Abstract
An evolutionary perspective can help unify disparate observations and make testable predictions. We consider an evolutionary model in relation to two mechanistic frameworks of cancer biology: multistage carcinogenesis and the hallmarks of cancer. The multistage model predicts that cancer risk increases with body size and longevity; however, this is not observed across species (Peto's paradox), but the paradox is resolved by invoking the evolution of additional genetic mechanisms to suppress cancer in large, long-lived species. It is when cancer cells overcome these defence mechanisms that they exhibit the hallmarks of cancer, driving the ongoing evolution of these defences, which in turn is expected to create the differences observed in the genetics of cancer across species and tissues. To illustrate the utility of an evolutionary model we examined some recently published data linking stem-cell divisions and cancer incidence across a range of tissues and show why the original analysis was faulty, and demonstrate that the data are consistent with a multistage model varying from three to seven mutational hits across different tissues. Finally, we demonstrate how an evolutionary model can both define patterns of inherited (familial) cancer and explain the prevalence of cancer in post-reproductive years, including the dominance of epithelial cancers.
Keywords: evolution, cancer, Peto's paradox, hallmarks, multistage carcinogenesis, mutation-selection balance
1. Introduction
A major goal of cancer research is to uncover the nature and number of genetic alterations leading to the development of cancer. The mechanistic approach that dominates cancer research has yielded many triumphs in this endeavour such as the discovery of the tumour suppressor genes and proto-oncogenes that initiate cancer when mutated. For many of these genes, their function and the molecular pathways within which they act have been meticulously worked out. This has aided the development of anti-cancer drug treatments aimed at targeting the faulty forms of these genes. However, the mechanistic approach has not been able to answer an important question: Why do differences exist in the genetics of cancer between tissues and species? In other words, why do the genetic discoveries for one cancer type not necessarily apply to all others?
Observations across human tissues suggest that many tissues are using different genes and a different number of genes to defend against cancer. The tissue-specificity of hereditary cancers suggests that the action of an individual gene is limited to specific tissues. A striking example of this is that inherited mutations in the BRCA1 gene lead to a high lifetime risk of cancer in women in breast and ovarian tissues, but not in other tissues. As for the number of genes involved, cancer has been shown to initiate following as few as two ‘driver’ mutations, one in each copy of a single gene (retinoblastoma) [1], or up to eight such mutations [2]. Understanding why these genetic differences exist can inform us as to why gene-specific drugs often have a limited applicability across cancer types.
Genetic differences also exist between species in the control of the same type of cancer. Hereditary mutations of Brca1 in mice, for example, do not lead to breast and ovarian cancer as they do in humans [3]. Human cells have also been shown to require additional mutations to be transformed in cell culture relative to mice cells [4]. Understanding why these genetic differences (and similarities) exist can inform our choice of animal model for studying specific cancer types, and may reveal novel therapeutic avenues [5].
The mechanistic approach has helped uncover the differences and similarities in the genetics of cancer. But we need to understand why this pattern exists. Hanahan & Weinberg [6] concluded their influential paper with ‘we continue to foresee cancer research as an increasingly logical science, in which myriad phenotypic complexities are manifestations of a small set of underlying organizing principles’. Here we argue that these underlying organizing principles must be based on the recognition that cancer suppression is an adaptive trait, that is often tissue specific, and that is continuously evolving by natural selection due to the (Darwinian) fitness loss caused by cancers. To understand the nature and number of genes involved in suppressing a given cancer, an evolutionary framework is necessary, and the logical starting point for this evolutionary framework is to build upon the well-established multistage model of carcinogenesis.
The multistage model of carcinogenesis was built on the observations of Nordling [7] and Armitage & Doll [8] that the age-specific incidence of cancer was consistent with the stepwise accumulation of six to seven mutational ‘hits’ within a cell. However, Knudson [1] found that retinoblastoma, a childhood cancer of retinal cells, conformed to a two-hit model of carcinogenesis, suggesting that the number of ‘hits’ required to initiate a cancer varies across cancer types. This conclusion raises the question of what drives such variation in the number of mutational ‘hits’ required for cancer initiation. A simple evolutionary model of multistage carcinogenesis demonstrated that the primary determining factors are the size of the tissue and the number of cell divisions it undergoes [9].
This evolutionary modelling of multistage carcinogenesis directly addressed a concern raised by Peto [10]: if cancers are initiated by a series of somatic mutations, then, relative to mice, humans, with their larger body size (more cells) and longer lifespans (more lifetime cell divisions) should have a cancer risk that is orders of magnitude greater than a mouse. This dilemma has become known as Peto's paradox [9], and recent data from humans and domestic dogs show conclusively that larger individuals within a species do indeed suffer more cancer [11], and, of course, it is well established that the rate of cancer increases with age. This problem of size and longevity was also noted by Cairns [12]. He suggested three possible mechanisms that may have evolved in larger, longer lived organisms to resolve the problem: restricting the number of stem cells, retaining the template strand of DNA in the stem cells during asymmetric divisions (immortal strand hypothesis), and compartmentalization of stem-cell populations to restrict competition between cells. The importance of Cairns' suggestions is that it invokes adaptive change. The starting point for developing an evolutionary framework is the recognition that cancer suppression is not a fixed property of cells or tissues, so we can expect genetic differences among species, and within species tissue-specific gene expression is likely to be an important factor in understanding cancer suppression mechanisms [13]. Evolutionary modelling allows us to quantify when we expect changes in the level of cancer suppression in a given tissue to be favoured by natural selection. Such models can provide testable hypotheses about the patterns of cancer suppression and cancer incidence that are expected.
2. Evolution and the hallmarks of cancer
Hanahan & Weinberg [14] presented six ‘hallmarks of cancer’ characteristic of tumour cells. A decade later Hanahan & Weinberg [6] re-examined their list and emphasized the role of four more items, two new potential hallmarks and two ‘enabling characteristics’ (table 1). They proposed that the expression of each hallmark reflects a breach of an anti-cancer defence, and as such the list provides a framework for dissecting the mechanics of specific cancers.
Table 1.
The six ‘hallmarks of cancer’ of Hanahan & Weinberg [14] in 2000, plus the four additional features added in 2011 [6] and their relationship to the evolution of cancer suppression.
| hallmark no. (year) | hallmarka | potential evolutionary response | examples of inter-specific differences |
|---|---|---|---|
| 1 (2000) | sustaining proliferative signalling | add more layers of necessary signals (more proto-oncogenes) | transformation of human fibroblasts requires additional inactivation of Ral-GEF relative to mice [4] |
| 2 (2000) | evading growth suppressors | recruit more tumour suppressor loci | transformation of human fibroblasts requires additional inactivation of TP53 relative to mice [4] |
| 3 (2000) | enabling replicative immortality | telomerase suppression | telomerase suppression increases with body size in rodents [15] |
| 4 (2000) | resisting cell death | evolve a more robust system | concerted necrotic cell death (IFN-β) in blind mole-rat [16] TP53 duplications in elephants [17] |
| 5 (2000) | inducing angiogenesis | add more layers of necessary signals required to induce angiogenesis | |
| 6 (2000) | activating invasion and metastasis | inhibiting cell invasion and metastasis | early contact inhibition in naked mole-rat [18] |
| 7 (2011) | deregulating cellular energetics | add more layers of protection preventing energetic reprogramming | |
| 8 (2011) | avoiding immune destruction | more effective immune surveillance | |
| 9 (2000, 2011) | genome instability and mutation | more effective DNA repair and genome surveillance | levels of endogenous retroviruses (ERVs) decrease with body size in mammals [19] |
| 10 (2011) | tumour-promoting inflammation | improve anti-inflammatory responses |
aHallmarks 9 and 10 are considered ‘enabling characteristics’.
Their goal in defining these hallmarks was to establish unifying principles. Without doubt this list is an important first step; however, we believe that defining unifying principles will require an evolutionary understanding of how these defences arose and why they differ among different human cancers. Each hallmark defines the breakdown of a set of defence mechanisms, and each can potentially be strengthened by natural selection (table 1). For example, ‘resisting cell death’ involves cells overcoming the machinery of apoptosis, which often means disabling TP53. It is therefore notable that there is evidence of multiple TP53 duplications in the large-bodied, long-lived elephant [17], a response that is consistent with the evolutionary expectation.
It is important to note that while we can predict a defensive response to increased cancer incidence, the precise mechanistic nature of that response cannot be predicted as it depends upon the genetic variation that is available in the population at the time selection is acting [11,20]. This aspect is predicted to give rise to the differences in the nature of the genes involved in cancer suppression across tissues that is being so strongly highlighted in recent tumour sequencing projects [21]. As dozens of tumour suppressor genes and proto-oncogenes exist [22], we expect that the same gene will not always be independently recruited in different tissues to suppress cancer. For example, the tissue-specificity of inherited mutations in BRCA1 may result from BRCA1 being evolutionarily recruited to suppress cancer in breast and ovarian tissues, but not for such a role in the non-susceptible tissues. Higher expression levels of tumour suppressors in their normal (non-cancerous) susceptible tissues relative to non-susceptible tissues would be expected, given the assumption either that the expression of these genes entails some cost, or that expression that is truly redundant (i.e. does not affect fitness) decays [23]. This pattern has been found for the protein product of the MLH1 and MSH2 tumour suppressors [24], and recently Muir & Nunney [13] found that across 15 tumour suppressors and eight proto-oncogenes there was a highly significant trend for the majority (more than 60%) of these genes to be most highly expressed in their susceptible tissue. As this tissue-specific expression of cancer inhibiting genes is detrimental to the development of a cancer, it is interesting to note that the pattern appears to be different in tumours, where genes important in housekeeping and other essential functions are favoured [25].
Perhaps, the strongest evolutionary predictions are that if individuals of a given species were selected in the past to become larger and/or longer lived (hence inducing an increased cancer risk), such animals will generally have evolved additional cancer defences [9]. Research based on comparative studies that exploit these predictions is still in its infancy; however, a notable exception is the comparison of rodents of different sizes and longevity [5]. The study of the cells of different rodent species in culture both supported the expectations of the evolutionary model and revealed novel species-specific anti-cancer strategies (table 1). Furthermore, interest in comparative approaches that exploit data from non-model species are rapidly increasing [26–29], so that we can anticipate further tests of the evolutionary model. The domestic dog is a species notable for having an extensive dataset and studies of breed-specific differences in cancer risk are already proving to be productive [28–30].
3. Evolution, stem-cell divisions and cancer risk
Recently, Tomasetti & Vogelstein [31] made two observations concerning the relationship between the incidence of specific cancers and the total number of stem-cell divisions in the healthy tissue in which it originates. First, they noted a strong linear relationship, on a log–log scale, between these two variables, and second, they proposed that it was possible to identify cancers more strongly influenced by environmental and inherited factors versus random (irreducible) risk using their ‘extra risk score’ (ERS) which is the centred x . y product of each point on the graph. Without worrying about the meaning of the ERS, we will show that their analysis is impossible to interpret in a causal manner because it ignores the basic theory of multistage carcinogenesis and the possibility of differences in the genetics of cancer across tissues. As a result, while a broad correlation between lifetime cancer risk and lifetime stem-cell divisions is expected, deviations from this relationship can arise for multiple reasons. However, we can use this example to illustrate one of the ways in which an evolutionary approach can be highly informative.
Total stem-cell divisions were calculated by Tomasetti & Vogelstein [31] as the number of stem cells (C) × the number of divisions expected per cell (K), with an additional correction for the growth phase of the tissue during embryonic and juvenile development. The implicit assumption underpinning the log–log relationship between cancer incidence (p) and the number of stem-cell divisions is that cancer is induced by a 1-hit model, i.e. every cell division has an equal chance of giving rise to a cancer, regardless of its history. Given that such a model is untenable, it is more appropriate to examine the relationship derived for a more realistic multistage model. In its simplest form (assuming suppression by a set of identical tumour suppressor loci and given low cancer rates), the expected relationship is (from Nunney [9])
| 3.1 |
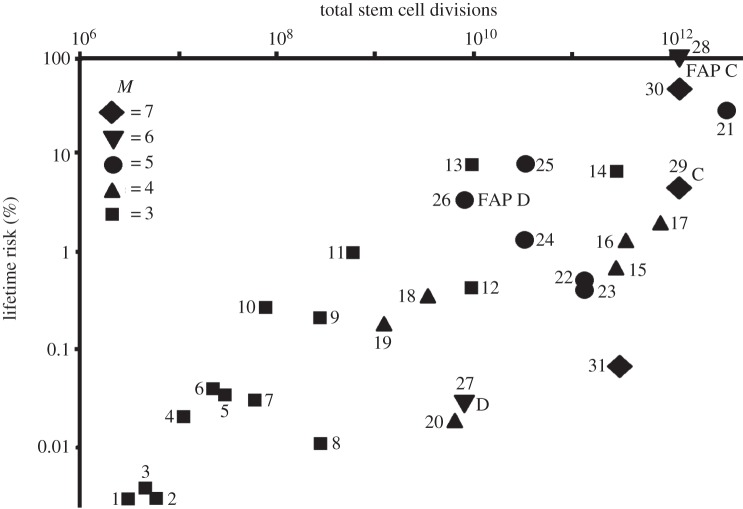
where u is the somatic mutation rate and M is the number of mutational ‘hits’ required to initiate cancer. An approximate correction for growth phase involves replacing K in equation (3.1) with K′ = K + k/2, where k (=ln C/ln 2) is the number of divisions required for the initial tissue growth [9]. An important feature of relationship (3.1) is that it formalizes the criticism noted above: the log–log relationship between p and CK is only expected if M = 1. In principle, one could fit the data to equation (3.1) to estimate an overall value of M, but only if all cancers in the dataset are suppressed by the same number of ‘hits’. Given the mix of sarcomas and carcinomas, it is very unlikely that M is constant [23]. As a result, it is not possible to get a statistical fit of the data to the model defined by equation (3.1) unless there is an a priori reason to group particular cancers by value of M; however, there are no reliable data that would allow us to do that grouping (note that a more justifiable tissue-based grouping was adopted by Noble et al. [32]). Instead, we estimated a value of M for each cancer type using equation (3.1), an approach that uses all of the available data. In addition, to fit the equation we needed to assume that the same somatic mutation rate underpins each mutational hit and assign it some value. In fact, a range of values was assigned by establishing the values of u that maintained 7 ≥ M ≥ 3 across all of the cancers. The lower limit on M was based on Nunney's [9,20] proposal that M ≥ 3 (except for retinoblastoma which has M = 2), noting that M = 3 was likely to be adequate for small tissues with minimal post-growth division, while the results of Armitage & Doll [8] suggested an upper limit of M = 7. This is broadly consistent with the recently proposed range of 8 ≥ M ≥ 2 [2] given the probable restriction of M = 2 to one or very few paediatric cancers. The range of u satisfying these criteria (with values of M rounded to the nearest integer) was surprisingly narrow: 7.5 × 10−6 to 8.9 × 10−6 oncogenic changes/division and the resulting distribution is shown in figure 1 (using u = 8.3 × 10−6), where the data points are identical to those shown in Tomasetti & Vogelstein [31]. Figure 1 illustrates several important issues. (i) The estimates are generally consistent with the expectation that smaller and/or slowly dividing tissues would evolve less protection (lower M) than large rapidly dividing tissues. The estimates for the five osteosarcomas (points 1–5) are all M = 3, while the colorectal and small intestine adenocarcinomas define the three cancers with M = 7 (points 29–31). (ii) The comparison of FAP-induced familial and non-familial cases of colorectal and duodenal cancer show the expected result that M is reduced by one (from 7 to 6 and 6 to 5, respectively). FAP (familial adenomatous polyposis) is typically dominant, resulting from the effects of a single defective copy of the APC gene, so it represents a single inherited mutational hit. (iii) The lungs appear to be protected by only three mutational steps, an estimate driven by the very slow rate of stem-cell division and it is consistent for smokers and non-smokers, even though smokers presumably experience a higher somatic mutation rate that was not taken into account (see below). A similar effect is apparent in comparing Lynch syndrome colorectal cancer with the non-familial form (M = 7 for both), even though Lynch syndrome causes a higher somatic mutation rate. (iv) The rate of somatic mutation (u) consistent with 7 ≥ M ≥ 3 is quite high at approximately 8 × 10−6 per daughter cell. For example, it is substantially higher than the estimate of 4 × 10−7 that accurately predicts the frequency of retinoblastoma [33], and that value is consistent with estimates of human somatic mutation rates of just under 10−9 per base per cell division [34].
Figure 1.
The relationship between total stem-cell divisions and lifetime cancer risk is complicated by variation in the number of mutational hits (M) predicted to be required to initiate a given cancer. The estimated value of M was calculated for each cancer and shown by the different symbols. Two pairs (FAP C versus C, and FAP D versus D) are comparisons of familial and non-familial colorectal (C) and duodenal (D) adenocarcinoma discussed in the text. Data are from Tomasetti & Vogelstein [28], and the cancers represented are: 1–5, osteosarcoma (pelvis, head, arms, legs and overall); 6, ovarian germ cell cancer; 7, thyroid medullary carcinoma; 8, medulloblastoma; 9, glioblastoma; 10, gallbladder adenocarcinoma; 11, thyroid follicular carcinoma; 12 and 13, lung adenocarcinoma (non-smokers, smokers); 14 and 15, hepatocellular carcinoma (hepatitis C virus, normal); 16, pancreatic ductal adenocarcinoma; 17, melanoma; 18, testicular germ cell cancer; 19, oesophageal squamous cell carcinoma; 20, pancreatic endocrine (islet cell) carcinoma; 21, basal cell carcinoma; 22 and 23, leukaemia (chronic lymphocytic, acute myeloid); 24 and 25, head and neck squamous cell carcinoma (no human papilloma virus, HPV); 26 and 27, duodenal adenocarcinoma (FAP, normal); 28–30, colorectal adenocarcinoma (FAP, normal, Lynch syndrome); 31, small intestinal adenocarcinoma.
There are at least three factors that are likely to contribute to an elevated estimate of the somatic mutation rate that initiates cancer. The first is the possibility that some initial mutational hits lead to increased cellular proliferation [35]. The result is an apparent increase in the somatic mutation rate for all subsequent mutations [9]. A second possibility is that some cells acquire somatic mutations that impair DNA repair, and if these cells are more likely to initiate cancer then again the apparent rate of somatic mutation is increased. Finally, there is a strong possibility that epigenetic changes contribute to cancer initiation, which would also increase the estimated somatic mutation rate.
Recent work of Tomasetti et al. [36] suggests that only three driver mutations (i.e. M = 3) are required to initiate lung and colorectal cancer. Their calculations are based on comparing the incidence of adenocarcinoma of the lung between smokers and non-smokers and of the colon between those with or without MLH1 silencing given estimates of their relative somatic mutation rates (u). As can be seen from equation (3.1), the effect of changes in u on cancer incidence scales as uM. In the case of lung cancer, the ratio was estimated at 3.23, predicting a 33.7-fold increase given M = 3. This estimate of M is in accord with the estimate based on stem-cell divisions (figure 1). In the case of colon cancer, they used a mutation rate ratio of about 8 to account for the 114-fold increase in incidence, which is consistent with M = 3 (or less). This estimate of M is substantially lower than the estimate of M = 7 based on stem-cell divisions (figure 1). It suggests that one or more of the parameters used in one or both estimates are inaccurate. In this context, it is notable that the estimates of the somatic mutation rate was highly variable: the ratio of the somatic mutation rates was 8.3 based on median values (two estimates), leading to the result M ≤ 3, whereas the same ratio based on means was 2.0, leading to the result M = 7, identical to the result based on stem-cell divisions.
Contrary to Tomasetti & Vogelstein [31], the recognition that M varies among the cancers means that the data shown in figure 1 cannot be used to identify stochastic versus environmental and familial differences in the causation of the cancers (except in the obvious within-cancer cases, such as lung cancer incidence in smokers versus non-smokers). In principle, it would be possible to compare cancers with the same value of M, using their deviation from the integer value estimated to measure environmental or familial influence. For example, for lung adenocarcinoma, the integer estimate for non-smokers and smokers is M = 3; however, for non-smokers the actual estimate is 3.0, whereas for smokers it is 2.7. Thus, smokers appear to have 10% less protection (suggesting a u increased 2.6 fold), reflecting the carcinogenic effect of smoking. However, variation in target sizes (and hence somatic mutation rates) plus other inaccuracies in the data immediately bring such a subtle approach into question when comparing between different cancers with the same integer value of M. As noted by others, the conclusion of Tomasetti & Vogelstein [31] that two-thirds of cancers are due to bad luck (i.e. random but unavoidable mutation) does not follow from the data [37]. The real value for a given cancer could be higher or lower, but we simply cannot predict from these data. Most environmental carcinogens probably act either directly or indirectly by increasing the somatic mutation rate, so that factoring out their contribution would require a knowledge of the minimum somatic mutation rate for each specific cancer.
4. Why does cancer persist?
One important contribution of an evolutionary approach to cancer suppression is to provide a more nuanced understanding of why early-onset familial cancer persists. In general, serious genetic diseases that significantly reduce an individual's fitness are rare in a population, maintained at a predictable level by a process called mutation-selection balance: natural selection eliminates deleterious alleles, but recurrent mutation creates new ones. The balance point for single gene disorders has long been known, and depends upon whether the mutant alleles are dominant or recessive, whether they are autosomal or sex-linked, and how much they affect an individual's fitness. This provides an accurate picture of mutation-selection balance for the only definitive case of cancer suppression by a single gene, retinoblastoma, which acts as an autosomal dominant [1]. For other cancers, more genes are involved making the mutation-selection balance calculation more complex; however, assuming that a cancer is regulated by a set of identical tumour suppressor genes, then it is possible to accurately approximate the frequency q of mutant alleles at each locus at mutation-selection balance by a cubic equation in q [23]. This approach predicts that the commonest early-onset (i.e. pre-reproductive) cancers will have the lowest proportion of familial cases, and conversely, that very rare cancers will be almost entirely inherited. This negative correlation arises because cancer suppression is inevitably imprecise since it depends on a small number of genes that are likely to be recruited one at a time. This integer property creates stepwise levels of suppression [23]. As a result, we can expect some cancers to be so well controlled that only individuals inheriting a mutation probably succumb to the disease; such cancers are largely familial. Others will be less effectively controlled, and hence more common because even individuals with the ‘best’ genotype succumb. Under this less effective control, selection against genotypes carrying mutant alleles is very strong, which together with the increased occurrence of sporadic cancers, results in a low proportion of familial cases [23]. For example, the most common solid tumour of children is neuroblastoma and most cases are sporadic—estimates of familial cases range from only a few per cent [38] up to about 25% [39].
Most cancers arise late in life when natural selection is less effective. In that respect there is an evolutionary analogy between late-onset cancer and senescence, and as such there are two broad types of hypothesis to account for late-life cancer. The first is the absence of selection to promote cancer suppression late in life: once reproduction is complete (noting that in this context reproduction includes any late-life ‘grandmother effect’ or similar behaviour that enhances the reproductive success of offspring; [40]), then natural selection cannot act (which can be termed the ‘obsolescence hypothesis’). The second concerns antagonistic pleiotropy, which is the possibility that cancer suppression later in life trades off with some benefit early in life, such as the suggestion that the negative effects of BRCA1/2 mutations trade-off with increases in fertility [41]. These two possibilities are generally hard to distinguish; however, antagonistic pleiotropy would generally be expected to result in significant rates of cancer prior to the cessation of reproduction.
The obsolescence hypothesis is based on the reality that natural selection will act on cancer suppression with declining effectiveness as reproduction proceeds to completion. As a result, all cancers are expected to be at a fairly low frequency until the post-reproductive period, but beyond that point cancers will increase. Viewed in terms of a multistage process, this means that cells will generally be one or more mutational steps away from cancer initiation when reproduction draws to a close. This result has an interesting consequence noted by Nunney [23]. Tissues that are relatively small and have a slow rate of division should remain largely cancer-free well beyond the reproductive threshold, as they will accumulate additional somatic mutations at a slow rate. By contrast, large rapidly dividing tissues are expected to rapidly accumulate additional mutations and initiate cancer. This is a strong prediction of the evolutionary model and is supported in humans by the observation that epithelial cancers increase markedly with age, a transition that DePinho [42] noted as an important trend needing an explanation. Epithelial cancers are generally in tissues that are large and divide relatively rapidly, as evidenced by the high numbers of ‘hits’ estimated for colorectal and small intestine adenocarcinomas (figure 1), so their late-life dominance is to be expected. Further illustration is provided by the profoundly different distribution in the types of cancers seen in children versus adults [43].
5. Conclusion
Evolutionary modelling has been very successful in beginning to develop a framework for understanding the similarities and differences among cancers, both within and between species. While the model of multistage carcinogenesis is an over-simplification, it provides testable hypotheses, and where necessary will undoubtedly be improved. In general, the match between the simple model and observation is very good: Peto's paradox can be resolved by adaptation; the hallmarks of cancer can be viewed as cells overcoming a set of evolved defences; the data on stem-cell divisions are consistent with a multistage model with the number of mutational hits varying from 3 to 7; and the dominance of epithelial cancers in old age is consistent with the evolutionary model. However, a value of models is also to highlight results that do not immediately fit expectation, hence we will end with an interesting conundrum: Varki & Varki [27] note that adenocarcinomas and similar epithelial cancers are not observed in old chimpanzees. This is contrary to expectation and suggests a very interesting starting point for some more detailed comparative oncology.
Acknowledgements
We would like to thank Michael Hochberg and two reviewers for their valuable comments.
References
- 1.Knudson A. 1971. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA 68, 820–823. ( 10.1073/pnas.68.4.820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339, 1546–1558. ( 10.1126/science.1235122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangarajan A, Weinberg RA. 2003. Comparative biology of mouse versus human cells: modeling human cancer in mice. Nat. Rev. Cancer 3, 952–959. ( 10.1038/nrc1235) [DOI] [PubMed] [Google Scholar]
- 4.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6, 171–183. ( 10.1016/j.ccr.2004.07.009) [DOI] [PubMed] [Google Scholar]
- 5.Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J. 2014. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540. ( 10.1038/nrg3728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144, 646–674. ( 10.1016/j.cell.2011.02.013) [DOI] [PubMed] [Google Scholar]
- 7.Nordling CO. 1953. A new theory on the cancer-inducing mechanism. Br. J. Cancer 7, 68–72. ( 10.1038/bjc.1953.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armitage P, Doll R. 1954. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br. J. Cancer 8, 1–12. ( 10.1038/bjc.1954.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunney L. 1999. Lineage selection and the evolution of multistage carcinogenesis. Proc. R. Soc. Lond. B 266, 493–498. ( 10.1098/rspb.1999.0664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peto R. 1977. Epidemiology, multistage models, and short-term mutagenicity tests. In The origins of human cancer, vol. 4 (eds Hiatt HH, Watson JD, Winsten JA.), pp. 1403–1428. Cold Spring Harbor Conf. on cell proliferation New York, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 11.Nunney L. 2013. The real war on cancer: the evolutionary dynamics of cancer suppression. Evol. Appl. 6, 11–19. ( 10.1111/eva.12018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairns J. 1975. Mutation selection and the natural history of cancer. Nature 255, 197–200. ( 10.1038/255197a0) [DOI] [PubMed] [Google Scholar]
- 13.Muir B, Nunney L. 2015 The expression of tumor suppressors and proto-oncogenes in tissues susceptible to their hereditary cancers. Brit. J. Cancer. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100, 57–70. ( 10.1016/S0092-8674(00)81683-9) [DOI] [PubMed] [Google Scholar]
- 15.Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. 2007. Telomerase activity coevolves with body mass not lifespan. Aging Cell 6, 45–52. ( 10.1111/j.1474-9726.2006.00262.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbunova V, Hine C, Tian X, Ablaeva J, Gudkov AV, Nevo E, Seluanov A. 2012. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl Acad. Sci. USA 109, 19 392–19 396. ( 10.1073/pnas.1217211109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caulin AF, Graham TA, Wang L-S, Maley CC. 2015. Solutions to Peto's paradox revealed by mathematical modeling and cross-species cancer gene analysis. Phil. Trans. R. Soc. B 370, 20140222 ( 10.1098/rstb.2014.0222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian X, et al. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349. ( 10.1038/nature12234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzourakis A, Magiorkinis G, Lim AG, Gupta S, Belshaw R, Gifford R. 2014. Larger mammalian body size leads to lower retroviral activity. PLoS Pathog. 10, e1004214 ( 10.1371/journal.ppat.1004214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunney L. 1999. Lineage selection: natural selection for long-term benefit. In Levels of selection in evolution (ed. Keller L.), pp. 238–252. Princeton, NJ: Princeton University Press. [Google Scholar]
- 21.Alexandrov LB, et al. 2013. Signatures of mutational processes in human cancer. Nature 500, 415–421. ( 10.1038/nature12477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nature Rev. Cancer 4, 177–183. ( 10.1038/nrc1299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunney L. 2003. The population genetics of multistage carcinogenesis. Proc. R. Soc. Lond. B 270, 1183–1191. ( 10.1098/rspb.2003.2351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plevová P, Sedláková E, Zapletalová J, Křepelová A, Skýpalová P, Kolář Z. 2005. Expression of the hMLH1 and hMSH2 proteins in normal tissues: relationship to cancer predisposition in hereditary non-polyposis colon cancer. Virchows Arch. 446, 112–119. ( 10.1007/s00428-004-1139-5) [DOI] [PubMed] [Google Scholar]
- 25.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. 2014. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 10, e1004239 ( 10.1371/journal.pgen.1004239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browning HM, Gulland FMD, Hammond JA, Colegrove KM, Hall AJ. 2015. Common cancer in a wild animal: the California sea lion (Zalophus californianus) as an emerging model for carcinogenesis. Phil. Trans. R. Soc. B 370, 20140228 ( 10.1098/rstb.2014.0228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varki NM, Varki A. 2015. On the apparent rarity of epithelial cancers in captive chimpanzees. Phil. Trans. R. Soc. B 370, 20140225 ( 10.1098/rstb.2014.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy MK, Page RL, Jensen WA, Olson PN, Haworth JD, Searfoss EE, Brown DE. 2015. The Golden Retriever Lifetime Study: establishing an observational cohort study with translational relevance for human health. Phil. Trans. R. Soc. B 370, 20140230 ( 10.1098/rstb.2014.0230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiffman JD, Breen M. 2015. Comparative oncology: what dogs and other species can teach us about humans with cancer. Phil. Trans. R. Soc. B 370, 20140231 ( 10.1098/rstb.2014.0231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis BW, Ostrander EA. 2014. Domestic dogs and cancer research: a breed-based genomics approach. ILAR J. 55, 59–68. ( 10.1093/ilar/ilu017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasetti C, Vogelstein B. 2015. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81. ( 10.1126/science.1260825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noble R, Kaltz O, Hochberg ME. 2015. Peto's paradox and human cancers. Phil. Trans. R. Soc. B 370, 20150104 ( 10.1098/rstb.2015.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hethcote HW, Knudson AG. 1978. Model for the incidence of embryonal cancers: application to retinoblastoma. Proc. Natl Acad. Sci. USA 75, 2453–2457. ( 10.1073/pnas.75.5.2453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch M. 2010. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl Acad. Sci. USA 107, 961–968. ( 10.1073/pnas.0912629107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowell PC. 1976. The clonal evolution of tumor cell populations. Science 194, 23–28. ( 10.1126/science.959840) [DOI] [PubMed] [Google Scholar]
- 36.Tomasetti C, Marchionnic L, Nowak MA, Parmigianie G, Vogelstein B. 2015. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc. Natl Acad. Sci. USA 112, 118–123. ( 10.1073/pnas.1421839112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Various authors. 2015. Letters on cancer risk. Science 347, 727–730. [Google Scholar]
- 38.Ogawa S, Takita J, Sanada M, Hayashi Y. 2011. Oncogenic mutations of ALK in neuroblastoma. Cancer Sci. 102, 302–308. ( 10.1111/j.1349-7006.2010.01825.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knapke S, Nagarajan R, Correll J, Kent D, Burns K. 2012. Hereditary cancer risk assessment in a pediatric oncology follow-up clinic. Pediatr. Blood Cancer 58, 85–89. ( 10.1002/pbc.23283) [DOI] [PubMed] [Google Scholar]
- 40.Brown JS, Aktipis A. 2015. Inclusive fitness effects can select for cancer suppression into old age. Phil. Trans. R. Soc. B 370, 20150160 ( 10.1098/rstb.2015.0160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith KR, Hanson HA, Hollingshaus MS. 2013. BRCA1 and BRCA2 mutations and female fertility. Curr. Opin. Obstet. Gynecol. 25, 207–213. ( 10.1097/GCO.0b013e32835f1731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DePinho RA. 2000. The age of cancer. Nature 408, 248–254. ( 10.1038/35041694) [DOI] [PubMed] [Google Scholar]
- 43.Downing JR, Wilson RK, Zhang J, Mardis ER, Pui C-H, Ding L, Ley TJ, Evans WE. 2012. The pediatric cancer genome project. Nat. Genet. 44, 619–622. ( 10.1038/ng.2287) [DOI] [PMC free article] [PubMed] [Google Scholar]