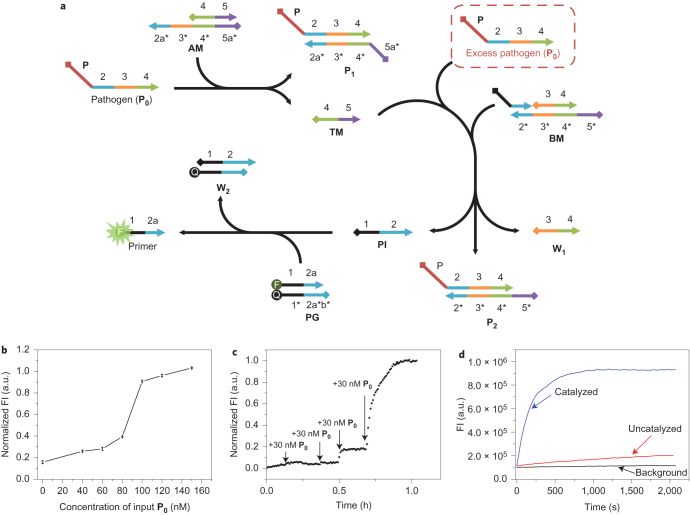
To ensure the proper operation of the entire network, signal transduction at each step was validated separately. To examine the precision of the threshold function of AM, we used a Förster resonance energy transfer (FRET)-based method to study the hybridization order of P0 with AM and BM. A fluorophore (fluorescein (FAM)) and quencher (DABCYL (DAB)) pair was coupled in the PG duplex to indicate the released amount of primer for the RCA reaction. Fluorescence restoration only occurs if the threshold level of AM is exceeded, after which the reaction proceeds to step 2, immune response, based on strand-displacement reactions that can release the quencher-labelled primer strand. Therefore, in the presence of 80 nM AM, 100 nM BM and 100 nM fluorophore/quencher-labelled PG, different concentrations of P0 (from 0 to 150 nM) were introduced to the system with fluorescence monitoring at 517 nm. As shown in Fig. 2b, the fluorescence restoration exhibited a sharp upturn at the P0 concentration of 80 nM, but little increase below 80 nM, which indicates that the reaction between P0 and BM had begun at the depletion point of AM. In other words, the threshold value of AM to P0 is adjustable by changing the concentration of AM. In this case, the depletion point, or threshold level, of AM was reached at 80 nM, indicating that the reaction between P0 and BM could initiate step 2, which, in humans, is the production of antibodies by B lymphocytes. To confirm this result further, the fluorescence kinetics of the above system was monitored. Initially, 80 nM AM, 100 nM BM and 100 nM PG were mixed in the buffer, and a small amount of P0 (30 nM) was added. As a result of the excess amount of AM and its reaction priority to P0 by the requirements of step 1, slow fluorescence restoration kinetics occurred. However, when more P0 (30 nM each time) was added continuously to the system, a sharp increase (after 90 nM in total) was seen in the fluorescence kinetics, which indicates that the reaction could proceed to step 2. Different threshold values were tested by changing the concentrations of AM and BM (Supplementary Fig. 1). Gel electrophoresis was also used to indicate the order of reactions (Supplementary Fig. 2). As the same recognition domain (2-3-4) is shared, similar experimental results were also observed by substituting P0 for another pathogen sequence, P0-SARS (severe acute respiratory syndrome coronavirus genomic DNA) (Supplementary Fig. 3). All the assays confirmed the effective threshold function of AM relative to the reaction between BM and P0, and thus provided a macroscopic mimicry of the ‘recognition and tolerance’ state of AIS.
Figure 2. Scheme and results that confirm the reaction priority between steps 1 (recognition and tolerance) and 2 (immune response).
a, Scheme of reaction steps that occur in steps 1 and 2. W2 is the side product from the displacement reaction of PI and PG. b, Plot of fluorescence restoration of the system with 80 nM AM, 100 nM BM and 100 nM FAM/DAB-labelled PG with different concentrations of the ssDNA pathogen P0. c, Kinetics experiments of the system with 80 nM AM, 100 nM BM and 100 nM FAM/DAB-labelled PG. At each time point, 30 nM P0 were added to the buffer separately. d, Kinetics experiments to study the catalytic effect of TM. The black curve represents the background kinetics with 80 nM AM, 100 nM BM, 100 nM FAM/DAB-labelled PG and no P0. The red curve shows the uncatalysed reaction kinetics with 100 nM BM, 100 nM FAM/DAB-labelled PG and 150 nM P0. The blue curve exhibits the catalysed reaction kinetics with 80 nM AM, 100 nM BM, 100 nM FAM/DAB-labelled PG and 150 nM P0. The experiments were performed at 25 °C in 50 mM Tris-HCl buffer that contained 10 mM MgCl2. a.u., arbitrary units.
Another significant function of AM is to produce catalytic ssDNA TM to facilitate the hybridization reaction between P0 and BM, which, according to Zhang et al.18, can accelerate the hybridization reaction by two to four orders of magnitude. ssDNA TM is not only important in facilitating the hybridization between P0 and BM, but it is also key to the formation of ‘mimetic memory cells’. Therefore, to prove the catalytic effect of TM, we continued to use our FRET-based reporting model, as described above. Accordingly, 80 nM AM, 100 nM BM and 100 nM FAM/DAB-labelled PG were incubated in buffer. Afterwards, 150 nM P0 were added to the system. We used an excess amount of P0 to AM to overcome the threshold value. TM is generated in step 1 as a result of the reaction between P0 and AM. Thus, P0, when it exceeds the threshold (as set by controlling the concentration of AM), in turn results in a catalytic reaction between excess P0 and TM by entropy that can quickly restore the fluorescence of the system. As a comparison, the same concentration of P0 was added directly to a solution with 100 nM BM and 100 nM FAM/DAB-labelled PG, but no AM. As shown in Fig. 2d, measurements of the fluorescence kinetics of the catalysed reaction exhibited an over 500-fold acceleration in contrast to that without catalysis18,20. Thus, as explained above, the released TM could serve as specific memory mimicry, which results in a stronger and faster immune response for the next exposure to the same pathogen. Afterwards, we also confirmed that the quantity of P* generated from enzymatic amplification for hybridization with domain P in P0, P1 and P2 could be determined by the amount of primer, thus making the enzymatic reaction controllable by upstream reactions in steps 1 and 2 (see the Supplementary Information). In addition, the restriction-enzyme-based digestion of the generated antigen–antibody complex was studied and is shown in the Supplementary Information.
Whole-system performance
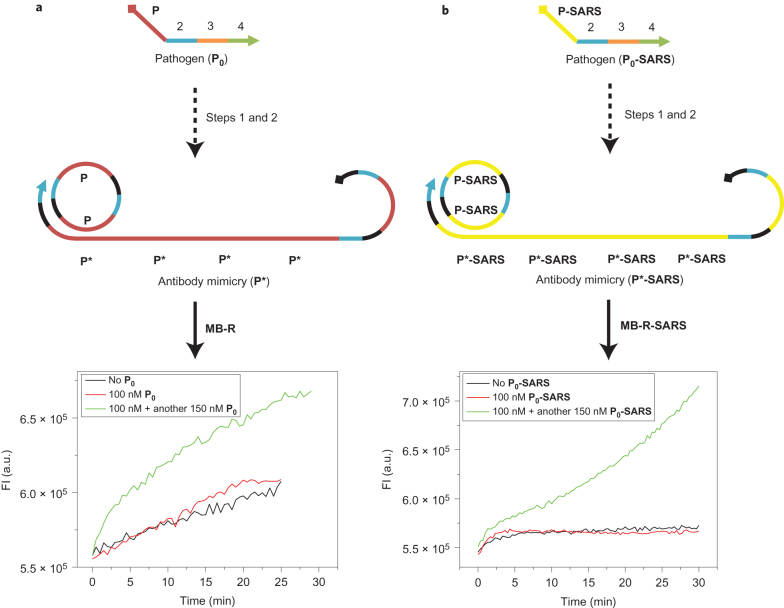
After confirming the proper function of each step, we further tested the performance of the entire system with the same pathogen strand input from the B. anthracis genome (P0). To confirm the tolerance of AIRS, a small amount of P0 (100 nM) was then added to the system, and fluorescence was monitored in real time. Next, we designed a DNA molecular beacon (MB-R) to report the amount of RCA product fluorescently by using the pathogen B. anthracis genomic sequence P as its loop. MB-R could be opened by the RCA products, that is P*, and thereby exhibit fluorescence restoration that represents different amounts of P*. Figure 3a shows that the fluorescence kinetics is similar to that without P0, which confirms that the system is not activated to produce P* when the amount of pathogen is below the level of immune tolerance (that is, a threshold of 200 nM). However, when another 150 nM P0 was added to the system to mimic a second exposure to the same pathogen, the fluorescence restoration showed a much faster kinetics behaviour, which indicates the production of a large amount of P* and a successful triggering of the immune response. In addition, the faster response during the second exposure also confirms the memory effect of this system for a specific pathogen. If P0-SARS is used as a pathogen input and a molecular beacon encoded with P0-SARS (MB-R-SARS) is used as the reporter, similar fluorescence-response trends are seen, as shown in Fig. 3b, which indicates that AIRS potentially works for a second pathogen input. We also used a different fluorescence-reporting system to indicate the amount of generated P*, which is shown in Supplementary Fig. 10.
Figure 3. Validation of signal transduction in the entire AIRS mimicry network by fluorescence.
a,b, The fluorescence kinetics of the antibody mimicry P*(P*-SARS) generation was monitored with different concentrations of P0 (a) and P0-SARS (b). Here we used double the amounts of the individual DNA components compared with those used for the data shown in Fig. 2 to ensure that the concentrations were consistent with the gel experiment.
Using a fluorescence reporter in independent experiments, AIRS was demonstrated to operate equally well with two different pathogen inputs. Therefore, we next studied if it could also work when two different inputs were presented simultaneously. To make this determination, P0 and P0-SARS were introduced together to a solution that contained all the components (300 nM AM, 300 nM BM, 300 nM PG, 50 nM CT-P0, 50 nM CT-P-SARS, 0.5 U μl–1 Phi29 and 250 μM dNTP). As in the experiment above, two different molecular beacons with the same fluorophore and quencher (MB-R and MB-R-SARS) were also added to the mixture to report the amounts of both RCA products fluorescently, that is P* and P*-SARS. As shown in Supplementary Fig. 14, as long as both pathogen inputs are below the level of immune tolerance (threshold of 300 nM), AIRS is not activated and fluorescence remains unchanged compared with the background signal. However, when the concentrations increase to a level higher than the threshold for either one, or both, pathogens, fluorescence is restored, which indicates that both pathogen inputs can generate corresponding antibody mimicry when they are mixed together. This confirms that AIRS has the capacity to respond simultaneously to two different pathogen inputs.
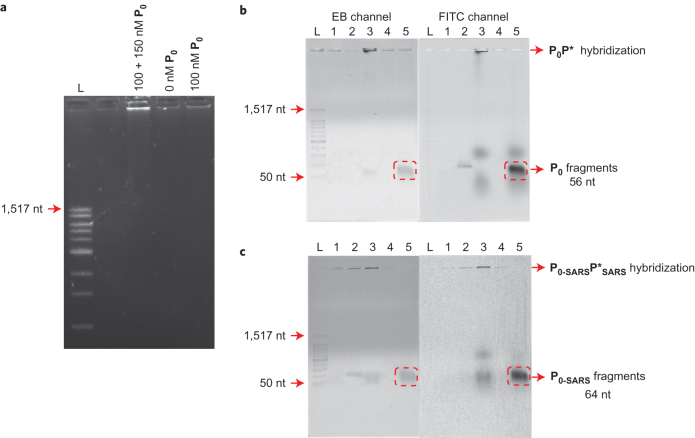
To characterize further the error-free operation of the AIRS system, we used gel electrophoresis to analyse the final amount of P*. As shown in Fig. 4a, only one sharp band of high molecular weight can be seen when the amount of pathogen exceeds the threshold level (100 + 150 nM), which is consistent with the previous fluorescence results. To study the binding between the pathogen input strand (P0) and the antibody mimicry strand (P*) in step 3, we modified P0 with a fluorophore (FAM). After the generation of P*, the hybridization between P0 and P* was confirmed to be present, as a band of high molecular weight could be seen in the fluorescein isothiocyanate (FITC)-imaging channel (Supplementary Fig. 13). With the addition of more FAM-labelled P0 (from one to 250 excess) to the system, the hybridization between excess P0 and generated P* was still observed, which indicates that AIRS could sufficiently bind the excess pathogen input strand (P0) by RCA. Finally, we incubated the restriction enzyme (2 U µl–1) together with all other components (200 nM AM, 200 nM BM, 200 nM PG, 50 nM CT, 0.5 U µl–1 Phi29 and 250 µM dNTP). After introducing an excess concentration of FAM-labelled P0 (500 nM) to surpass the immune tolerance of the system, a digestion fragment band with a FAM modification (56 nt) was discovered under the FITC channel, which leads to the conclusion that the pathogen had been successfully (Fig. 4b) digested. Interestingly, if P0 is substituted by a double-nucleotide polymorphism input (P0-mis), antibody mimicry P* can still be generated with an excess concentration of P0-mis. However, the hybridization between P0-mis and P* cannot be digested by the SspI restriction enzyme, which demonstrates that the AIRS system is sensitive to the recognition of nucleotide polymorphism of the pathogen input (Supplementary Fig. 11). AIRS has been designed with two layers of selectivity, which makes it possible for the system to differentiate P0 from a double-nucleotide polymorphism input, P0-mis. To explain, the relatively weaker binding ability between P0-mis and P* when compared to that between P0 and P* makes the P0-mis–P* duplex less stable. Additionally, a P0-mis–P* mismatch duplex cannot form an effective recognition binding site with the restriction enzyme SspI (AAT-ATT), and thereby reduce the enzyme's digestion rate, which, in turn, results in much less cleavage product from the input P0-mis (Supplementary Fig. 11). In contrast, because the hybridization of P0-SARS with P* creates a recognition binding site identical to that of the restriction enzyme SspI, similar gel electrophoresis results were found (Fig. 4c), which again confirms that AIRS can work for different pathogen inputs.
Figure 4. Characterization of error-free operations of the entire AIRS system by gel electrophoresis.
a, Analysis of antibody mimicry P* generation by agarose gel (1.5%). L, 100-base-pair ladder. The image was taken under an ethidium bromide (EB) imaging channel (λexcitation = 520 nm, λemission = 605 nm). b, Validation of the AIRS analogue system performance with pathogen strand P0 by agarose gel (1%). M is the mixture of original present components in the AIRS. Here M = mixture of 200 nM AM, 200 nM BM, 200 nM PG, 50 nM CT, 0.5 U µl–1 Phi29 and 250 µM dNTP; incubation time, one hour. Lane 1, M + 0 nM FAM-labelled P0; lane 2, M + 100 nM FAM-labelled P0; lane 3, M + 500 nM FAM-labelled P0; lane 4, M + 100 nM FAM-labelled P0 + 2 U µl–1 SspI; lane 5, M + 500 nM FAM-labelled P0 + 2 U µl–1 SspI; L, 1 kb ladder. c, Validation of the AIRS analogue system performance with the pathogen strand P0-SARS by agarose gel (1%). M = mixture of 200 nM AM, 200 nM BM, 200 nM PG, 50 nM CT-SARS, 0.5 U µl–1 Phi29 and 250 µM dNTP; incubation time, one hour. Lane 1, M + 0 nM FAM-labelled P0-SARS; lane 2, M + 100 nM FAM-labelled P0-SARS; lane 3, M + 500 nM FAM-labelled P0-SARS; lane 4, M + 100 nM FAM-labelled P0-SARS + 2 U µl–1 SspI; lane 5, M + 500 nM FAM-labelled P0-SARS + 2 U µl–1 SspI; L, 1 kb ladder.
Finally, as noted above, the specificity of the system mainly depends on the restriction enzyme and sequence of CT (see the Supplementary Information). When we challenged the system by introducing FAM-labelled P0-SARS as the pathogenic input, but CT for B. anthracis as the amplification template, no obvious fluorescent band of high molecular weight appeared with either small or large amounts of P0-SARS under the FITC channel because of the lack of hybridization between P0-SARS and P* specifically generated for B. anthracis (Supplementary Fig. 12).
Conclusions
We began by hypothesizing the feasibility of designing a network of chemical reactions able to function in a manner similar to the vertebrate AIS, using DNA–DNA and DNA–enzyme interactions as simplified artificial analogues to mimic the macroscopic behaviours of the vertebrate adaptive immune system. Accordingly, the designed artificial AIRS was, de novo, demonstrated to mimic superficially three basic steps of the host immune response, namely recognition and tolerance, immune response, and killing and memory, but without the presence any living component to achieve functionality. Moreover, even though the designed system is entirely synthetic, biocompatibility was shown by its ability to use only biological molecules (DNA and enzyme). Consequently, as the cellular state can be controlled by DNA or RNA, it could be possible to encode cellular functions to the system output (that is, the DNA and/or RNA strand)5,21,22,23,24,25,26, which makes it possible to scale up this prototype to increasingly intelligent molecular nanocontrollers and nano-operators. However, to achieve this, our current construction design will need to be improved and integrated with other chemistries.
Methods
Preparation of DNA CT
A 7.2 µl sample of 20 µM CT (with a phosphate group on its 5′ end) was placed in 4 µl CircLigase II 10x reaction buffer that contained 330 mM Tris-acetate (pH 7.5), 0.66 M KAc, 5 mM dithiothreitol (DTT) and 2.5 mM MnCl2. Afterwards, 2.8 µl CircLigase II (Epicentre) was mixed with the buffer and diluted to 40 µl with water. The mixture was incubated at 60 °C for three hours. Then 20 U Exonuclease I and 200 U Exonuclease III (New England Biotech) were added to the mixture and incubated at 37 °C for one hour. The enzyme was denatured by heating the solution to 90 °C for 20 minutes. Finally, the CT product was purified by denatured PAGE and desalted with NAP-5 columns.
Validation of signal transduction in each step and the entire system by fluorescence
When the reaction priority between step 1 and step 2 was tested, purified AM (50 nM), BM (50 nM) and PG (with FAM and DAB, 50 nM) were mixed in 1 × TAE (Tris base, acetic acid and EDTA)–Mg buffer to a total volume of 100 µl and the fluorescence was monitored for different concentrations of P0 (or P0-SARS or P0-mis) (Supplementary Fig. 1). Fluorescence intensities (FIs) were tested after incubation for one hour at room temperature. When the signal transduction in the entire system was tested by fluorescence, purified AM (200 nM), BM (200 nM), PG (no FAM or DAB, 200 nM), 50 nM CT (or CT-SARS) and 500 nM MB-R (or 200 nM DNA hairpin A1, 200 nM DNA hairpin A2 and 250 nM fluorescent reporter R12, (for another fluorescence-reporting method described in Supplementary Fig. 9) were incubated in 1 × TAE–Mg2+ buffer (50 mM Tris-HCl, 10 mM MgCl2). The fluorescence kinetics was started when different amounts of P0 were added to the solution. To check the fluorescence kinetics of the system on a second exposure to P0, 100 nM P0 were first added to a buffer that contained purified AM (200 nM), BM (200 nM), PG (no FAM or DAB, 200 nM) and 50 nM CT. Then another 150 nM P0 together with 500 nM MB-R (or 200 nM A1, 200 nM A2 and 250 nM R12) were mixed together in the solution simultaneously.
Electrophoresis analysis of the system by agarose gel
A 10 µl system with purified AM (200 nM), BM (200 nM), PG (200 nM), CT (50 nM), Phi29 (0.5 U µl–1) and dNTP (250 µM) was chosen for this experiment. For the pathogen-digestion experiment, the concentration of restriction enzyme SspI was set at 2 U µl–1. The reaction was carried out in 1 × NEB buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM (NH4)2SO4 and 4 mM DTT) with different amounts of P0 input at 30 °C for three hours. The agarose gel was run at 4 °C for one hour under 100 V.
Supplementary information
Supplementary information (PDF 1381 kb)
Acknowledgements
We thank C. Yang, X. Fang, S. Benner, J. Jiang, X. Xiong and K. Williams for discussion and manuscript review. This work is supported by the National Key Scientific Program of China (2011CB911000), by National Natural Science Foundation of China grants (NSFC 21221003 and NSFC 21327009), by the China National Instrumentation Program 2011YQ03012412 and by the National Institutes of Health (GM079359 and CA133086).
Author Contributions
D.H., C.W. and M.Y. contributed equally to this work. D.H. and W.T. conceived the idea and designed the experiment. D.H., C.W., M.Y. and T.Z. performed the experiment. M.Y., T.Z., T.C., C.W., L.Q., S.W., H.L. and Z.Z. provided experimental advice, performed some experiments and analysed the data. D.H., M.Y. and W.T. analysed the data and wrote the manuscript. W.T. supervised the project.
Competing interests
The authors declare no competing financial interests.
Footnotes
Da Han, Cuichen Wu and Mingxu You: These authors contributed equally to this work
References
- 1.Bottomly K. 1984: all idiotypes are equal, but some are more equal than others. Immunol. Rev. 1984;79:45–61. doi: 10.1111/j.1600-065X.1984.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 2.Krishnan Y, Simmel FC. Nucleic acid based molecular devices. Angew Chem. Int. Ed. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- 3.Simmel FC. Programming the dynamics of biochemical reaction networks. ACS Nano. 2013;7:6–10. doi: 10.1021/nn305811d. [DOI] [PubMed] [Google Scholar]
- 4.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 5.Han D, et al. A logical molecular circuit for programmable and autonomous regulation of protein activity using DNA aptamer–protein interactions. J. Am. Chem. Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johne R, Muller H, Rector A, van Ranst M, Stevens H. Rolling-circle amplification of viral DNA genomes using Phi29 polymerase. Trends Microbiol. 2009;17:205–211. doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Franco E, et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA. 2011;108:E784–E793. doi: 10.1073/pnas.1100060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc. Natl Acad. Sci. USA. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou C, et al. Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Mol. Syst. Biol. 2010;6:350. doi: 10.1038/msb.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padirac A, Fujii T, Rondelez Y. Bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA. 2012;109:E3212–E3220. doi: 10.1073/pnas.1212069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 13.Fujii T, Rondelez Y. Predator–prey molecular ecosystems. ACS Nano. 2013;7:27–34. doi: 10.1021/nn3043572. [DOI] [PubMed] [Google Scholar]
- 14.Padirac A, Fujii T, Estevez-Torres A, Rondelez Y. Spatial waves in synthetic biochemical networks. J. Am. Chem. Soc. 2013;135:14586–14592. doi: 10.1021/ja403584p. [DOI] [PubMed] [Google Scholar]
- 15.Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nature Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 16.Zhao WA, Ali MM, Brook MA, Li YF. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008;47:6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 17.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 19.McManus SA, Li YF. Turning a kinase deoxyribozyme into a sensor. J. Am. Chem. Soc. 2013;135:7181–7186. doi: 10.1021/ja311850u. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Ellington AD. Shaping up nucleic acid computation. Curr. Opin. Biotech. 2010;21:392–400. doi: 10.1016/j.copbio.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 2011;7:466. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soengas MS, Gutierrez C, Salas M. Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J. Mol. Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 23.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 24.Hood L, Heath JR, Phelps ME, Lin BY. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 25.Simmel FC. Towards biomedical applications for nucleic acid nanodevices. Nanomedicine. 2007;2:817–830. doi: 10.2217/17435889.2.6.817. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information (PDF 1381 kb)