Abstract
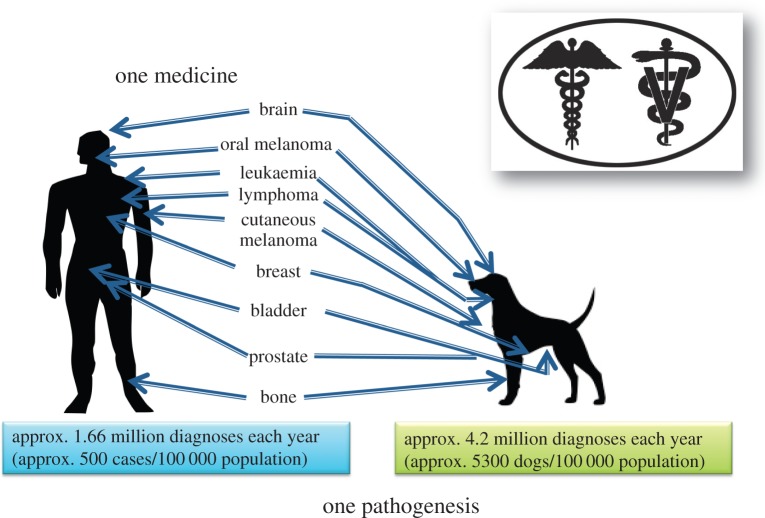
Over 1.66 million humans (approx. 500/100 000 population rate) and over 4.2 million dogs (approx. 5300/100 000 population rate) are diagnosed with cancer annually in the USA. The interdisciplinary field of comparative oncology offers a unique and strong opportunity to learn more about universal cancer risk and development through epidemiology, genetic and genomic investigations. Working across species, researchers from human and veterinary medicine can combine scientific findings to understand more quickly the origins of cancer and translate these findings to novel therapies to benefit both human and animals. This review begins with the genetic origins of canines and their advantage in cancer research. We next focus on recent findings in comparative oncology related to inherited, or genetic, risk for tumour development. We then detail the somatic, or genomic, changes within tumours and the similarities between species. The shared cancers between humans and dogs that we discuss include sarcoma (osteosarcoma, soft tissue sarcoma, histiocytic sarcoma, hemangiosarcoma), haematological malignancies (lymphoma, leukaemia), bladder cancer, intracranial neoplasms (meningioma, glioma) and melanoma. Tumour risk in other animal species is also briefly discussed. As the field of genomics advances, we predict that comparative oncology will continue to benefit both humans and the animals that live among us.
Keywords: comparative oncology, genomics, genetics, cancer, canine, human
1. Introduction
Comparative oncology is a quickly expanding field that examines both cancer risk and tumour development across species. Characterized by interdisciplinary collaboration, its goal is advancement of both human and animal health. Nowhere has this been more evident than in the investigation and comparison of canine and human tumours. The study of naturally occurring cancers in the domestic dog provides a suitable model for advancement of the understanding, diagnosis and management of cancer in humans [1–4]. There are over 70 million pet dogs in the USA, residing in over 40 million households [5,6]. With over 100 million vet visits each year in the USA, our dogs provide a powerful resource for closely monitored health data. Canine cancers occur spontaneously, and have similar clinical presentation and pathophysiology to equivalent human cancers. As such, they closely parallel the natural progression of human cancer to a greater extent than is observed in induced cancer animal models [7–9]; genomic analysis of canine tumours has revealed shared features between both species, providing fresh insight into the genetic basis of tumour development [10–12]. Additionally, society's practice of dog breeding has unwittingly created a high-risk model for breed-specific disease due to consanguinity and inbreeding [13,14]. The restricted genetic variation in many purebred dog breeds allows for easier identification of the genetic basis of disease, including cancers, and the shorter lifespan of dogs facilitates timely and efficient evaluation of new approaches to cancer diagnosis, treatment and prevention. This review will discuss the domestic dog as a model for comparative oncology, with a focus on both inherited risk and tumour genomics, along with a brief assessment of other species relevant to the field of study.
(a). On the origin of dogs (and their cancer risk)
While there are probably over 400 breeds of dog recognized worldwide, the number of breeds recognized by the dog fancy in different countries varies. For example, in the USA the American Kennel Club (AKC) now recognizes 184 breeds of dog (including 21 varieties) and the United Kennel Club (UKC) recognizes over 300 breeds. The characteristics of each dog of a specific breed have been refined and maintained to meet the stringent breed standards and be considered a contender for selection as a champion.
Over 15 000 years ago, man's relationship with the dog was quite different, being one in which dogs were selected primarily for their function. Dogs helped humans to survive, using their fast pace to aid in hunting of animals and their strength and boldness to serve as protectors. Over the course of subsequent millennia, the relationship between man and dog continued to be one of co-dependency. Around the time of the industrial revolution, we started to breed dogs as much for their form as their function, in an industrializing society. In just the past few hundred years, intensive selection and breeding of dogs for key desirable traits resulted in the development of what we regard today as the major characteristics of the breed. When considered as a species, the level of genetic variation sampled across all dog breeds today is perhaps as extensive as for human populations [15]. In individual pure breeds, however, the level of genetic diversity is now variably restricted [16]. The process of breed formation over just the past 2–3 centuries has been estimated to have resulted in sevenfold greater reduction in genetic diversity than did the thousands of years of early domestication [17]. This is compounded further by the use of popular sires and gene pool decline during the twentieth century. Since many of the key phenotypes are characteristics of the particular breed, their presence in the breed had been positively selected, resulting in high frequency of the genes that cause these specific phenotypes. With such intense selection, it is perhaps not surprising that there are now over several hundred inherited diseases recognized in dogs. While some diseases have simple inheritance patterns, others, including cancers, are likely to be more complex. The genetic background of some purebred dogs may predispose the breed to a higher risk for specific cancers, or cancers in general. It is this increased risk of cancer in purebred dogs that may be leveraged to accelerate the process of cancer gene discovery from a comparative perspective.
The histological and clinical presentation of numerous canine cancers closely parallels that of the corresponding cancers in humans. The extended lifespan of the dog, combined with the shared environment and development of spontaneous cancers, places the dog in a unique position to better reflect cancer development and progression than traditional rodent models. As is the case for human and rodent models, the advancement of genomic technologies and the development of canine custom reagents and resources have facilitated studies of both somatic and inherited genomic variation in the domestic dog. As discussed in §2, canine cancers share evolutionarily conserved genomic changes that are found in their human counterparts. Man's best friend is already providing scientists with an opportunity to generate data beneficial to both species [3,4,10,11,18,19].
2. Germline and cancer risk
Several purebreds of dog are known to have a high incidence and elevated risk of specific cancer subtypes, sometimes even more than one subtype. The high degree of expectation has led veterinarians and pathologists to associate specific cancers with certain breeds of dogs (table 1). Such breed-specific risk reflects the underlying genetics of the different breeds. The patterns of specific cancers found within dog breeds is very reminiscent of the human cancer predisposition syndromes, whereby inherited genetic mutations in humans lead to very specific cancer risks in related children and families [21–25]. In humans, knowledge of specific cancer risk leads to designated early cancer screening approaches to decrease morbidity and mortality [24–29]. High-risk breeds of dog can be thought of as if they carry a hereditary cancer syndrome, although breed-specific screening in asymptomatic dogs has not yet become standard of care. Interestingly, several of the human cancer predisposition genes have been found in the constitutional (germline) DNA of dogs with cancer; this includes BRCA1/BRCA2 germline mutations in dogs [30,31] which leads to hereditary breast and ovarian cancer syndrome in humans and TP53 germline mutations in dogs [32] which lead to Li–Fraumeni syndrome in humans with multiple different cancers.
Table 1.
| cancer subtype | dog breed |
|---|---|
| lymphoma (unspecified) — B-cell lymphoma — T-cell lymphoma |
Old English sheepdog, boxer, pointer, golden retriever, Rottweiler, St Bernard, Scottish terrier, bulldog — Irish wolfhound, Siberian husky, shih tzu, Airedale terrier, Cavalier King Charles spaniel, Yorkshire terrier — boxer, cocker spaniel, basset hound |
| osteosarcoma | large and giant breeds, such as Irish wolfhound, Scottish deerhound, Great Dane, BMD, mastiff, St Bernard, Irish setter, golden retriever, Rottweiler, Dobermann pinscher, greyhound |
| soft tissue tumours | larger dogs, such as boxer, BMD, Airedale terrier, Great Dane, St Bernard, basset hound, golden retriever—all with twice as many as the general canine population |
| hemangiosarcoma | German shepherd, BMD, golden retriever, flat-coated retriever, Portuguese water dog, Labrador retriever, boxer, Skye terrier, Australian shepherd |
| hs/malignant histiocytosis | BMD, flat-coated retriever, Rottweiler, golden retriever |
| mast cell tumours | boxer, pug, Labrador retriever, golden retriever, vizsla |
| meningiomas | mesocephalic (medium) and dolichocephalic (long)-nosed breeds, e.g. Labrador, golden retriever, collies |
| gliomas (including glioblastoma multiforme) | brachiochephalic (short-nosed) breeds, including boxers, bulldogs and terriers |
| testicular seminoma | Norwegian elkhound |
| nasal cavity carcinoma | golden retriever, beagle, Boston terrier, rough collie, Belgian shepherd |
| UC | Scottish terrier, beagle, West Highland white terrier, Shetland sheepdog, American Eskimo dog, standard schnauzer |
| lower urinary tract carcinoma | Airedale terrier, beagle |
| squamous cell carcinoma (digit) | STPO, giant schnauzer |
| melanoma — oral melanoma — cutaneous melanoma |
— poodles — schnauzers, beauce shepherds |
In addition to known deleterious mutations, more common single nucleotide polymorphisms (SNPs) and copy number variations (CNVs) (or combination of SNPs and CNVs) have been associated with disease risk in specific dog breeds. The approach of using genome-wide association studies (GWAS) in humans to identify disease-risk alleles has been relatively successful, with an explosion in published GWAS data over the past decade [33,34], although admittedly, revealing varying levels of clinically significant disease risk [35,36]. In dogs, the rewards of GWAS can be more readily realized owing to the genetic homogeneity readily found within dog breeds [15,37]. Indeed, this has been the case for several recent GWAS findings in canine cancer.
One of the first cancer GWAS to evaluate a canine cancer explored the risk of histiocytic sarcoma (HS) in the Bernese mountain dog (BMD) [38]. This exceedingly rare sarcoma in humans occurs frequently in a few breeds of dog, including BMD, flat-coated retriever, Rottweiler and golden retriever. The lifetime risk of developing a HS in BMDs is 15–25% [38,39]. The authors performed the first GWAS in 111 BMD cases and 117 BMD controls from North America, as well as a second GWAS in 125 BMD cases and 117 BMD controls from Europe. Independent and combined analyses identified a significant risk allele on dog chromosome (CFA) 11 (North American BMD: Praw = 1.41 × 10−9, European BMD: Praw = 1.50 × 10−6, combined BMD: Praw = 1.11–10−13). Follow-up fine mapping and targeted sequencing revealed a shared haplotype in 96% of affected BMDs that included MTAP and part of CDKN2A (two very well-described cancer genes in humans). The exact mechanism leading to HS is still under investigation. Humans with germline MTAP mutations have been reported and these individuals develop diaphyseal medullary stenosis with malignant fibrous histiocytoma (a soft tissue sarcoma) [40]; humans with germline CDKN2A mutations develop melanoma and pancreatic cancer [41,42]. Clearly, the story of HS risk in dogs is only just beginning, and when combined with tumour genomics (see §3), there is still much to be learned to the benefit of both dogs and their human owners.
The same group also investigated the very specific increased risk for squamous cell carcinoma of the digit (SCCD) found to be especially prevalent in standard poodles (STPOs) [43]. This aggressive cancer causes lytic bone lesions of a focal nature, but interestingly, SCCD almost always occurs in dark-coated STPOs and rarely in their light-coated counterparts. The authors performed a GWAS, comparing 31 SCCD cases in STPOs to 34 unrelated black STPO controls. The GWAS identified a statistically significant peak SNP localized to CFA 15 (Praw = 1.60 × 10−7). Further genotype mapping discovered a minimal region of less than 30 kilobases (kb) that contained the KIT Ligand (KITLG) gene locus with a CNV with at least four copies due to a tandem repeat. Since both the light-coloured and dark-coloured STPOs carry this KITLG tandem repeat, a subsequent GWAS compared light and dark STPOs (N = 24 versus 24) and identified the MC1R locus to be the only difference between the groups (p = 2.52 × 10−15). The study authors concluded that mutations found with MC1R may actually be cancer protective for light-coloured STPOs, and that MC1R may be interacting with KITLG CNVs as a genomic modifier. In humans, KITLG is not associated with cancer risk, although rare germline KIT mutations have been associated with familial gastrointestinal stromal tumours (GIST), a type of gastrointestinal sarcoma [44]. Interestingly, there is precedent for the role of MC1R as a modifier of disease, as germline MC1R variants in humans affect pigmentation [45,46] and seemingly function as genomic modifiers for several human diseases [47–51], in one study conferring risk for BRAF-mutant melanoma [52].
A third canine cancer GWAS has been performed to identify risk of osteosarcoma. Using a study cohort comprising 492 dogs as 261 cases and 231 controls across three breeds (greyhounds, Rottweilers and Irish wolfhounds), the GWAS identified 33 different inherited risk loci that could explain 55–85% of the phenotype variance in each breed [53]. The strongest association within the greyhounds was within a 150 kb segment upstream from CDKN2A/B, one of the most highly rearranged genes detected in canine osteosarcoma tumour cells. This study revealed a polygenic series of germline-risk factors that collectively highlighted specific pathways as drivers of disease. Consistent with the biology of the tumour, the candidate regions were enriched for genes in bone differentiation and growth pathways. A GWAS in human osteosarcoma also was recently published by Savage et al. [54], but with different findings than in the dog; they analysed 941 individuals with osteosarcoma and 3291 cancer-free adult controls of European ancestry and found a very high statistical association with disease risk in GRM4 located at 6p21.3 (p = 8.1 × 10−9) and two SNPs in the gene desert at 2p25.2 (p = 1.0 × 10−8 and 2.9 × 10−7). As Machiela & Chanock [37] discuss, although GRM4, which is involved in intracellular signalling and inhibition of the cyclic AMP signalling cascade, was not found to be associated itself with canine osteosarcoma, GRIK4, another glutamate receptor, was significantly associated with osteosarcoma risk in greyhounds; also, the SNPs close to GRM4 were fixed in Rottweilers with osteosarcoma [53]. This comparative approach may provide novel insight and validation of signalling pathways associated with osteosarcoma risk in both humans and dogs. More comparison work is currently underway.
The most recent canine cancer GWAS explored the genetic risk for haematologic malignancy in the dog, specifically B-cell lymphoma and haemangiosarcoma in golden retrievers [55]. These cancers occur at high rates in this breed (B-cell lymphoma 6% and haemangiosarcoma 20%), and the investigators looked at 148 haemangiosarcoma cases versus 41 B-cell lymphoma cases versus 172 cancer-free controls. When the results from each GWAS were combined, two associated loci were identified on CFA 5 and contributed nearly 20% of the risk for haemangiosarcoma and B-cell lymphoma (4.6 × 10−7 and 2.7 × 10−6, respectively). Whole genome sequencing (WGS) of nine cases and controls discovered risk haplotypes without coding changes. The authors investigated gene expression in B-cell lymphoma tumours and concluded that these germline-risk alleles affect T-cell regulatory pathways and immune-mediated responses. Based on their canine GWAS findings, the authors conclude that the immune system and malignant cells may interact in tumour risk and development [55], a hypothesis that now can be explored in other haematologic malignancy models and even primary tumours in humans. In support of this concept, recent human GWAS in diffuse large B-cell lymphoma [56], follicular lymphoma [57] and marginal zone lymphoma [58] have all demonstrated a role for immune recognition and immune function, with lymphoma risk strongly linked to HLA and other regions [56–58].
A striking and recurrent theme in the analysis of canine cancer GWAS data compared to comparable human GWAS is the ability to analyse a much lower number of canine cases and controls to identify risk factors compared to human studies of identical cancers. The actual number of cases and controls depends on a variety of factors, but, in general, it is evident that the restricted level of genetic heterogeneity in purebred dogs provides opportunities for an efficient means to identify risk loci for cancers that may be of comparative value to human studies.
3. Somatic and tumour genomics
Identification of risk for developing cancer is a key factor in health management of the individual and the population. Once constitutional changes to the canine genome have been determined, these data offer a powerful approach to screen and stratify populations by risk. Within purebred lines, knowledge of risk will play a key role in selecting more informed breeding programmes designed to reduce carefully the frequency of deleterious alleles in the population. The inherited risk of a cancer may not be directly related to the genome dysregulation associated with initiation and/or propagation of tumour cells, and so be of little to no value in making decisions about patient care once a cancer develops. Genome-wide assessment of changes at the somatic level is therefore another key area of active research in comparative oncology. Identification of recurrent genome changes in cancer cells is a key approach to identifying potential targets for therapeutic intervention. The introduction of molecular genetic tools has revolutionized the way we are able to interrogate cancer cells to identify specific changes in gene dosage, organization and regulation. Numerical and structural changes to the genome have been identified in over 65 000 cases of human cancer, representing over 70 different types of cancer (see http://cgap.nci.nih.gov/Chromosomes/Mitelman). Many of these recurrent chromosome aberrations initially were associated with histopathological or immunological subgroups, leading to their use as diagnostic signatures. More recently the cytogenetic status of tumour cells has been demonstrated to be of established clinical value for prognosis, guiding therapy and assessing remission for a range of cancers, including ovarian cancer [59], colorectal carcinoma [60,61], gliomas [62], melanoma [63,64] and breast carcinoma [65]. From the perspective of comparative oncology, we may consider all animals to be differentially organized collections of the same collection of ancestrally related genes. As such, assessment of changes to genome architecture in canine cancers has been an active area of research, aimed at identifying regions of genome aberration shared between canine and human cancers, suggestive of a conversed mechanism of pathogenesis.
(a). Sarcomas
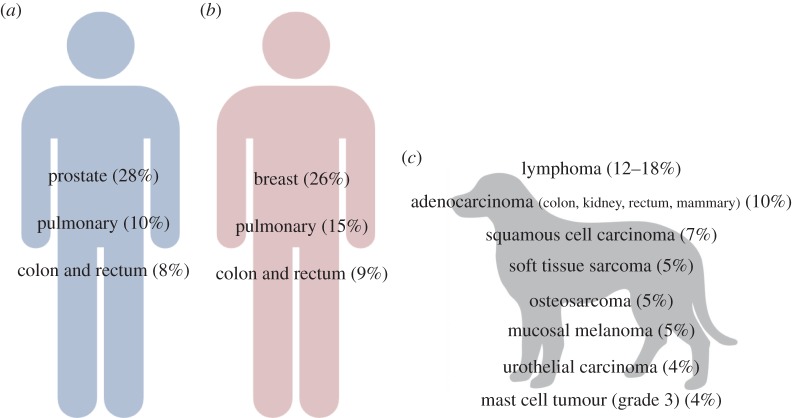
In the USA, it is estimated that approximately 500 new cases of cancer are diagnosed per 100 000 of the population each year (http://seer.cancer.gov), whereas in dogs the estimate is approximately 5300 cancer diagnoses/100 000 population (AVMA, 2011). These figures are striking in that they indicate that 2.5× the number of cancers are diagnosed in pet dogs each year and that this represents an incidence of over 10× that of the human population with which they live. Approximately 50% of all human cancer-related deaths in the USA in 2014 were the result of just four types of cancer, all of which were carcinomas; pulmonary, prostate and colon carcinoma in males, and breast, pulmonary and colon carcinoma in females (figure 1). While accurate numbers of cancers in pet dogs are not known, extrapolation from extensive academic records suggests that almost 60% of the estimated 1 500 000 diagnosed malignant cancers in dogs in the USA each year are represented by a combination of sarcomas and carcinomas (figure 1). The apparent lower numbers of pulmonary and colon carcinoma in pet dogs may in part be due to the human-specific influences (diet and smoking) affecting rates of these prevalent cancers, both of which are associated with factors not necessarily shared with pet dogs. Considering some of the most prevalent cancers in human, the corresponding canine cancers are highly evident also (figure 2) indicating the potential for the role of the dog as an appropriate biological model system.
Figure 1.
Frequency of cancer-related deaths in human and dog. Approximately 50% of all human cancer deaths in the USA each year are a consequence of four cancers, two of which are common to males (a) and females (b) (human data from (http://www.cancer.org)). (c) The estimated annual frequencies of death-related malignant cancers in pet dogs in the USA are shown (extrapolated canine data from US academic accessions). (Online version in colour.)
Figure 2.
The remarkable similarity in cancers shared by human and dog. The cancers shown are found in both human and canine populations, and several ongoing studies are highlighting the similarities at the genomic level. The incidence of cancers in both species is shown, highlighting the more than 2.5×number of cancers diagnoses in pet doges each year. Comparative oncology is a growing trans-disciplinary field that harnesses these data, adding evidence to support a shared pathogenesis. (Online version in colour.)
(i). Osteosarcoma
Osteosarcoma is the most common human bone tumour in adolescents and young adults. This aggressive bone tumour has been associated with germline TP53 mutations (Li–Fraumeni syndrome), among other hereditary cancer syndromes [66]. With the introduction of combination chemotherapy and multimodal approaches, the cure for human osteosarcoma has risen to nearly 70%, although it still remains at less than 20% for relapsed or recurrent disease [67,68]. Although relatively rare in humans, with up to 75 000 cases of canine osteosarcoma diagnoses each year in the USA, the rate is as high as 75× that of humans. Breeds considered at high risk of developing canine osteosarcoma tend to be some of larger and giant breeds, including the Rottweiler, Great Pyrenees, mastiff, Dobermann pinscher, Irish wolfhound, Scottish deerhound. Given its frequency in canines, the dog model of naturally occurring osteosarcoma has offered an unparalleled opportunity to understand the genomic origins of osteosarcoma, to learn about the role of metastasis in disease and to pilot new investigational drugs in trials that would otherwise take too long to accrue in humans [12,69]. Several years ago, Scott et al. [70] took advantage of the genetic homogeneity in dogs to identify molecular subtypes of osteosarcoma based on genome-wide gene expression profiling in a cohort of high-risk breeds of dogs with osteosarcoma (N = 79). In this study, the authors were able to divide samples into two distinct subgroups based on gene expression that also correlated with overall survival. Similar to human osteosarcoma, the two canine groups involved ‘G2/M transition and DNA damage checkpoint’ and ‘microenvironment-interaction’. As the authors conclude, the genomic findings in canine osteosarcoma are of benefit for both dogs and humans, and further investigation ‘may enhance prognosis and prediction, and identify relevant therapeutic targets' [70].
In a parallel study, Angstadt et al. [71] used high-resolution oligonucleotide aCGH to interrogate copy number alterations (CNAs) in canine osteosarcoma (N = 23, various breeds) and compared the data directly with cases of human osteosarcoma (N = 15). The authors demonstrated that even though osteosarcoma is a cancer with enormous genomic instability, the identification of common regions of conserved genomic alterations between species provides a means to narrow the search for genomic drivers versus passengers. This study identified shared regions of CNAs that were less than 500 kb in size between dog and humans and then interrogated those regions for orthologous osteosarcoma-associated genes. Genes with known association in osteosarcoma were revealed (CDC5L, MYC, RUNX2 and CDKN2A/CDKN2B) as well as new genes that had not been described previously in osteosarcoma (ADAM15, CTC1, MEN1, CDK7, along with several more genes) [71].
Primarily a cancer of children and young adults, osteosarcoma is rare, with fewer than 1000 cases each year diagnosed in the USA. As such, a major challenge to the progress of human osteosarcoma clinical trials is low patient accrual. The high number of canine osteosarcomas diagnosed each year, combined with the remarkable motivation of dog owners to enroll their pets in clinical trials offers a tremendous opportunity to advance studies for this cancer. The net result is that both biological and clinical studies can be performed with pet dogs that would otherwise take an inordinately long period of time to accomplish in humans. In the study of osteosarcoma progression and metastasis, key advances have been made by the Comparative Oncology Program at the National Institutes of Health (NIH) through the study of dogs, including the recognition of the role of ezrin [12,72–75]. Furthermore, early clinical trials are now ongoing in dogs in conjunction with the Children's Oncology Group in North America, an approach that will quickly ascertain the effectiveness of osteosarcoma therapy, owing primarily to the much larger number of dogs with osteosarcoma compared with humans [12,72,76–78]. With time and additional canine trials, researchers believe the success of this approach will become readily apparent.
(ii). Soft tissue sarcoma
In 2013, Caserto published a comprehensive review of canine and human rhabdomyosarcoma (RMS) with an emphasis on comparing the classification and pathogenesis of this soft tissue sarcoma [79]. Not surprisingly, there were numerous similarities between RMS in both species, including their histological classification. Caserto described the ability to diagnose RMS in dogs using immunohistochemical (IHC) stains for desmin, α-actins, myogenin and MyoD1, in addition to electron microscopic identification of sarcomeric structures [79]. More recently, Milovancev et al. [80] have described non-RMS soft tissue sarcomas in dogs and their comparison to human non-RMS tumours. In this study, veterinary and human pathologists interpreted 32 clinically archived, canine soft tissue sarcomas. There was general agreement on the diagnoses and their similarities to human tumours, including low-grade spindle cell sarcomas (N = 13/32), undifferentiated sarcoma (N = 32), liposarcoma with atypical desmin-positive epithelioid cells (N = 5/32), spindle cell sarcoma with myxoid features (N = 5/32) and myxofibrosarcoma (N = 2/32). The comparative genomics of soft tissue sarcomas, both RMS and non-RMS, is a field that will continue to grow as technology advances and more of these rare tumours are collected in canines.
(iii). Histiocytic sarcoma
Canine HS is a very aggressive and rapidly fatal sarcoma of dendritic cell origin with a tendency to metastasize to the spleen and lungs. This cancer is very commonly reported in just two less abundant breeds of dog, the BMD and flat-coated retriever, and also in large numbers of three of the most popular breeds, including the Rottweiler, golden retriever and Labrador retriever [81–83]. The BMD is extraordinarily susceptible to this sarcoma, with as many as 25% of the breed diagnosed at a young age [38,39]. In humans, the equivalent of HS is not quite so clear; human HS is a very rare tumour, presents in lymph nodes, skin, and the gastrointestinal tract, often will be misdiagnosed as non-Hodgkin lymphoma (NHL), and is considered a diagnosis of exclusion [84,85]. As is the case with osteosarcoma, comparative oncology of HS offers a unique opportunity to leverage data from the canine disease to learn more about a poorly understood human cancer [86].
Hedan et al. [86] used molecular cytogenetic profiling with aCGH in spontaneously occurring HS from BMDs and flat-coated retrievers (N = 104 total) [86]. The data indicated a high level of genome disruption and instability, and the investigators identified that many of the highly recurrent CNAs were shared between both breeds, suggesting a common underlying pathogenesis. Specially, recurrent deletions were detected in commonly described human tumour suppressor genes, including CDKN2A/B, RB1 and PTEN [86]. Interestingly, the GWAS of canine HS in BMDs (see above) localized a possible HS-risk allele also to CDKN2A (in addition to MTAP) [38]. Hedan et al. [86] also identified several private CNAs unique and recurrent to each breed, perhaps suggesting a difference in initiation and/or progression leading to a more universal HS genomic phenotype once the tumour develops. Molecular work on canine HS continues, and this hopefully will inform our understanding and treatment approaches to human HS.
(iv). Haemangiosarcoma
Angiosarcomas and haemangiosarcomas are an aggressive group of tumours that have been studied extensively in both canines and humans. Initially thought to be of endothelial cell origin [87,88], there is now suggestion that these aggressive sarcomas of blood vessels may be of haematopoietic origin [89–91]. In humans, angiosarcomas are a very rare, heterogeneous subgroup of soft tissue sarcoma, representing much less than 1% of all tumours. These malignant vascular tumours occur spontaneously as primary tumours and also as secondary tumours following radiation therapy or in the context of chronic lymphoedema [92,93]. Almost half of angiosarcomas occur in the head and neck, though these represent less than 0.1% of all head and neck malignancies [94]. Insidious in nature, clinical symptoms of angiosarcomas often do not manifest until the disease is well advanced. In the dog, haemangiosarcomas frequently involve major vascular organs, (spleen, liver, heart), and also can be subcutaneous. As in humans the tumours are indolent and often remain undetected until the point at which the vascular mass ruptures and the dog suffers internal bleeding that can result in death. While haemangiosarcomas can affect any breed, those of notably high risk include the golden retriever, German shepherd, Portuguese water dog and Australian shepherd.
Gorden et al. [95] performed genome-wide expression profiling of a small number of primary canine haemangiosarcomas (N = 6 golden retrievers, N = 1 Rottweiler, N = 1 golden retriever × Great Pyrenees, N = 1 Portuguese water dog) and determined three distinct haemangiosarcoma subtypes associated with: angiogenesis (group 1), inflammation (group 2) and adipogenesis (group 3) [95]. The authors investigated haemangiosarcoma cell lines and discovered expression of multiple and distinct markers for early endothelial, haematopoietic and myeloid cells along with several phagocytic and adipogenic functional experiments [95], further supporting the multipotent potential and origin of this blood vessel sarcoma. Genome-wide DNA copy number profiling of canine haemangiosarcoma has also been performed using high-resolution oaCGH [96]. In this study, primary intra-abdominal haemangiosarcomas (N = 75; golden retriever (n = 40), Australian shepherd (n = 10), German shepherd (n = 10), flat-coated retriever (n = 9) and BMD (n = 6)) were assessed and the data revealed a relatively low rate of CNAs with small amplitudes. Despite the presence of multiple passenger alterations, potential recurrent driver alternations were seen in CDKN2A, VEGFA and SKI. Interestingly, VEGFA gains were observed at nearly half the rate in golden retrievers compared with the other breeds (22 versus 40%) [96]. These data support an alternative origin of tumorigenesis among breeds and multiple haemangiosarcoma molecular subtypes owing to alternative activating pathways as demonstrated in the gene expression studies. Further work comparing canine haemangiosarcoma to human angiosarcoma is now awaited.
(b). Haematological malignancies
(i). Lymphoma
Haematological malignancies are common forms of cancer in both dogs and humans. In 2014, approximately 71 000 new cases of human NHL were diagnosed in the USA (http://www.cancer.org). In the domestic dog, key opinion leaders estimate that over 250 000 cases of canine lymphoma (comparable to NHL) were diagnosed in the same period. The high degree of similarity between human and canine lymphoma has been extremely helpful in understanding this disease in both species [97–99]. The high similarity in pathologic presentation of canine and human lymphoma allows for use of World Health Organization (WHO) criteria for accurate and reproducible classification of the canine tumours [100,101]. In human NHL, the vast majority (approx. 90%) of cases are of B-cell origin. In the domestic dog, the proportions of B- and T-cell lymphomas across all dogs is estimated to be approximately 2 : 1, but the immunophenotype prevalence in individual breeds is highly variable [102]. A large study of canine lymphoma characterized 608 cases based on cytomorphological, histomorphological and immunological criteria and importantly included both epidemiological and clinical data [103]; the majority of tumours (76%) were classified as high-grade malignant lymphomas, highlighting the clinical importance of understanding both lymphoma risk and designing novel therapeutic approaches. As is the case across all breeds, approximately two-thirds of the canine lymphomas in this cohort were classified as B cell (CD3–, CD79a+) with one-third classified as T-cell lymphomas (CD3+, CD79a–) [103].
Diffuse large B-cell lymphoma (DLBCL) has been one subtype of canine lymphoma that has been very well studied with genomic profiling. Richards et al. used IHC and gene expression profiling on canine DLBCLs (N = 49) and identified similar profiles to human DLBCL, including activation of NF-κB pathway genes and immunoglobulin heavy chain alterations [104]. Although some differences with human lymphoma existed (lack of BCL6 and MIM1/IRF4 protein expression), the authors concluded they could identify germinal centre and post-germinal centre subtypes in canine DLBCL, including different survival times, and that their canine findings reflect human DLBCL [104]. Using a bivariate mixture model based on two-species data, Su et al. [105] were also able to use gene expression profiling to distinguish both the germinal centre B-cell-like DLBCL and the activated B-cell-like DLBCL, including different clinical outcomes based on survival. Another study used gene expression profiling on 35 canine lymphoma samples to define three major groups: (i) low-grade T-cell lymphoma, composed entirely by T-zone lymphoma; (ii) high-grade T-cell lymphoma, consisting of lymphoblastic T-cell lymphoma and peripheral T-cell lymphoma not otherwise specified and (iii) B-cell lymphoma, consisting of marginal B-cell lymphoma, diffuse large B-cell lymphoma and Burkitt lymphoma [106]. Similar to the DLBCL studies, the genomic subtypes were associated with different clinical outcomes. Remarkably, the authors distilled their gene expression profiling into four genes whose expression could reliably predict lymphoma subtype and survival (CD28, ABCA5, CCDC3 and SMOC2) [106]. These findings now need to be confirmed in larger studies in human lymphomas to assess their universal clinical utility.
(ii). Leukaemia
Leukaemia is another haematological malignancy with shared high incidence in dogs and humans. Genomic studies in canine leukaemia are underway, although recent results display similar mechanisms driving leukaemogenesis. RB1 deletions in chronic lymphocytic leukaemia and BCR–ABL fusion in chronic myeloid leukaemia (CML) were among the first cytogenetic aberrations detected in canine cancers that mirror the corresponding human cancers [107]. The BCR–ABL tyrosine kinase translocation (the so-called ‘Raleigh chromosome’ in dogs and ‘Philadelphia chromosome’ in humans) has since been demonstrated to be present in additional subtypes [108,109] and proven useful for monitoring cytogenetic remission in CMLs [110]. Another canine study included acute lymphoblastic leukaemia (ALL)/acute undifferentiated leukaemia (AUL) (N = 11) and chronic lymphocytic leukaemia (CLL) (N = 12) and demonstrated increased c-KIT expression in the ALL/AUL samples [111], offering the possibility of using tyrosine kinase inhibitors as a treatment option in canine leukaemia, an approach similar to that used for human leukaemia with tyrosine kinase-affected pathways.
(c). Bladder cancer
Bladder cancer, also called transitional cell carcinoma (TCC), or urothelial carcinoma (UC) is yet another tumour that affects both humans and their pet dogs [112,113]. In humans, environmental exposure, including tobacco smoke, is a major risk factor for UC. In dogs, however, the risk for UC appears to be mostly genetic through predisposition [13]. Similar to osteosarcoma, the molecular genetics/genomics of canine UC reflect human UC, and the knowledge gained through canine therapeutic trials in UC benefits both dogs and humans [114]. A recent study explored aCGH in canine UC (N = 31), compared canine data with comparable results in human TCC (N = 285) and discovered both large chromosomal and focal regions of alterations shared between the species even among a large amount of background genomic instability [18]. Given the strong environmental risk for UC in humans and the prominent genetic risk in dogs, the study of shared genomic events leading to UC tumorigenesis will prove important for both prevention and treatment across species. The introduction of sea lion genomic data for sea lion specific UC will further hone candidate genes for clinical targeting (see §4).
(d). Intracranial malignancies
The clinical and histological presentation of intracranial malignancies affecting human and dog are highly comparable, allowing similar diagnostic criteria to be used from both species [115,116]. In support of the role of the pet dog as a model for such cancers, canine and human intracranial tumours share key histopathological features that are absent in rodent models [117]. There is considerable potential to exploit the dog as a preclinical model for development and evaluation of novel brain tumour therapies [118]. It is widely accepted that short-nosed (brachycephalic) breeds, such as boxer, pug and bulldog, are predisposed to gliomas [7,119,120], while longer nosed (dolichocephalic) breeds, such a collies, golden retriever and Labrador retriever, are more predisposed to meningiomas [11].
(i). Meningioma
While meningiomas represent approximately 25% of adult primary intracranial tumours in human patients [116], they are estimated to comprise almost 40% of all canine intracranial neoplasms [121,122], which largely reflect the enormous popularity of several medium/long-nosed dog breeds. The elevated incidence of meningiomas in the dog, as compared to human populations is of particular concern in veterinary medicine and simultaneously offers a higher caseload to investigate meningioma biology. Meningiomas in both species are highly comparable and share similar phenotypes and gene expression profiles [116,121,123]. Genomic profiling of canine meningiomas for DNA copy number by Thomas et al. [11] indicated that canine meningiomas share a limited extent of whole chromosome aneuploidy with their human counterpart. Importantly, the authors hypothesized that by considering the DNA copy number data from both species, the shared aberrations should be the focus of research; for example, in this study they were able to reduce the size of conserved genomic segments by as much as 50-fold. Interrogation of the minimally shared regions revealed genes of interest that are now being investigated for their functional significance in meningioma biology.
(ii). Glioma
Intracranial gliomas are the most common and lethal primary brain tumours in both the human and canine populations. These tumours are particularly detrimental to the paediatric age group, accounting for 80% of malignant brain tumours in children less than 18 years old. In human adults, gliomas progress from low-grade (I–II) tumours to high-grade (III–IV) tumours and are often rapidly fatal once detected, with long-term survival in grade IV glioblastoma multiforme (GBM) averaging less than 1 year [124]. The molecular genetics of human GBMs have been studied in depth [125,126], with three main pathways identified as RTK/RAS/PI-3K, p53 and RB signalling [125]. Human patients often present with neurological symptoms such as headaches and seizures, and despite the vast amount of genomic data and novel therapies now available, GBMs recur quickly and aggressively after resection and often lead to death [124]. Several of the human cancer predisposition syndromes include gliomas in their clinical spectrum [127–133], supporting the genetic risk for development of these tumours.
Among canines, gliomas are the second most common brain tumour behind meningiomas and occur with the highest frequency in brachycephalic dog breeds [7,119,120]. Published and anecdotal incidences of brain tumours in boxers, mostly gliomas, range from 5 to 25% incidence [119,120], while some individual pedigrees can have even higher presentation rates. Similar to humans, an untreated glioma in a dog will rapidly progress to a stage IV GBM. Previous cytogenomic studies of canine intracranial malignancies by Thomas et al. [11] revealed genomic architecture similar to the human counterparts. Similar to humans, survival in dogs with gliomas is extremely poor despite treatment ranging from chemotherapy, radiation therapy, hyperthermia, to gene and vaccine therapy [134–138]: most dogs (and humans) will present with neurological symptoms such as seizures [139] and unfortunately die within months of diagnosis.
Genomic analyses of human gliomas reveal increasing molecular complexity as the clinical tumour stage progresses, along with accumulation of specific driver mutations [125,126,140]. Our own genomic data demonstrate very distinct patterns of genome-wide instability when measuring CNAs in human GBMs (JD Schiffman 2015, unpublished data). Genomic analysis of canine GBMs reveals a very similar pattern [11]. The comparison of genomic changes in canine versus human GBMs, and early- versus late-stage tumours, will permit the continued identification of drivers versus passenger mutations.
(e). Melanoma
Human melanomas are the most common malignant skin cancers, often occurring in sun-exposed areas due to UV exposure [141,142], although the rarer mucosal melanomas also can occur in people [143–145]. In humans, as described previously (§2), germline mutations in CDKN2A can lead to familial melanoma with presentation at a young age regardless of sun exposure [42]. Dogs also develop melanoma, and similarly to all the other cancers discussed in this review, these melanocytic canine tumours will very closely resemble human melanoma, emphasizing the beneficial role of the canine preclinical model in studying both UV and non-UV pathways in melanoma [20,146]. Furthermore, the dog has been extremely useful for clinical trials and has contributed to a phase I study for DNA vaccination with xenogeneic human tyrosinase for advanced malignant melanoma [147–151]. Poorman et al. [152] used aCGH profiling to compare cutaneous melanomas (often benign) with the more aggressive oral mucosal form. Distinct patterns of CNAs emerged in the malignant tumours including recurrent gains of dog CFA 13 and 17 and loss of CFA 22, whereas the more benign tumours were more copy number neutral, presenting fewer CNAs (except for recurrent gain of CFA 20q15.3–17) [152]; this pattern resembles that reported with human melanoma, where malignant melanoma can be differentiated from benign nevi using genomic microarray based on number of CNAs [153]. Canine mucosal melanomas display specific and unique ‘sigmoidal patterns' of copy number loss followed immediately by a gain on CFA 30q14, a characteristic feature conserved on HSA 15 in human mucosal melanoma [154]. In addition, both species show numerous other CNAs including frequent gain of c-MYC and deletion of CDKN2A [152]. Other recent studies have shown microRNA (mi)R-203 to be a common tumour-suppressive miRNA in both human and canine melanoma cell experiments [152]. Clearly, the study of canine melanoma, especially mucosal melanoma, offers an opportunity to understand melanoma biology and rapidly translate that information into both veterinary and medical clinical care.
4. Other animals besides dogs (sea lions, whales, bats and naked mole rats)
In addition to canines, many other animals offer their own unique opportunity to better understand the universal processes involved in tumorigenesis. Some of these animals, such as dogs, are more prone to develop cancer, while others seem to be resistant to its development. By studying both ends of the cancer-risk spectrum, it is hoped that the knowledge gleaned will help with both the treatment and prevention of cancer.
For over half a century, marine mammals have been widely accepted as highly suited sentinels for assessing the health of the world's oceans. The typical lifespan of marine mammals is sufficient to allow assessment of the development of numerous chronic diseases, including cancers [155,156]. The number of marine mammals diagnosed with cancers has been increasing over the past 30 years. Of the 500 live adult and sub-adult free-ranging California sea lions (CSLs) that strand on the west coast of the USA each year, 200–300 are admitted to the Marine Mammal Center in Sausalito, California for rehabilitation. Necropsy data indicate that approximately 20% have neoplasms, of which 85% are aggressive, widely metastatic genitourinary carcinomas [157,158]. While the gamma herpes virus, otarine herpesvirus-1 was initially thought to play a key role in urogenital tumour pathogenesis [159,160], this virus has been reported in the genital secretions of healthy sea lions [161], questioning the role of the virus in cancer pathogenesis. Of key concern for all top-trophic predators (including the sea lion) is the bioaccumulation of high levels of persistent fat-soluble organic contaminants (OCs), including PCB and DDT. Accumulation of high concentrations of these and other persistent OCs are known to cause immune suppression, hormonal and metabolic disruption and genotoxicity leading to cancer [162]. Such compounds remain as major contaminants along the west coast of the USA and continue to pose a serious health threat to the CSL and other marine mammals. Assessment of the levels of DDT and PCB in CSLs with and without genitourinary carcinoma revealed that there are statistically higher levels of both of these OCs in the cancer patients [162]. These cancers, which are not a feature of captive bred and maintained CSLs, are progressive and cause slow painful death in wild animals. Initial genomic investigation of these aggressive urothelial neoplasms, as part of the larger ongoing work by the Sea Lion Cancer Consortium has revealed that they share conserved features with both canine and human counterparts (M Breen 2015, personal communication). We propose that a more detailed investigation of shared cancers using this multi-species approach will highlight genes associated with uroethial carcinogenesis in the context of risk related to both genetics and environmental exposure.
On the opposite side of tumour risk, bats may be protected against cancer (although anecdotal, this seems to be a common consensus in the field) [163]. Naked mole rats, another cancer-free rodent, with an impressive 30-year lifespan, have evolved hypersensitivity to cellular contact inhabitation mediated through alternative INK4a/b splicing and very high molecular weight hyaluronan [164–166]. Bowhead whales, the longest living mammal with a lifespan of over 200 years and reportedly very low cancer rates, recently were described to harbour genomic alterations associated with cancer, ageing, cell cycle and DNA repair (e.g. ERCC1 and PCNA) [167]. Elephants represent another large mammal that appears to be protected from cancer [168,169], and various studies are underway to explain the molecular basis for this phenomenon known as Peto's paradox (large and long-lived animals that appear to be cancer resistant) [170–175].
The study of comparative oncology truly embraces all cancer risks, great and small, including humans and all types of animals, wild and domesticated. Working in a transdisciplinary setting, colleagues provide expertise across the basic sciences, medical oncology, tumour biology, pharmacology, evolutionary biology, epidemiology, patient care, drug development, clinical trials and a series of other key disciplines. Whether the primary research focus of the individual is to seek benefit for the human or animal patient, the combined goals of the field are to advance our overall understanding of oncology and translate this towards improving the health and welfare of all animals affected by cancer.
Competing Interests
We declare we have no competing interests.
Funding
We gratefully acknowledge support of Skippy Frank Fund for Life Sciences and Translational Research/Rockefeller Philanthropy Advisors (awarded to M.B./J.D.S.). J.D.S. receives support through the Primary Children's Hospital (PCH) Pediatric Cancer Program funded by the Intermountain Healthcare Foundation and the PCH Foundation. J.D.S. hold the Edward B. Clark, MD Chair in Pediatric Research at the University of Utah, and M.B. holds the Oscar J. Fletcher Distinguished Professorship of Comparative Oncology Genetics at North Carolina State University.
References
- 1.Khanna C, et al. 2006. The dog as a cancer model. Nat. Biotechnol. 24, 1065–1066. ( 10.1038/nbt0906-1065b) [DOI] [PubMed] [Google Scholar]
- 2.Rowell JL, McCarthy DO, Alvarez CE. 2011. Dog models of naturally occurring cancer. Trends Mol. Med. 17, 380–388. ( 10.1016/j.molmed.2011.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez CE. 2014. Naturally occurring cancers in dogs: insights for translational genetics and medicine. ILAR J. 55, 16–45. ( 10.1093/ilar/ilu010) [DOI] [PubMed] [Google Scholar]
- 4.Ostrander EA, Franklin H. 2012. Both ends of the leash—the human links to good dogs with bad genes. N. Engl. J. Med. 367, 636–646. ( 10.1056/NEJMra1204453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US Pet Ownership Statistics 2015. See http://www.avma.org/KB/Resources/Statistics/Pages/Market-research-statistics-US-pet-ownership.aspx (28 April 2015).
- 6.AVMA. 2012. US Pet Ownership and Demographics Sourcebook. Schaumburg, IL: American Veterinary Medical Association. [Google Scholar]
- 7.Stoica G, Levine J, Wolff J, Murphy K. 2011. Canine astrocytic tumors: a comparative review. Vet. Pathol. 48, 266–275. ( 10.1177/0300985810389543) [DOI] [PubMed] [Google Scholar]
- 8.Pinho SS, Carvalho S, Cabral J, Reis CA, Gartner F. 2012. Canine tumors: a spontaneous animal model of human carcinogenesis. Transl. Res. 159, 165–172. ( 10.1016/j.trsl.2011.11.005) [DOI] [PubMed] [Google Scholar]
- 9.Antuofermo E, Miller MA, Pirino S, Xie J, Badve S, Mohammed SI. 2007. Spontaneous mammary intraepithelial lesions in dogs—a model of breast cancer. Cancer Epidemiol. Biomarkers Prev. 16, 2247–2256. ( 10.1158/1055-9965.EPI-06-0932) [DOI] [PubMed] [Google Scholar]
- 10.Cadieu E, Ostrander EA. 2007. Canine genetics offers new mechanisms for the study of human cancer. Cancer Epidemiol. Biomarkers Prev. 16, 2181–2183. ( 10.1158/1055-9965.EPI-07-2667) [DOI] [PubMed] [Google Scholar]
- 11.Thomas R, et al. 2009. ‘Putting our heads together’: insights into genomic conservation between human and canine intracranial tumors. J. Neurooncol. 94, 333–349. ( 10.1007/s11060-009-9877-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paoloni M, et al. 2009. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics 10, 625 ( 10.1186/1471-2164-10-625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson JM. 2013. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013, 941275 ( 10.1155/2013/941275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis BW, Ostrander EA. 2014. Domestic dogs and cancer research: a breed-based genomics approach. ILAR J. 55, 59–68. ( 10.1093/ilar/ilu017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819. ( 10.1038/nature04338) [DOI] [PubMed] [Google Scholar]
- 16.Vonholdt BM, et al. 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902. ( 10.1038/nature08837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray MM, Granka JM, Bustamante CD, Sutter NB, Boyko AR, Zhu L, Ostrander EA, Wayne RK. 2009. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics 181, 1493–1505. ( 10.1534/genetics.108.098830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro SG, et al. 2015. Canine urothelial carcinoma: genomically aberrant and comparatively relevant. Chrom Res. 23, 311–331. ( 10.1007/s10577-015-9471-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas R, et al. 2011. Refining tumor-associated aneuploidy through ‘genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk Lymphoma. 52, 1321–1335. ( 10.3109/10428194.2011.559802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillard M, et al. 2014. Naturally occurring melanomas in dogs as models for non-UV pathways of human melanomas. Pigment Cell Melanoma Res. 27, 90–102. ( 10.1111/pcmr.12170) [DOI] [PubMed] [Google Scholar]
- 21.Schiffman JD, Geller JI, Mundt E, Means A, Means L, Means V. 2013. Update on pediatric cancer predisposition syndromes. Pediatr Blood Cancer. 60, 1247–1252. ( 10.1002/pbc.24555) [DOI] [PubMed] [Google Scholar]
- 22.Malkin D, Nichols KE, Zelley K, Schiffman JD. 2014. Predisposition to pediatric and hematologic cancers: a moving target. Am. Soc. Clin. Oncol. Educ. Book 34, e44–e55. ( 10.14694/EdBook_AM.2014.34.e44) [DOI] [PubMed] [Google Scholar]
- 23.Testa JR, Malkin D, Schiffman JD. 2013. Connecting molecular pathways to hereditary cancer risk syndromes. Am. Soc. Clin. Oncol. Educ. Book 33, 81–90. ( 10.1200/EdBook_AM.2013.33.81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knapke S, Zelley K, Nichols KE, Kohlmann W, Schiffman JD. 2012. Identification, management, and evaluation of children with cancer-predisposition syndromes. Am. Soc. Clin. Oncol. Educ. Book 32, 576–584. [DOI] [PubMed] [Google Scholar]
- 25.Garber JE, Offit K. 2005. Hereditary cancer predisposition syndromes. J. Clin. Oncol. 23, 276–292. ( 10.1200/JCO.2005.10.042) [DOI] [PubMed] [Google Scholar]
- 26.Villani A, Tabori U, Schiffman J, Shlien A, Beyene J, Druker H, Novokmet A, Finlay J, Malkin D. 2011. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol. 12, 559–567. ( 10.1016/S1470-2045(11)70119-X) [DOI] [PubMed] [Google Scholar]
- 27.Smith RA, Manassaram-Baptiste D, Brooks D, Doroshenk M, Fedewa S, Saslow D, Brawley OW, Wender R. 2015. Cancer screening in the United States, 2015: a review of current American Cancer Society guidelines and current issues in cancer screening. Cancer J. Clin. 65, 30–54. ( 10.3322/caac.21261) [DOI] [PubMed] [Google Scholar]
- 28.Samadder NJ, Jasperson K, Burt RW. 2014. Hereditary and common familial colorectal cancer: evidence for colorectal screening. Dig. Dis. Sci. 60, 734–747. ( 10.1007/s10620-014-3465-z) [DOI] [PubMed] [Google Scholar]
- 29.Jasperson KW, et al. 2014. Role of rapid sequence whole-body MRI screening in SDH-associated hereditary paraganglioma families. Fam. Cancer 13, 257–265. ( 10.1007/s10689-013-9639-6) [DOI] [PubMed] [Google Scholar]
- 30.Borge KS, Borresen-Dale AL, Lingaas F. 2011. Identification of genetic variation in 11 candidate genes of canine mammary tumour. Vet. Comp. Oncol. 9, 241–250. ( 10.1111/j.1476-5829.2010.00250.x) [DOI] [PubMed] [Google Scholar]
- 31.Enginler SO, Akis I, Toydemir TS, Oztabak K, Haktanir D, Gunduz MC, Kırşan I, Fırat I. 2014. Genetic variations of BRCA1 and BRCA2 genes in dogs with mammary tumours. Vet. Res. Commun. 38, 21–27. ( 10.1007/s11259-013-9577-7) [DOI] [PubMed] [Google Scholar]
- 32.Veldhoen N, Watterson J, Brash M, Milner J. 1999. Identification of tumour-associated and germ line p53 mutations in canine mammary cancer. Br. J. Cancer 81, 409–415. ( 10.1038/sj.bjc.6690709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung CC, Chanock SJ. 2011. Current status of genome-wide association studies in cancer. Hum. Genet. 130, 59–78. ( 10.1007/s00439-011-1030-9) [DOI] [PubMed] [Google Scholar]
- 34.Stadler ZK, Vijai J, Thom P, Kirchhoff T, Hansen NA, Kauff ND, Robson M, Offit K. 2010. Genome-wide association studies of cancer predisposition. Hematol. Oncol. Clin. North Am. 24, 973–996. ( 10.1016/j.hoc.2010.06.009) [DOI] [PubMed] [Google Scholar]
- 35.Panagiotou OA, Evangelou E, Ioannidis JP. 2010. Genome-wide significant associations for variants with minor allele frequency of 5% or less—an overview: a HuGE review. Am. J. Epidemiol. 172, 869–889. ( 10.1093/aje/kwq234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ku CS, Loy EY, Pawitan Y, Chia KS. 2010. The pursuit of genome-wide association studies: where are we now? J. Hum. Genet. 55, 195–206. ( 10.1038/jhg.2010.19) [DOI] [PubMed] [Google Scholar]
- 37.Machiela MJ, Chanock SJ. 2014. GWAS is going to the dogs. Genome Biol. 15, 105 ( 10.1186/gb4166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shearin AL, et al. 2012. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. Biomarkers Prev. 21, 1019–1027. ( 10.1158/1055-9965.EPI-12-0190-T) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abadie J, et al. 2009. Epidemiology, pathology, and genetics of histiocytic sarcoma in the Bernese mountain dog breed. J. Hered. 100(Suppl. 1), S19–S27. ( 10.1093/jhered/esp039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho-Vanegas O, et al. 2012. Primate genome gain and loss: a bone dysplasia, muscular dystrophy, and bone cancer syndrome resulting from mutated retroviral-derived MTAP transcripts. Am. J. Hum. Genet. 90, 614–627. ( 10.1016/j.ajhg.2012.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstein AM, et al. 1995. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N. Engl. J. Med. 333, 970–974. ( 10.1056/NEJM199510123331504) [DOI] [PubMed] [Google Scholar]
- 42.Hussussian CJ, et al. 1994. Germline p16 mutations in familial melanoma. Nat. Genet. 8, 15–21. ( 10.1038/ng0994-15) [DOI] [PubMed] [Google Scholar]
- 43.Karyadi DM, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, Parker HG, Wayne RK, Ostrander EA. 2013. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 9, e1003409 ( 10.1371/journal.pgen.1003409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishida T, et al. 1998. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat. Genet. 19, 323–324. ( 10.1038/1209) [DOI] [PubMed] [Google Scholar]
- 45.John PR, Ramsay M. 2002. Four novel variants in MC1R in red-haired South African individuals of European descent: S83P, Y152X, A171D, P256S. Hum. Mutat. 19, 461–462. ( 10.1002/humu.9030) [DOI] [PubMed] [Google Scholar]
- 46.Lalueza-Fox C, et al. 2007. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science 318, 1453–1455. ( 10.1126/science.1147417) [DOI] [PubMed] [Google Scholar]
- 47.King RA, Willaert RK, Schmidt RM, Pietsch J, Savage S, Brott MJ, Fryer JP, Summers CG, Oetting WS. 2003. MC1R mutations modify the classic phenotype of oculocutaneous albinism type 2 (OCA2). Am. J. Hum. Genet. 73, 638–645. ( 10.1086/377569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koppula SV, Robbins LS, Lu D, Baack E, White CR, Jr, Swanson NA, Cone RD. 1997. Identification of common polymorphisms in the coding sequence of the human MSH receptor (MCIR) with possible biological effects. Hum. Mutat. 9, 30–36. () [DOI] [PubMed] [Google Scholar]
- 49.Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bouwes Bavinck JN. 2001. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J. Invest. Dermatol. 117, 294–300. ( 10.1046/j.0022-202x.2001.01421.x) [DOI] [PubMed] [Google Scholar]
- 50.Jones FI, Ramachandran S, Lear J, Smith A, Bowers B, Ollier WE, Jones P, Fryer AA, Strange RC. 1999. The melanocyte stimulating hormone receptor polymorphism: association of the V92M and A294H alleles with basal cell carcinoma. Clin. Chim. Acta 282, 125–134. ( 10.1016/S0009-8981(99)00017-0) [DOI] [PubMed] [Google Scholar]
- 51.Duffy DL, et al. 2004. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum. Mol. Genet. 13, 447–461. ( 10.1093/hmg/ddh043) [DOI] [PubMed] [Google Scholar]
- 52.Landi MT, et al. 2006. MC1R germline variants confer risk for BRAF-mutant melanoma. Science 313, 521–522. ( 10.1126/science.1127515) [DOI] [PubMed] [Google Scholar]
- 53.Karlsson EK, et al. 2013. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 14, R132 ( 10.1186/gb-2013-14-12-r132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savage SA, et al. 2013. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 45, 799–803. ( 10.1038/ng.2645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonomura N, et al. 2015. Genome-wide association study identifies shared risk loci common to two malignancies in golden retrievers. PLoS Genet. 11, e1004922 ( 10.1371/journal.pgen.1004922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerhan JR, et al. 2014. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat. Genet. 46, 1233–1238. ( 10.1038/ng.3105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skibola CF, et al. 2014. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am. J. Hum. Genet. 95, 462–471. ( 10.1016/j.ajhg.2014.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vijai J, et al. 2015. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat. Commun. 6, 5751 ( 10.1038/ncomms6751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pejovic T, et al. 1992. Prognostic impact of chromosome aberrations in ovarian cancer. Br. J. Cancer 65, 282–286. ( 10.1038/bjc.1992.56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bardi G, Fenger C, Johansson B, Mitelman F, Heim S. 2004. Tumor karyotype predicts clinical outcome in colorectal cancer patients. J. Clin. Oncol. 22, 2623–2634. ( 10.1200/JCO.2004.11.014) [DOI] [PubMed] [Google Scholar]
- 61.Brosens RP, et al. 2011. Deletion of chromosome 4q predicts outcome in stage II colon cancer patients. Cell Oncol. (Dordr.) 34, 215–223. ( 10.1007/s13402-011-0042-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wemmert S, et al. 2005. Patients with high-grade gliomas harboring deletions of chromosomes 9p and 10q benefit from temozolomide treatment. Neoplasia 7, 883–893. ( 10.1593/neo.05307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ganguly A, Richards-Yutz J, Ewens KG. 2014. Molecular karyotyping for detection of prognostic markers in fine needle aspiration biopsy samples of uveal melanoma. Methods Mol. Biol. 1102, 441–458. ( 10.1007/978-1-62703-727-3_23) [DOI] [PubMed] [Google Scholar]
- 64.Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, Gaiser T. 2013. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res. 73, 1454–1460. ( 10.1158/0008-5472.CAN-12-0928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.A'Hern RP, Jamal-Hanjani M, Szasz AM, Johnston SR, Reis-Filho JS, Roylance R, Swanton C. 2013. Taxane benefit in breast cancer: a role for grade and chromosomal stability. Nat. Rev. Clin. Oncol. 10, 357–364. ( 10.1038/nrclinonc.2013.67) [DOI] [PubMed] [Google Scholar]
- 66.Calvert GT, Randall RL, Jones KB, Cannon-Albright L, Lessnick S, Schiffman JD. 2012. At-risk populations for osteosarcoma: the syndromes and beyond. Sarcoma 2012, 152382 ( 10.1155/2012/152382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.HaDuong JH, Martin AA, Skapek SX, Mascarenhas L. 2015. Sarcomas. Pediatr. Clin. North Am. 62, 179–200. ( 10.1016/j.pcl.2014.09.012) [DOI] [PubMed] [Google Scholar]
- 68.Kansara M, Teng MW, Smyth MJ, Thomas DM. 2014. Translational biology of osteosarcoma. Nat. Rev. Cancer 14, 722–735. ( 10.1038/nrc3838) [DOI] [PubMed] [Google Scholar]
- 69.Fenger JM, London CA, Kisseberth WC. 2014. Canine osteosarcoma: a naturally occurring disease to inform pediatric oncology. ILAR J. 55, 69–85. ( 10.1093/ilar/ilu009) [DOI] [PubMed] [Google Scholar]
- 70.Scott MC, et al. 2011. Molecular subtypes of osteosarcoma identified by reducing tumor heterogeneity through an interspecies comparative approach. Bone 49, 356–367. ( 10.1016/j.bone.2011.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Angstadt AY, Thayanithy V, Subramanian S, Modiano JF, Breen M. 2012. A genome-wide approach to comparative oncology: high-resolution oligonucleotide aCGH of canine and human osteosarcoma pinpoints shared microaberrations. Cancer Genet. 205, 572–587. ( 10.1016/j.cancergen.2012.09.005) [DOI] [PubMed] [Google Scholar]
- 72.Khanna C, et al. 2014. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin. Cancer Res. 20, 4200–4209. ( 10.1158/1078-0432.CCR-13-2574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren L, Khanna C. 2014. Role of ezrin in osteosarcoma metastasis. Adv. Exp. Med. Biol. 804, 181–201. ( 10.1007/978-3-319-04843-7_10) [DOI] [PubMed] [Google Scholar]
- 74.McCleese JK, Bear MD, Kulp SK, Mazcko C, Khanna C, London CA. 2013. Met interacts with EGFR and Ron in canine osteosarcoma. Vet. Comp. Oncol. 11, 124–139. ( 10.1111/j.1476-5829.2011.00309.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong SH, Osborne T, Ren L, Briggs J, Mazcko C, Burkett SS, Khanna C. 2011. Protein kinase C regulates ezrin-radixin-moesin phosphorylation in canine osteosarcoma cells. Vet. Comp. Oncol. 9, 207–218. ( 10.1111/j.1476-5829.2010.00249.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Withrow SJ, Khanna C. 2009. Bridging the gap between experimental animals and humans in osteosarcoma. Cancer Treat Res. 152, 439–446. ( 10.1007/978-1-4419-0284-9_24) [DOI] [PubMed] [Google Scholar]
- 77.Rankin KS, Starkey M, Lunec J, Gerrand CH, Murphy S, Biswas S. 2012. Of dogs and men: comparative biology as a tool for the discovery of novel biomarkers and drug development targets in osteosarcoma. Pediatr. Blood Cancer 58, 327–333. ( 10.1002/pbc.23341) [DOI] [PubMed] [Google Scholar]
- 78.Davis LE, et al. 2013. A case study of personalized therapy for osteosarcoma. Pediatr. Blood Cancer 60, 1313–1319. ( 10.1002/pbc.24512) [DOI] [PubMed] [Google Scholar]
- 79.Caserto BG. 2013. A comparative review of canine and human rhabdomyosarcoma with emphasis on classification and pathogenesis. Vet. Pathol. 50, 806–826. ( 10.1177/0300985813476069) [DOI] [PubMed] [Google Scholar]
- 80.Milovancev M, Hauck M, Keller C, Stranahan LW, Mansoor A, Malarkey DE. 2015. Comparative pathology of canine soft tissue sarcomas: possible models of human non-rhabdomyosarcoma soft tissue sarcomas. J. Comp. Pathol. 152, 22–27. ( 10.1016/j.jcpa.2014.09.005) [DOI] [PubMed] [Google Scholar]
- 81.Moore PF. 2014. A review of histiocytic diseases of dogs and cats. Vet. Pathol. 51, 167–184. ( 10.1177/0300985813510413) [DOI] [PubMed] [Google Scholar]
- 82.Affolter VK, Moore PF. 2002. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet. Pathol. 39, 74–83. ( 10.1354/vp.39-1-74) [DOI] [PubMed] [Google Scholar]
- 83.Moore PF, Affolter VK, Vernau W. 2006. Canine hemophagocytic histiocytic sarcoma: a proliferative disorder of CD11d+ macrophages. Vet. Pathol. 43, 632–645. ( 10.1354/vp.43-5-632) [DOI] [PubMed] [Google Scholar]
- 84.Takahashi E, Nakamura S. 2013. Histiocytic sarcomaa: an updated literature review based on the 2008 WHO classification. J. Clin. Exp. Hematop. 53, 1–8. ( 10.3960/jslrt.53.1) [DOI] [PubMed] [Google Scholar]
- 85.Hornick JL, Jaffe ES, Fletcher CD. 2004. Extranodal histiocytic sarcoma: clinicopathologic analysis of 14 cases of a rare epithelioid malignancy. Am. J. Surg. Pathol. 28, 1133–1144. ( 10.1097/01.pas.0000131541.95394.23) [DOI] [PubMed] [Google Scholar]
- 86.Hedan B, Thomas R, Motsinger-Reif A, Abadie J, Andre C, Cullen J, Breen M. 2011. Molecular cytogenetic characterization of canine histiocytic sarcoma: a spontaneous model for human histiocytic cancer identifies deletion of tumor suppressor genes and highlights influence of genetic background on tumor behavior. BMC Cancer 11, 201 ( 10.1186/1471-2407-11-201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cioffi A, Reichert S, Antonescu CR, Maki RG. 2013. Angiosarcomas and other sarcomas of endothelial origin. Hematol. Oncol. Clin. North Am. 27, 975–988. ( 10.1016/j.hoc.2013.07.005) [DOI] [PubMed] [Google Scholar]
- 88.Young RJ, Brown NJ, Reed MW, Hughes D, Woll PJ. 2010. Angiosarcoma. Lancet Oncol. 11, 983–991. ( 10.1016/S1470-2045(10)70023-1) [DOI] [PubMed] [Google Scholar]
- 89.Antonescu C. 2014. Malignant vascular tumors—an update. Mod. Pathol. 27(Suppl. 1), S30–S38. ( 10.1038/modpathol.2013.176) [DOI] [PubMed] [Google Scholar]
- 90.Spiguel A. 2014. Soft tissue sarcomas. Cancer Treat Res. 162, 203–223. ( 10.1007/978-3-319-07323-1_10) [DOI] [PubMed] [Google Scholar]
- 91.Liu L, Kakiuchi-Kiyota S, Arnold LL, Johansson SL, Wert D, Cohen SM. 2013. Pathogenesis of human hemangiosarcomas and hemangiomas. Hum. Pathol. 44, 2302–2311. ( 10.1016/j.humpath.2013.05.012) [DOI] [PubMed] [Google Scholar]
- 92.Italiano A, et al. 2012. The miR-17–92 cluster and its target THBS1 are differentially expressed in angiosarcomas dependent on MYC amplification. Genes Chromosomes Cancer 51, 569–578. ( 10.1002/gcc.21943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fletcher C, Unni K, Mertens FE. 2002. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press. [Google Scholar]
- 94.Sturgis EM, Potter BO. 2003. Sarcomas of the head and neck region. Curr. Opin. Oncol. 15, 239–252. ( 10.1097/00001622-200305000-00011) [DOI] [PubMed] [Google Scholar]
- 95.Gorden BH, et al. 2014. Identification of three molecular and functional subtypes in canine hemangiosarcoma through gene expression profiling and progenitor cell characterization. Am. J. Pathol. 184, 985–995. ( 10.1016/j.ajpath.2013.12.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomas R, Borst L, Rotroff D, Motsinger-Reif A, Lindblad-Toh K, Modiano JF, Breen M. 2014. Genomic profiling reveals extensive heterogeneity in somatic DNA copy number aberrations of canine hemangiosarcoma. Chromosome Res. 22, 305–319. ( 10.1007/s10577-014-9406-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richards KL, Suter SE. 2015. Man's best friend: what can pet dogs teach us about non-Hodgkin's lymphoma? Immunol. Rev. 263, 173–191. ( 10.1111/imr.12238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ito D, Frantz AM, Modiano JF. 2014. Canine lymphoma as a comparative model for human non-Hodgkin lymphoma: recent progress and applications. Vet. Immunol. Immunopathol. 159, 192–201. ( 10.1016/j.vetimm.2014.02.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marconato L, Gelain ME, Comazzi S. 2013. The dog as a possible animal model for human non-Hodgkin lymphoma: a review. Hematol. Oncol. 31, 1–9. ( 10.1002/hon.2017) [DOI] [PubMed] [Google Scholar]
- 100.Valli VE, et al. 2011. Classification of canine malignant lymphomas according to the World Health Organization criteria. Vet. Pathol. 48, 198–211. ( 10.1177/0300985810379428) [DOI] [PubMed] [Google Scholar]
- 101.Vezzali E, Parodi AL, Marcato PS, Bettini G. 2010. Histopathologic classification of 171 cases of canine and feline non-Hodgkin lymphoma according to the WHO. Vet. Comp. Oncol. 8, 38–49. ( 10.1111/j.1476-5829.2009.00201.x) [DOI] [PubMed] [Google Scholar]
- 102.Modiano JF, et al. 2005. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 65, 5654–5661. ( 10.1158/0008-5472.CAN-04-4613) [DOI] [PubMed] [Google Scholar]
- 103.Ponce F, Marchal T, Magnol JP, Turinelli V, Ledieu D, Bonnefont C, Pastor M, Delignette ML, Fournel-Fleury C. 2010. A morphological study of 608 cases of canine malignant lymphoma in France with a focus on comparative similarities between canine and human lymphoma morphology. Vet. Pathol. 47, 414–433. ( 10.1177/0300985810363902) [DOI] [PubMed] [Google Scholar]
- 104.Richards KL, et al. 2013. Gene profiling of canine B-cell lymphoma reveals germinal center and postgerminal center subtypes with different survival times, modeling human DLBCL. Cancer Res. 73, 5029–5039. ( 10.1158/0008-5472.CAN-12-3546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su Y, Nielsen D, Zhu L, Richards K, Suter S, Breen M, Motsinger-Reif A, Osborne J. 2013. Gene selection and cancer type classification of diffuse large-B-cell lymphoma using a bivariate mixture model for two-species data. Hum. Genomics 7, 2 ( 10.1186/1479-7364-7-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frantz AM, et al. 2013. Molecular profiling reveals prognostically significant subtypes of canine lymphoma. Vet. Pathol. 50, 693–703. ( 10.1177/0300985812465325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Breen M, Modiano JF. 2008. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans--man and his best friend share more than companionship. Chromosome Res. 16, 145–154. ( 10.1007/s10577-007-1212-4) [DOI] [PubMed] [Google Scholar]
- 108.Figueiredo JF, Culver S, Behling-Kelly E, Breen M, Friedrichs KR. 2012. Acute myeloblastic leukemia with associated BCR-ABL translocation in a dog. Vet. Clin. Pathol. 41, 362–368. ( 10.1111/j.1939-165X.2012.00450.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perez ML, Culver S, Owen JL, Dunbar M, Kow K, Breen M, Milner RJ. 2013. Partial cytogenetic response with toceranib and prednisone treatment in a young dog with chronic monocytic leukemia. Anti-Cancer Drugs 24, 1098–1103. ( 10.1097/CAD.0000000000000018) [DOI] [PubMed] [Google Scholar]
- 110.Culver S, Ito D, Borst L, Bell JS, Modiano JF, Breen M. 2013. Molecular characterization of canine BCR-ABL-positive chronic myelomonocytic leukemia before and after chemotherapy. Vet. Clin. Pathol. 42, 314–322. ( 10.1111/vcp.12055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giantin M, Aresu L, Arico A, Gelain ME, Riondato F, Martini V, Comazzi S, Dacasto M. 2013. Evaluation of tyrosine-kinase receptor c-KIT (c-KIT) mutations, mRNA and protein expression in canine leukemia: might c-KIT represent a therapeutic target? Vet. Immunol. Immunopathol. 152, 325–332. ( 10.1016/j.vetimm.2013.01.003) [DOI] [PubMed] [Google Scholar]
- 112.Higuchi T, Burcham GN, Childress MO, Rohleder JJ, Bonney PL, Ramos-Vara JA, Knapp DW. 2013. Characterization and treatment of transitional cell carcinoma of the abdominal wall in dogs: 24 cases (1985–2010). J. Am. Vet. Med. Assoc. 242, 499–506. ( 10.2460/javma.242.4.499) [DOI] [PubMed] [Google Scholar]
- 113.Park JC, Hahn NM. 2014. Bladder cancer: a disease ripe for major advances. Clin. Adv. Hematol. Oncol. 12, 838–845. [PubMed] [Google Scholar]
- 114.Knapp DW, Ramos-Vara JA, Moore GE, Dhawan D, Bonney PL, Young KE. 2014. Urinary bladder cancer in dogs, a naturally occurring model for cancer biology and drug development. ILAR J. 55, 100–118. ( 10.1093/ilar/ilu018) [DOI] [PubMed] [Google Scholar]
- 115.Kimmelman J, Nalbantoglu J. 2007. Faithful companions: a proposal for neurooncology trials in pet dogs. Cancer Res. 67, 4541–4544. ( 10.1158/0008-5472.CAN-06-3792) [DOI] [PubMed] [Google Scholar]
- 116.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109. ( 10.1007/s00401-007-0243-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Candolfi M, et al. 2007. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J. Neurooncol. 85, 133–148. ( 10.1007/s11060-007-9400-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Candolfi M, et al. 2007. Optimization of adenoviral vector-mediated transgene expression in the canine brain in vivo, and in canine glioma cells in vitro. Neuro Oncol. 9, 245–258. ( 10.1215/15228517-2007-012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Snyder JM, Shofer FS, Van Winkle TJ, Massicotte C. 2006. Canine intracranial primary neoplasia: 173 cases (1986–2003). J. Vet. Intern. Med. 20, 669–675. [DOI] [PubMed] [Google Scholar]
- 120.Song RB, Vite CH, Bradley CW, Cross JR. 2013. Postmortem evaluation of 435 cases of intracranial neoplasia in dogs and relationship of neoplasm with breed, age, and body weight. J. Vet. Intern. Med. 27, 1143–1152. ( 10.1111/jvim.12136) [DOI] [PubMed] [Google Scholar]
- 121.Koestner A, Higgins R. 2002. Tumors of the nervous system. In Tumors in domestic animals (ed. Meuten D.), pp. 697–738, 4th edn Iowa: Iowa State Press. [Google Scholar]
- 122.Koestner A, Higgins R. 2015. Tumors of the nervous system. In Tumors in domestic animals (ed. Stanley MD.), p. 697 Oxford, UK: Blackwell. [Google Scholar]
- 123.Thomson SA, et al. 2005. Microarray analysis of differentially expressed genes of primary tumors in the canine central nervous system. Vet. Pathol. 42, 550–558. ( 10.1354/vp.42-5-550) [DOI] [PubMed] [Google Scholar]
- 124.Omuro A, DeAngelis LM. 2005. Glioblastoma and other malignant gliomas: a clinical review. JAMA 310, 1842–1850. ( 10.1001/jama.2013.280319) [DOI] [PubMed] [Google Scholar]
- 125.Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. ( 10.1038/nature07385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schiffman JD, et al. 2010. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 70, 512–519. ( 10.1158/0008-5472.CAN-09-1851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Villani A, Malkin D, Tabori U. 2012. Syndromes predisposing to pediatric central nervous system tumors: lessons learned and new promises. Curr. Neurol. Neurosci. Rep. 12, 153–164. ( 10.1007/s11910-011-0244-5) [DOI] [PubMed] [Google Scholar]
- 128.Bakry D, et al. 2014. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the Constitutional Mismatch Repair Deficiency Consortium. Eur. J. Cancer 50, 987–996. ( 10.1016/j.ejca.2013.12.005) [DOI] [PubMed] [Google Scholar]
- 129.Sadetzki S, et al. 2013. Description of selected characteristics of familial glioma patients—results from the Gliogene Consortium. Eur. J. Cancer 49, 1335–1345. ( 10.1016/j.ejca.2012.11.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Robertson LB, et al. 2010. Survey of familial glioma and role of germline p16INK4A/p14ARF and p53 mutation. Fam. Cancer 9, 413–421. ( 10.1007/s10689-010-9346-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schiffman JD, Chun N, Fisher PG, Dahl GV, Ford JM, Eggerding FA. 2008. Identification of a novel p53 in-frame deletion in a Li-Fraumeni-like family. Pediatr. Blood Cancer 50, 914–916. ( 10.1002/pbc.21247) [DOI] [PubMed] [Google Scholar]
- 132.Bainbridge MN, et al. 2015. Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl Cancer Inst. 107, 384 ( 10.1093/jnci/dju384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MH, Eeles RA, Thomas DM, Mitchell G. 2014. Li–Fraumeni syndrome: cancer risk assessment and clinical management. Nat. Rev. Clin. Oncol. 11, 260–271. ( 10.1038/nrclinonc.2014.41) [DOI] [PubMed] [Google Scholar]
- 134.Brearley MJ, Jeffery ND, Phillips SM, Dennis R. 1999. Hypofractionated radiation therapy of brain masses in dogs: a retrospective analysis of survival of 83 cases (1991–1996). J. Vet. Intern. Med. 13, 408–412. () [DOI] [PubMed] [Google Scholar]
- 135.Fulton LM, Steinberg HS. 1990. Preliminary study of lomustine in the treatment of intracranial masses in dogs following localization by imaging techniques. Semin. Vet. Med. Surg. (Small Anim). 5, 241–245. [PubMed] [Google Scholar]
- 136.Pluhar GE, et al. 2010. Anti-tumor immune response correlates with neurological symptoms in a dog with spontaneous astrocytoma treated by gene and vaccine therapy. Vaccine 28, 3371–3378. ( 10.1016/j.vaccine.2010.02.082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spugnini EP, Thrall DE, Price GS, Sharp NJ, Munana K, Page RL. 2000. Primary irradiation of canine intracranial masses. Vet. Radiol. Ultrasound 41, 377–380. ( 10.1111/j.1740-8261.2000.tb02091.x) [DOI] [PubMed] [Google Scholar]
- 138.Thrall DE, et al. 1999. Use of whole body hyperthermia as a method to heat inaccessible tumours uniformly: a phase III trial in canine brain masses. Int. J. Hyperthermia 15, 383–398. ( 10.1080/026567399285576) [DOI] [PubMed] [Google Scholar]
- 139.Bagley RS, Gavin PR. 1998. Seizures as a complication of brain tumors in dogs. Clin. Tech. Small Anim. Pract. 13, 179–184. ( 10.1016/S1096-2867(98)80039-X) [DOI] [PubMed] [Google Scholar]
- 140.Kloosterman WP, Koster J, Molenaar JJ. 2014. Prevalence and clinical implications of chromothripsis in cancer genomes. Curr. Opin. Oncol. 26, 64–72. ( 10.1097/CCO.0000000000000038) [DOI] [PubMed] [Google Scholar]
- 141.Azoury SC, Lange JR. 2014. Epidemiology, risk factors, prevention, and early detection of melanoma. Surg. Clin. North Am. 94, 945–962. ( 10.1016/j.suc.2014.07.013) [DOI] [PubMed] [Google Scholar]
- 142.Mayer JE, Swetter SM, Fu T, Geller AC. 2014. Screening, early detection, education, and trends for melanoma: current status (2007–2013) and future directions. J. Am. Acad. Dermatol. 71, 599.e1–599.e12. ( 10.1016/j.jaad.2014.05.046) [DOI] [PubMed] [Google Scholar]
- 143.Manigandan T, Sagar GV, Amudhan A, Hemalatha VT, Babu NA. 2014. Oral malignant melanoma: a case report with review of literature. Contemp. Clin. Dent. 5, 415–418. ( 10.4103/0976-237X.137978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lazarev S, Gupta V, Hu K, Harrison LB, Bakst R. 2014. Mucosal melanoma of the head and neck: a systematic review of the literature. Int. J. Radiat. Oncol. Biol. Phys. 90, 1108–1118. ( 10.1016/j.ijrobp.2014.03.042) [DOI] [PubMed] [Google Scholar]
- 145.Sondak VK, Messina JL. 2014. Unusual presentations of melanoma: melanoma of unknown primary site, melanoma arising in childhood, and melanoma arising in the eye and on mucosal surfaces. Surg. Clin. North Am. 94, 1059–1073. ( 10.1016/j.suc.2014.07.010) [DOI] [PubMed] [Google Scholar]
- 146.Simpson RM, et al. 2014. Sporadic naturally occurring melanoma in dogs as a preclinical model for human melanoma. Pigment Cell Melanoma Res. 27, 37–47. ( 10.1111/pcmr.12185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Grosenbaugh DA, et al. 2011. Safety and efficacy of a xenogeneic DNA vaccine encoding for human tyrosinase as adjunctive treatment for oral malignant melanoma in dogs following surgical excision of the primary tumor. Am. J. Vet. Res. 72, 1631–1638. ( 10.2460/ajvr.72.12.1631) [DOI] [PubMed] [Google Scholar]
- 148.Manley CA, et al. 2011. Xenogeneic murine tyrosinase DNA vaccine for malignant melanoma of the digit of dogs. J. Vet. Intern. Med. 25, 94–99. ( 10.1111/j.1939-1676.2010.0627.x) [DOI] [PubMed] [Google Scholar]
- 149.Bergman PJ, et al. 2006. Development of a xenogeneic DNA vaccine program for canine malignant melanoma at the Animal Medical Center. Vaccine 24, 4582–4585. ( 10.1016/j.vaccine.2005.08.027) [DOI] [PubMed] [Google Scholar]
- 150.Bergman PJ, et al. 2003. Long-term survival of dogs with advanced malignant melanoma after DNA vaccination with xenogeneic human tyrosinase: a phase I trial. Clin. Cancer Res. 9, 1284–1290. [PubMed] [Google Scholar]
- 151.Liao JC, et al. 2006. Vaccination with human tyrosinase DNA induces antibody responses in dogs with advanced melanoma. Cancer Immun. 6, 8. [PMC free article] [PubMed] [Google Scholar]
- 152.Poorman K, et al. 2015. Comparative cytogenetic characterization of primary canine melanocytic lesions using array CGH and fluorescence in situ hybridization. Chromosome Res. 23, 171–186. ( 10.1007/s10577-014-9444-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chandler WM, Rowe LR, Florell SR, Jahromi MS, Schiffman JD, South ST. 2012. Differentiation of malignant melanoma from benign nevus using a novel genomic microarray with low specimen requirements. Arch. Pathol. Lab. Med. 136, 947–955. ( 10.5858/arpa.2011-0330-OA) [DOI] [PubMed] [Google Scholar]
- 154.Curtin JA, et al. 2005. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147. ( 10.1056/NEJMoa050092) [DOI] [PubMed] [Google Scholar]
- 155.Reddy M, Dierauf L, Gulland F. 2001. Marine mammals as sentinels of ocean health. In CRC handbook of marine mammal medicine (eds Leslie AD, Gulland FMD.), pp. 3–13, 2nd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 156.Browning HM, Gulland FMD, Hammond JA, Colegrove KM, Hall AJ. 2015. Common cancer in a wild animal: the California sea lion (Zalophus californianus) as an emerging model for carcinogenesis. Phil. Trans. R. Soc. B 370, 20140228 ( 10.1098/rstb.2014.0228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gulland FM, Trupkiewicz JG, Spraker TR, Lowenstine LJ. 1996. Metastatic carcinoma of probable transitional cell origin in 66 free-living California sea lions (Zalophus californianus), 1979 to 1994. J. Wildl. Dis. 32, 250–258. ( 10.7589/0090-3558-32.2.250) [DOI] [PubMed] [Google Scholar]
- 158.Lipscomb TP, et al. 2000. Common metastatic carcinoma of California sea lions (Zalophus californianus): evidence of genital origin and association with novel gammaherpesvirus. Vet. Pathol. 37, 609–617. ( 10.1354/vp.37-6-609) [DOI] [PubMed] [Google Scholar]
- 159.Buckles EL, et al. 2006. Otarine herpesvirus-1, not papillomavirus, is associated with endemic tumours in California sea lions (Zalophus californianus). J. Comp. Pathol. 135, 183–189. ( 10.1016/j.jcpa.2006.06.007) [DOI] [PubMed] [Google Scholar]
- 160.King DP, Hure MC, Goldstein T, Aldridge BM, Gulland FM, Saliki JT, Buckles EL, Lowenstine LJ, Stott JL. 2002. Otarine herpesvirus-1: a novel gammaherpesvirus associated with urogenital carcinoma in California sea lions (Zalophus californianus). Vet. Microbiol. 86, 131–137. ( 10.1016/S0378-1135(01)00497-7) [DOI] [PubMed] [Google Scholar]
- 161.Buckles EL, et al. 2007. Age-prevalence of otarine herpesvirus-1, a tumor-associated virus, and possibility of its sexual transmission in California sea lions. Vet. Microbiol. 120, 1–8. ( 10.1016/j.vetmic.2006.10.002) [DOI] [PubMed] [Google Scholar]
- 162.Ylitalo GM, et al. 2005. The role of organochlorines in cancer-associated mortality in California sea lions (Zalophus californianus). Mar. Pollut. Bull. 50, 30–39. ( 10.1016/j.marpolbul.2004.08.005) [DOI] [PubMed] [Google Scholar]
- 163.Natalie A. 2015. No time for bats to rest easy. New York Times. See http://www.nytimes.com/2015/01/13/science/no-time-for-bats-to-rest-easy.html?_r=0 (3 January 2015).
- 164.Tian X, Azpurua J, Ke Z, Augereau A, Zhang ZD, Vijg J, Gladyshev VN, Gorbunova V, Seluanov A. 2015. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc. Natl Acad. Sci. USA 112, 1053–1058. ( 10.1073/pnas.1418203112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tian X, et al. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349. ( 10.1038/nature12234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J. 2014. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540. ( 10.1038/nrg3728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Keane M, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122. ( 10.1016/j.celrep.2014.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Caulin AF, Maley CC. 2011. Peto's paradox: evolution's prescription for cancer prevention. Trends Ecol. Evol. 26, 175–182. ( 10.1016/j.tree.2011.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Roche B, Hochberg ME, Caulin AF, Maley CC, Gatenby RA, Misse D, Thomas F. 2012. Natural resistance to cancers: a Darwinian hypothesis to explain Peto's paradox. BMC Cancer 12, 387 ( 10.1186/1471-2407-12-387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nunney L. 2013. The real war on cancer: the evolutionary dynamics of cancer suppression. Evol. Appl. 6, 11–19. ( 10.1111/eva.12018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peto R, Roe FJ, Lee PN, Levy L, Clack J. 1975. Cancer and ageing in mice and men. Br. J. Cancer 32, 411–426. ( 10.1038/bjc.1975.242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nunney L. 1999. Lineage selection and the evolution of multistage carcinogenesis. Proc. R. Soc. Lond. B 266, 493–498. ( 10.1098/rspb.1999.0664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Noble R, Kaltz O, Hochberg ME. 2015. Peto's paradox and human cancers. Phil. Trans. R. Soc. B 370, 20150104 ( 10.1098/rstb.2015.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dang CV. 2015. A metabolic perspective of Peto's paradox and cancer. Phil. Trans. R. Soc. B 370, 20140223 ( 10.1098/rstb.2014.0223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Caulin AF, Graham TA, Wang L-S, Maley CC. 2015. Solutions to Peto's paradox revealed by mathematical modelling and cross-species cancer gene analysis. Phil. Trans. R. Soc. B 370, 20140222 ( 10.1098/rstb.2014.0222) [DOI] [PMC free article] [PubMed] [Google Scholar]