Abstract
The role of different czcRS genes in metal resistance and the cross-link between czcRS and czcCBA in Pseudomonas putida X4 were studied to advance understanding of the mechanisms by which P. putida copes with metal stress. Similar to P. putida KT2440, two complete czcRS1 and czcRS2 two-component systems, as well as a czcR3 without the corresponding sensing component were amplified in P. putida X4. The histidine kinase genes czcS1 and czcS2 were inactivated and fused to lacZ by homologous recombination. The lacZ fusion assay revealed that Cd2+ and Zn2+ caused a decrease in the transcription of czcRS1, whereas Cd2+ treatment enhanced the transcription of czcRS2. The mutation of different czcRSs showed that all czcRSs are necessary to facilitate full metal resistance in P. putida X4. A putative gene just downstream of czcR3 is related to metal ion resistance, and its transcription was activated by Zn2+. Data from quantitative real-time polymerase chain reaction (qRT-PCR) strongly suggested that czcRSs regulate the expression of czcCBA, and a cross-link exists between different czcRSs.
Keywords: czcRS, two-component system, metal resistance, Pseudomonas putida
1. Introduction
Heavy metal pollution leads to serious ecological and health problems due to the toxic effects of metal ions and their accumulation throughout the food chain. Even the essential metal ions, Zn2+, Cu2+, and Co2+ could be toxic at high concentrations [1,2,3]. Many microorganisms naturally possess the ability to inhabit metal-polluted areas. An advanced understanding of the mechanisms by which microbes cope with metal stress will facilitate the rational design of strategies for detection and bioremediation of heavy metal-polluted water and soil systems.
To cope with metal stresses, bacteria develop various detoxification mechanisms. The most effective way is the extrusion of excessive metal ions out of the cell by active transport. A number of studies over the past years have been devoted to the protein families that export metal ions. The protein families include P-type ATPases driven by ATP hydrolysis, cation diffusion facilitator family transporters, which act as chemiosmotic ion-proton exchangers, and the resistance nodulation division (RND) family of transporters that mediate proton-driven efflux [4]. Usually, more than one kind of efflux protein is found in metal resistance bacteria [5,6]. CzcCBA is a CBA transporter. This structural gene region encodes outer membrane factors CzcC, membrane fusion protein CzcB, and CzcA protein of the resistance-nodulation-cell division protein family [4]. The transcriptional responses of these exporting genes are controlled by different regulators, including MerR family regulators [7], the ArsR/SmtB family [8], and two-component systems [9,10,11].
Many adaptive bacterial responses to environmental changes are governed by two-component signal transduction systems [12]. In typical two-component systems, environmental variation is first sensed by a histidine sensor kinase capable of autophosphorylation. The phosphoryl group is then transferred to an aspartic acid residue of the response regulator. Usually, the phosphorylated response regulator binds to DNA, resulting in either activation or repression of target genes. Caille et al. [9] observed that in P. aeruginosa PAO1, Zn2+ and Cu2+ enhance the expression of czcRS. Phosphorylated CzcR then activates the expression of czcCBA operon encoding an efflux pump specific for zinc, cadmium, and cobalt. The copRS two-component system has been identified as key genes involved in copper resistance [13]. In Cupriavidus metallidurans CH34, the regulatory genes of czcCBA are arranged in an upstream region consisting of czcN and czcI, and a downstream region consisting of czcD, czcR, czcS and czcE [14]. CzcRS and a periplasmic copper-binding protein designated CzcE [15], exert metal-dependent control of czcNICBA expression via regulation of czcNp activity [16]. Grosse et al. [10] demonstrated that uninduced C. metallidurans CH34 with a mutation in czcS contains more czcCBA message and resumes growth faster when challenged. These pieces of evidence demonstrate the participation of two-component systems in the regulation of metal extrusion.
P. putida is a ubiquitous saprophytic bacterium, which has been extensively studied for the biodegradation of organic pollutants [17]. Although the genomes of P. putida contain several czcRS operons [5], no research has been reported regarding the simultaneous function of different czcRSs in the same strain. Our previous study indicated that P. putida X4 has a high tolerance and absorption capacity for metal ions [18]. In this study, PCR with primers designed from the homologous sequence in genome sequenced P. putida were used to detect the presence of czcRS two-component systems and the czcCBA operon. Because two czcRS two-component systems and one czcR3 without its corresponding sensor gene were detected, we examined their different roles and how they interacted with the czcCBA operon.
2. Results
2.1. Minimum Inhibitory Concentrations (MICs) of the Wild-Type and Mutant Strains
The MIC values of the wild-type and mutant strains are shown in Table 1. Determination of heavy metal resistance showed that the strain X4 displayed a high level of Cd2+, Co2+, Cu2+, and Zn2+ resistance. The functionalities of czcS1, czcS2, and czcH were studied by determining the resistance of gene-deficient strains to different divalent heavy metals. ΔS1 tended to be sensitive to Cd2+, Co2+, and Zn2+ ions. The MIC of Co2+ and Zn2+ declined to 0.5 and 2.25 mmol·L−1. The czcS2 mutation decreased the strain’s resistance to Cd2+ and Co2+. Both czcS1 and czcS2 were required for generating full cadmium resistance as the ΔS1S2 strains with both interrupted czcS1 and czcS2 could tolerate only 1.25 mmol·L−1 Cd2+, which is lower than that tolerated by single gene mutant strains. Small but reproducible repression was observed when ΔS1 and ΔS2 were exposed to copper ions. A putative open reading frame (ORF), designated as czcH in this study, just downstream of czcR3, was identified as necessary to generate full metal tolerance. The resistance of ΔczcH to Cd2+, Co2+, and Zn2+ were lower than that of the wild-type strains. The results presented above demonstrate that czcS1, czcS2, and czcH are necessary for the wild-type strain to generate full resistance to Cd2+, Co2+, Cu2+ and Zn2+.
Table 1.
Minimum inhibitory concentrations (MICs) of cadmium, cobalt, copper, and zinc in various derivatives of P. putida X4.
| Strains | MIC (mmol·L−1) a | |||
|---|---|---|---|---|
| Cd2+ | Co2+ | Cu2+ | Zn2+ | |
| X4 | 4.5 | 4.0 | 5.0 | 8.5 |
| X4 with pvlt31 | 4.5 | 4.0 | 5.0 | 8.5 |
| ΔS1 | 3.0 | 0.5 | 4.25 | 2.25 |
| ΔS2 | 2.0 | 1.5 | 4.75 | 8.5 |
| ΔczcH | 2.0 | 1.75 | 5.0 | 7.0 |
| ΔS1S2 | 1.25 | 0.5 | 4.25 | 5.0 |
| ΔS1c | 4.25 | 3.5 | 4.5 | 7.5 |
| ΔS2c | 4.25 | 4.0 | 4.75 | 8.5 |
| ΔczcHc | 3.75 | 3.5 | 5.0 | 8.25 |
a MICs were recorded after 60 h of growth at 30 °C on Tris-buffered medium containing metals salts from 0.5 to 10 mmol·L−1, with 0.25 mmol·L−1 interval. MIC values are the averages of four determinations.
2.2. Genetic Complementation of Mutant Genes
Complementation studies were carried out to confirm that the decreased resistance of strains ΔS1, ΔS2, and ΔczcH to divalent metal ions was caused by the mutation of target genes. For the complementation analysis, the complete ORF of czcS1, czcS2 and czcH was cloned into the broad-host-range vector pVLT31, resulting in plasmids pVLTS1, pVLTS2 and pVLTH. Strains ΔS1c, ΔS2c and ΔczcHc were obtained by mating the complementation plasmid into P. putida ΔS1, ΔS2 and ΔczcH, respectively. The MICs of ΔS1c, ΔS2c and ΔczcHc were tested in Tris-buffered medium with 0.1 mmol·L−1 IPTG using the method described above. The complementation data are shown in Table 1. More than 85% of the tolerance to the tested metals was recorded for ΔS1c and ΔczcHc. The resistance of ΔS2c to Co2+ and Cd2+ were recorded at 100% and 94%, respectively. At the same time, the MICs of control strains did not improve.
2.3. Cd2+ and Zn2+ Repressed the Transcription of czcRS1, Induced the Transcription of czcRS2 and czcH, Respectively
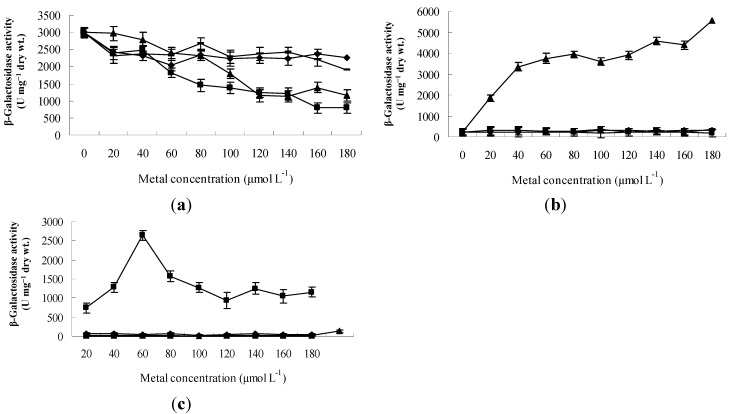
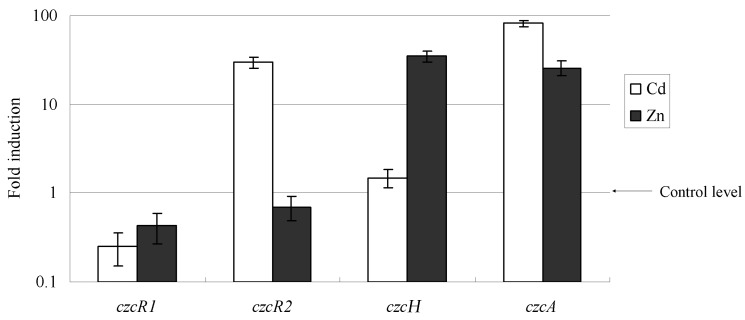
Expression patterns and inducer specificities were determined by measuring the changes of LacZ activity and verified by qRT-PCR. All promoters were tested for their inducibility in the presence of 0–180 μmol·L−1 Zn2+, Co2+, Cu2+, and Cd2+. The expression patterns and inducer specificities of czcS1 are shown in Figure 1a. The czcS1 is transcribed without any metal ions. The transcription of czcS1 dropped to 40% in the presence of 180 μmol·L−1 Zn2+, then to 34% after a 180 μmol·L−1 Cd2+ treatment. Compared with Zn2+ and Cd2+, Cu2+ and Co2+ moderately decreased the transcription of czcS1. The transcription of czcS2 was only induced by Cd2+; LacZ activity in ΔS2 increased about 24-fold after exposure to 180 μmol·L−1 Cd2+ (Figure 1b). The czcH gene was significantly induced by Zn2+, but was not affected by Co2+, Cu2+ and Cd2+. The highest level of czcH transcription was found at 100 μmol·L−1 Zn2+ and it was decreased when the Zn2+ concentration was higher than 100 μmol·L−1 (Figure 1c). These observations reveal that transcription from the promoters of czcS1, czcS2 and czcH are different, and that the main inducers or repressors are Zn2+ and Cd2+. qRT-PCR data confirmed the inducibility presented above (Figure 2); in the wild-type strain, addition of 300 μmol·L−1 Cd2+ or Zn2+ to the culture resulted in down-regulation of czcRS1 and up-regulation of czcRS2 and czcH, respectively (30-, and 35-fold). Besides, the transcription of czcCBA was strongly induced by Cd2+ (90-fold) and Zn2+ (47-fold) to keep the equilibrium of these metal ions in the cells.
Figure 1.
The expression patterns and inducer specificities of czcS1, czcS2, and czcH. LacZ activity were measured in permeabilized cells after induction with different concentrations of Cu2+ (◆), Zn2+ (■), Cd2+ (▲), and Co2+ (-) for four hours. Error bars represent the standard deviations of three determinations. (a) Response of the promoter czcRS1p to different metal ions; (b) Response of the promoter czcRS2p to different metal ions; and (c) Response of the promoter czcHp to different metal ions.
Figure 2.
Transcription of czcR1, czcR2, czcH, and czcA genes analyzed by qRT-PCR in the wild-type X4 strain grown in the presence of 300 μM Zn2+ or Cd2+. The amount of mRNA is represented relative to the X4 strain cultivated in the absence of metal ions. Error bars represent the standard deviations of three determinations.
2.4. The Transcription Variation of Mutant Strains
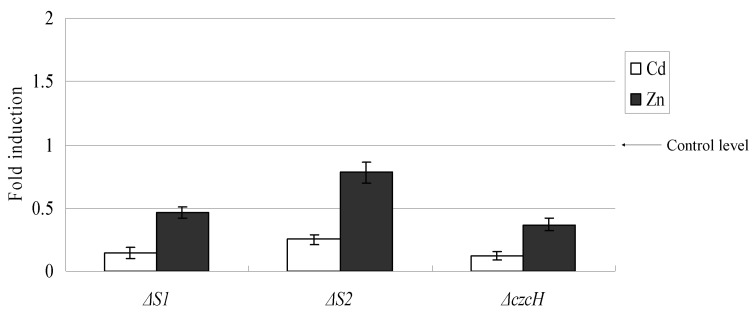
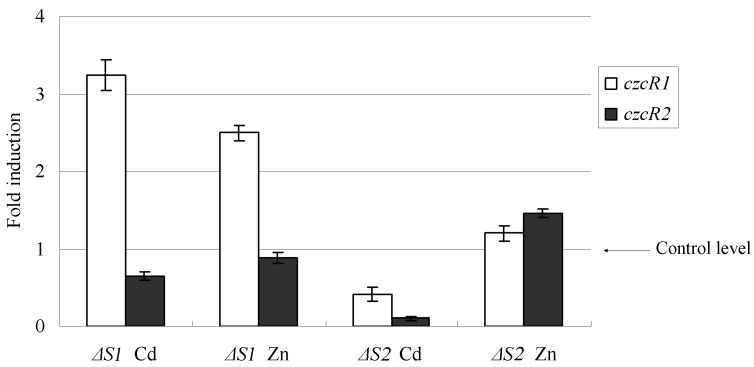
After treatment with Cd2+ or Zn2+, transcription of czcA was determined by qRT-PCR in the wild-type X4 strain and mutant strains. As shown in Figure 3, the mutation of czcS1 and czcS2 reduced the mRNA amount of czcA to 14%, and 23% that of the wild-type, respectively, in the presence of Cd2+. A similar trend was detected upon induction with Zn2+. The mutation in czcH also caused repressed transcription of the czcA. These findings show that all these three genes are required to maintain the expression of czcA at a high level. The transcript variation of czcR1 and czcR2 is shown in Figure 4. Compared with the wild-type, interruption of czcS1 enhanced the transcription of czcR1 by 3.22- and 2.5-fold, in the presence of Cd2+ and Zn2+, respectively. Mutation of czcS2 reduced the czcR1 and czcR2 mRNA to 40% and 2% of the wild type, while cultured with Cd2+. These transcription variations indicated that czcS1 in wild-type act negatively on the transcription of czcR1, and that functional czcS2 is needed to keep transcription of czcR1 and czcR2 at a relatively high level.
Figure 3.
The effects of the mutation in czcS1, czcS2, and czcH on the transcription of czcA. Transcription of czcA was analyzed by qRT-PCR in the wild-type X4 strain and mutant strains grown in the presence of 300 μM Zn2+ or Cd2+. The amount of mRNA is represented relative to the wild-type X4 strain cultivated in the same concentration of metal ions. Error bars represent the standard deviations of three determinations.
Figure 4.
The effects of the mutation in czcS1 and czcS2 on the transcription of czcR1and czcR2. Transcription of czcR1 and czcR2 was analyzed by qRT-PCR in the wild-type X4 strain and mutant strains grown with 300 μM Zn2+ or Cd2+. The amount of mRNA is represented relative to the wild-type X4 strain cultivated in the same concentration of metal ions. Error bars represent the standard deviations of three determinations.
2.5. Deposition of Strains and Nucleotide Sequences
P. putida X4 was deposited in the China Center for Type Culture Collection (CCTCC, http://www.cctcc.org/) with accession number CCTCCM209319. The nucleotide sequences isolated in this study are available in the NCBI Genbank database: czcR1 (HQ676126), czcR2 (HQ676125), czcS1 (HQ676127), czcS2 (HQ676128), czcA (HQ676129), czcH (dsgR3, HQ676130), czcR3 (HQ676124). The similarities of deduced amino acid sequences of czcR and czcS genes are available in Table S1.
3. Discussion
In the present study, we demonstrate the roles played by the czcRS two-component systems in the control of czcCBA transcription as well as their different inducibility. To our knowledge, this is the first report on the functional difference of czcRS two-component systems in P. putida. Our results clearly show that all czcRS two-component systems are needed for P. putida X4 to generate full metal resistance. The reductions in MICs of Zn2+, Co2+, and Cd2+ in the czcS mutant strains were less than eight-fold. It cannot be excluded that czcCBA is also induced by other regulators, or that other transporters present in X4 may contribute to the intrinsic resistance of this organism to heavy metals. The amplified czcA in this study which is induced by Zn2+, and Cd2+ in wild type revealed a >99% nucleotide identity with the czcCBA1 operon in P. putida KT2400. The czcCBA1 operon in P. putida KT2440 plays a major role in generating metal resistance to Zn2+, Cd2+, and possibly Pb2+ [6]. The czcCBA found in C. metallidurans CH34 confers resistance to Zn2+, Co2+, and Cd2+ [10]. Similar systems have been found in P. aeruginosa PAO1 and P. aeruginosa CMG103 [9,19].
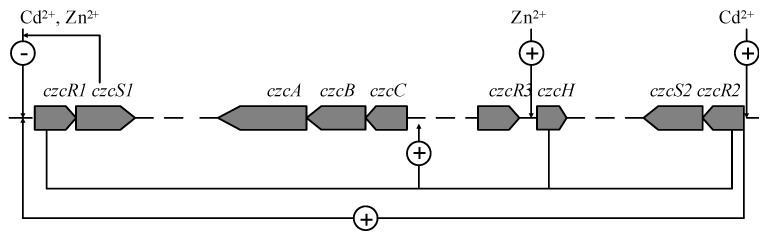
Plasmid pVIK112 was constructed by Kalogeraki et al. [20] to simultaneously disrupt and create fusions to target genes of diverse bacteria. The sequence of the fragment amplified in this study and the complete genome analysis of P. putida KT2440 [5] revealed that the czcRS in P. putida form separate operons and czcSs is located at the end of the operons. Therefore, a minimal polar effect was generated after single cross-over with pVIK112. The remarkable increase in metal resistance in the complementation strains supports this idea. Combining the data from the β-galactosidase activity assay and the transcription variation of czcRs and czcCBA, the cross-interaction pathway among Zn2+, Cd2+, czcRS and czcCBA was depicted (Figure 5). The control of czcCBA expression through czcRS has been studied in P. aeruginosa and C. metallidurans [9,10,11]. Compared with previous studies, the regulation of czcCBA in P. putida was different. P. putida evolved two czcRS operons. Simultaneously, other regulation genes, such as czcN and czcI were not found in the P. putida sequenced genome. The expression of the CzcCBA efflux pump was induced by two czcRS operons with different inducibilities. The czcH gene also influenced the transcription of czcCBA. The function of czcRS1, czcRS2 and czcH in P. putida X4 may be complementary. The transcription of czcRS1 was inhibited in the presence of Zn2+ and Cd2+, which may caused the reduced transcription of czcCBA. However, a high level of efflux pump CzcCBA expression is essential for coping with high concentrations of metal ions (Figure 2). The low expression trend of czcCBA resulting from the CzcR1 may be compensated for by the induction effect caused by czcRS2 and czcH, which were induced with Cd2+ and Zn2+, respectively. Complete czcRS1 negatively feedbacks on its expression, which creates a common regulating pathway in prokaryotes for maintaining the expression of transcription factors at appropriate levels [21]. Complete czcRS2 has a positive effect on the transcription of itself. This positive autoregulation loop could allow the cells to respond rapidly to the presence of small amounts of heavy metals in the environment [11].
Figure 5.
Model representing the cross-interaction among Zn2+, Cd2+, czcRS, and czcA. Based on the data from the LacZ fusion assay and qRT-PCR, the cross-talk among Zn2+, Cd2+, czcRS, and czcA was depicted. Zn2+ and Cd2+ repressed the transcription of czcRS1, and induced the transcription of czcRS2 and czcH, respectively. The czcRs and czcH gene could induce czcCBA transcription, leading to resistance to cobalt, zinc, and cadmium. Data from qRT-PCR also revealed a cross-interaction between czcRS2 and czcRS1.
In conclusion, there was a clear distinction between the different czcRSs. czcRS1 is constitutively expressed and possibly acts as a housekeeping resistance mechanism against Zn2+, Co2+, and Cd2+ in P. putida X4. The transcription of czcRS1 was repressed by Cd2+ and Zn2+, whereas the transcription of czcRS2 was induced only by Cd2+. Both czcS1 and czcS2 act positively on the transcription of czcA. Although czcH plays an important role in the metal resistance of P. putida X4, the underlying mechanism merits further studies. Different czcRS genes, along with other regulators, are needed to make precise adjustments to the gene expression of P. putida to adapt to changes in metal ions in the environment.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
Detailed information regarding the bacterial strains and plasmids are listed in Table 2. P. putida X4 was isolated from the waste soil around the Academy of Hubei Agricultural Sciences, Wuhan, China [22]. For construction and maintenance of plasmids, Escherichia coli strains DH5α and C118λpir were used.
Table 2.
Bacterial strains and plasmids used in this study.
| Relevant Characteristics a | Reference | ||
|---|---|---|---|
| Strains | E. coli | ||
| DH5α | supE44 ΔlacU169 (Ф80dlacZΔM15) recA1 endA1 hsdR17 thi-1 gyrA96 relA1 | Invitrogen | |
| C118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE (Am) recA1 λpir | [23] | |
| MM294 | endA thiA hsdR17 supE44 | [24] | |
| P. putida | |||
| X4 | Apr, Cor, Znr, Cdr, Cur, wild-type | [22] | |
| ΔS1 | X4ΔczcS1 | This study | |
| ΔS1c | X4ΔczcS1, with plasmid pVLTS1 | This study | |
| ΔS2 | X4ΔczcS2 | This study | |
| ΔS2c | X4ΔczcS2, with plasmid pVLTS2 | This study | |
| ΔczcH | X4ΔczcH | This study | |
| ΔczcHc | X4ΔczcH, with plasmid pVLTH | This study | |
| ΔS1S2 | X4ΔczcS1czcS2 | This study | |
| Plasmid | pRK2073 | RK2 helper plasmid, Sper | [24] |
| pTA2 | Cloning vector, Ampr | TOYOBO | |
| pVIK112 | LacZYA, Kmr, suicide vector | [20] | |
| pVIks1 | S1 fragment in pVIK112 | This study | |
| pVIKs2 | S2 fragment in pVIK112 | This study | |
| pVIKH | czcH fragment in pVIK112 | This study | |
| pRRT | Contains a Tetr gene instead of Kmr cassette | This study | |
| pRRTS2 | S2 fragment in pRRT | This study | |
| pVLT31 | lacI, Tetr | [25] | |
| pVLTS1 | pVLT31 with S1 ORF | This study | |
| pVLTS2 | pVLT31 with S2 ORF | This study | |
| pVLTH | pVLT31 with czcH ORF | This study | |
a Cor, Znr, Cdr, and Cur indicate resistance to cobalt, zinc, cadmium, and copper, respectively; Ampr, Kmr, Tetr, and Sper stand for resistance to ampicillin, kanamycin, tetracycline, and spectinomycin, respectively.
Bacteria were routinely grown in Luria–Bertani medium at 37 °C (E. coli) or 30 °C (P. putida). When required, antibiotics were added at the following final concentrations: for E. coli, 100 µg/mL ampicillin, 50 μg/mL kanamycin, and 12.5 μg/mL tetracycline; and for P. putida, 50 μg/mL kanamycin and 12.5 μg/mL tetracycline. Tris-buffered medium [26] containing 0.1% glucose was used for testing metal resistance and promoter inducibility. Analytical grade reagents CuSO4·5H2O, CdCl2·H2O, ZnCl2, and CoCl2·6H2O were prepared into 1.0 M stock solutions and sterilized by filtration.
4.2. Determination of MIC Values
The minimum inhibitory concentrations (MICs) of trace metals were determined as previously described [26]. Briefly, bacteria cultures were spread onto the Tris-buffered medium containing metal salts from 0.5 to 10 mmol·L−1 (0.25 mmol·L−1 interval). The MICs were defined as the lowest concentration of metal salts at which no CFU were observed after 60 h of incubation at 30 °C.
4.3. DNA Manipulation
All primer pairs used in this study were shown in Table 3. Axygene mini-prep, PCR purification, and gel purification kits (Axygene, Union City, CA, USA) were chosen for DNA separation and purification. PCRs were carried out using Taq DNA polymerase. The plasmids were constructed using standard recombinant DNA techniques and introduced into E. coli by transformation. Plasmids derived from pVIK112 [20] were introduced into P. putida by triparental conjugation. The helper strain was E. coli MM294 with the helper plasmid pRK2073 [24]. The essential DNA fragment was verified by DNA sequencing.
Table 3.
Primers used in this study.
| Purpose | Primer Pair | Sequence (5ʹ-3ʹ) a | Product Length (bp) |
|---|---|---|---|
| For homologous recombination | S1 | gag cag acc tgg aag taa aga | 1113 |
| ggt aga acc gct caa aca a | |||
| S2 | cgt agg cta tgt act tga ggc g | 920 | |
| tgt cgt tga tga tgc ggt tg | |||
| czcH | cac agg gca ttc agg gac caa cgc acg gga taa gag | 496 | |
| gcc cgt tgc acc aca gat | |||
| For qRT-PCR | Qr1 | aca acg gtg tag atg ctc tgc | 121 |
| cgg ctg gtc tta cgg atg g | |||
| Qr2 | gcc gca acg acc agc aac | 144 | |
| gac gca tca gca ggt gta gc | |||
| Qr3 | atg atg ctg acg gcg aga ag | 162 | |
| gcg aat gac ctc tac gga tgc | |||
| QczcA | cca ctg agc acg acc aag g | 128 | |
| aag gtg aag gaa gag gaa ggc | |||
| QrpsL | ctg cgt aaa gta tgc cgt gtg | 174 | |
| gcc cga agt atc cag aga gc | |||
| For complementation experiment | CS1 | cgg ggt acc taa gaa gga gat ata cca tga ggc cat tca gcc tgg | 1455 |
| cta gtc tag att aag cgg cgg tca ttg c | |||
| CS2 | cgg ggt acc taa gaa gga gat ata ccttg aaa aac gcc agc ctg tc | 1419 | |
| cta gtc tag atc act cgg cag gaa aca cca | |||
| CczcH | cgg ggt acc taa gaa gga gat ata cca tga ggt ata gca ttg att atc agc a | 360 | |
| cta gtc tag att ata aga agg cga gcg ag | |||
| For tetracycline resistance gene | Tet | cga cct gca gaa aat agg cgt atc acg agg | 1560 |
| cag cct gca gtc tgc taa cca gta agg caa cga gg |
a underlined are restriction sites.
4.4. Homologous Recombination for Construction of czcS Mutants and lacZ Fusion Reporter Strains
The suicide fusion vector pVIK112 was selected for the mutation and construction of a lacZ fusion reporter system. The plasmid pVIKS1 containing a czcS1 fragment was constructed as follows: the czcS1 fragment was cloned into pTA2 and its orientation was confirmed by sequencing. After digestion with XbaI and KpnI, the restriction fragment was cloned into pVIK112 to create pVIKS1. The same procedure was then employed to create plasmids pVIKS2 and pVIKH. These plasmids were introduced into P. putida X4 by triparental conjugation. To generate double czcS mutation strain ΔS1S2, another suicide plasmid pRRT was constructed by replacing lacZYA and the kanamycin resistance gene cassette in pVIK112 into a tetracycline resistance gene by PstI digestion. After ligating with czcS2 restriction fragment from pTA2, the plasmid was applied to generate double mutation based on the ΔS1.
4.5. β-Galactosidase Activity Assay
The activity of β-galactosidase was measured according to the method by Nies [27] using o-nitrophenyl-d-galactoside (4 mg/mL) as the substrate. The unit 1 U was defined as the activity that forms 1 nmol of o-nitrophenol per minute at 30 °C.
4.6. Complementation Experiment
Complementation experiments were carried out to confirm that the phenotype variation is caused by the czcS mutant. Two czcS and the czcH open reading frames (ORFs) were amplified using the total DNA from P. putida X4 as the template. After digestion with XbaI and KpnI, the amplified fragments were ligated into the broad-host-range expression vector pVLT31, which contains the lacI and tac promoter [25], yielding pVLTS1, pVLTS2, and pVLTH. The yielded plasmids were mated into mutant strains ΔS1, ΔS2, and ΔczcH respectively via conjugation. The MICs of the different transconjugants were checked as described above with IPTG induction. Control strains were constructed by transformed pVLT31 into ΔS1, ΔS2, and ΔczcH.
4.7. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
After Cd2+ or Zn2+ treatment, the total RNA for qRT-PCR was extracted using the RNA bacteria protect solution and the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The residual DNA was eliminated by treatment with 20 U RQ1 RNase-free DNase (Promega, Madison, WI, USA) followed by phenol-chloroform extraction. The RNA was precipitated with ethanol and resuspended in RNase-free water. For cDNA synthesis, 3 μg of RNA was reverse transcribed using random hexamer primers and MLV reverse transcriptase (Promega) according to the supplier’s instructions. Reverse transcriptase was inactivated by incubation at 70 °C for 15 min and the obtained cDNAs were stored at −20 °C until utilization. The cDNAs were quantitatively measured with a Bio-Rad iCycler machine (Bio-Rad, Berkeley, CA, USA) using a Sybr Green Quantitect kit (Qiagen). The primer pairs used for qRT-PCR were designed using the Primer3 [28] program. To check for contaminated DNA, control reactions without reverse transcriptase were analyzed using rpsL primer set. The pure RNA samples were obtained when no amplification of the non-template control was detected. The cDNA samples were diluted 10-fold and 1 μL of the dilution served as the PCR template, which was performed in duplicate for each gene and sample. To offset the difference in the amount of cDNAs, the ribosomal rpsL gene was chosen as the reference gene. Results are presented as the ratios of gene expression between the target gene (target) and the reference gene (rpsL), which were obtained according to the following equation: ratio = (Etarget gene)ΔCt target (wild type − test strain)/(ErpsL)ΔCt rpsL (wild type − test strain) [29], where E is the real-time PCR efficiency of a given gene and Ct is the crossing point of the amplification curve with the threshold. An effect on gene transcription was considered significant when the corresponding ratios were ≥2.0 or ≤0.5.
Acknowledgments
This work was supported by National High Technology Research and Development Program of China (“863” Program, 2012AA101402), the National Natural Science of Foundation of China (41230854) and the Program for Changjiang Scholars and Innovative Research Team in University of China (IRT1247). The authors gratefully acknowledge the help of Steven Vinans from Cornell University, Ithaca, NY, USA, for providing plasmid pVIK112.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/08/17005/s1.
Author Contributions
Wenli Chen and Qiaoyun Huang conceived the experiment; Pulin Liu performed the experiment and wrote the paper; Xi Chen contributed qRT-PCR data analysis and English revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Blindauer C.A., Harrison M.D., Parkinson J.A., Robinson A.K., Turner-Cavfet J.S., Robinson N.J., Sadler P.J. A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity. Proc. Natl. Acad. Sci. USA. 2001;98:9593–9598. doi: 10.1073/pnas.171120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fantino J.R., Py B., Fontecave M., Barras F. A genetic analysis of the response of Escherichia coli to cobalt stress. Environ. Microbiol. 2010;12:2846–2857. doi: 10.1111/j.1462-2920.2010.02265.x. [DOI] [PubMed] [Google Scholar]
- 3.Solioz M., Stoyanov J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 4.Nies D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 5.Cánovas D., Cases I., de Lorenzo V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 2003;5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 6.Leedjarv A., Ivask A., Virta M. Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 2008;190:2680–2689. doi: 10.1128/JB.01494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 8.Wu J., Rosen B.P. Metalloregulated expression of the ars operon. J. Biol. Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- 9.Caille O., Rossier C., Perron K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 2007;189:4561–4568. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Große C., Grass G., Anton A., Franke S., Santos A.N., Lawley B., Brown N.L., Nies D.H. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perron K., Caille O., Rossier C., van Delden C., Dumas J.L., Köhler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 12.Casino P., Rubio V., Marina A. The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 2010;6:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Teitzel G.M., Geddie A., de Long S.K., Kirisits M.J., Whiteley M., Parsek M.R. Survival and growth in the presence of elevated copper: Transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Lelie D., Schwuchow T., Schwidetzky U., Wuertz S., Baeyens W., Mergeay M., Nies D.H. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 15.Petit-Haertlen I., Girard E., Sarret G., Hazemann J., Gourhant P., Kahn R., Coves J. Evidence for conformational changes upon copper binding to Cupriavidus metallidurans CzcE. Biochemistry. 2010;49:1913–1922. doi: 10.1021/bi100001z. [DOI] [PubMed] [Google Scholar]
- 16.Grosse C., Anton A., Hoffmann T., Franke S., Schleuder G., Nies D.H. Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch. Microbiol. 2004;182:109–118. doi: 10.1007/s00203-004-0670-8. [DOI] [PubMed] [Google Scholar]
- 17.Timmis K.N. Pseudomonas putida: A cosmopolitan opportunist par excellence. Environ. Microbiol. 2002;4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 18.Fang L.C., Cai P., Li P.X., Wu H.Y., Liang W., Rong X., Chen W., Cai P. Microcalorimetric and potentiometric titration studies on the adsorption of copper by P. putida and B. thuringiensis and their composites with minerals. J. Hazard. Mater. 2010;181:1031–1038. doi: 10.1016/j.jhazmat.2010.05.118. [DOI] [PubMed] [Google Scholar]
- 19.Hassan M.T., van der Lelie D., Springael D., Romling U., Ahmed N., Mergeay M. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene. 1999;238:417–425. doi: 10.1016/S0378-1119(99)00349-2. [DOI] [PubMed] [Google Scholar]
- 20.Kalogeraki V.S., Winans S.C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 21.Stekel D.J., Jenkins D.J. Strong negative self regulation of prokaryotic transcription factors increases the intrinsic noise of protein expression. BMC Syst. Biol. 2008;2:6. doi: 10.1186/1752-0509-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y.J., Chen W.L., Huang Q.Y. Construction of two lux-tagged Hg2+-specific biosensors and their luminescence performance. Appl. Microbiol. Biotechnol. 2008;79:363–370. doi: 10.1007/s00253-008-1442-1. [DOI] [PubMed] [Google Scholar]
- 23.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:334–337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G.Y., Li Y.G., Zhou J.C. Biological characteristics of plasmids of Mesorhizobium huakuii HN3015 from Astragalus sinicus. World J. Microbiol. Biotechnol. 2007;23:845–851. doi: 10.1007/s11274-006-9308-0. [DOI] [Google Scholar]
- 25.De Lorenzo V., Eltis L., Kessler B., Timmis K.N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 26.Mergeay M., Nies D.H., Schlegel H.G., Gerits J., Charles P., van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid bound resistance to heavy metals. J. Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nies D.H. CzcR and CzcD, gene-products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST©) for group wise comparison and stastical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.