Abstract
A significant portion of the mammalian genome encodes numerous transcripts that are not translated into proteins, termed long non-coding RNAs. Initial studies identifying long non-coding RNAs inferred these RNA sequences were a consequence of transcriptional noise or promiscuous RNA polymerase II activity. However, the last decade has seen a revolution in the understanding of regulation and function of long non-coding RNAs. Now it has become apparent that long non-coding RNAs play critical roles in a wide variety of biological processes. In this review, we describe the current understanding of long non-coding RNA-mediated regulation of cellular processes: differentiation, development, and disease.
Keywords: development, differentiation, disease, gene expression, long non-coding RNAs, skeletal muscle
Abbreviations
- Bvht
braveheart
- CDT
C-terminal domain
- ceRNAs
competing endogenous RNAs
- ciRS-7
circular RNA sponge for miR-7
- DBE-T
D4Z4-binding element
- DMD
Duchenne muscular dystrophy
- ES
embryonic stem
- Fendrr
Foxf1a called fetal-lethal non-coding developmental regulatory RNA
- FSHD
facioscapulohumeral muscular dystrophy
- iPS
induced pluripotent stem
- lncRNAs
long non-coding RNAs
- Malat1
metastasis associated lung adenocarcinoma transcript 1
- MEF2
myocyte enhancer factor-2
- Mesp1
mesoderm progenitor 1
- MRFs
myogenic regulatory factors
- ncRNAa
non-coding RNA activating
- Neat2
nuclear-enriched abundant transcript 2
- PRC2
polycomb group repressive complex 2
- RNAP II
RNA polymerase II
- SINE
short interspersed element
- SR
serine arginine
- SRA
steroid receptor activator
- SRY
sex-determining region Y
- YAM 1-4
YY1-associated muscle 1-4
Introduction
Since the discovery of the structure of DNA and the genetic code, the primary paradigm for gene expression has been that of a DNA blueprint encoding RNA messengers, which are then translated into functional proteins. This paradigm became strongly embedded into the collective consciousness of molecular biology with the coining of the term “The Central Dogma of Molecular Biology”by Francis Crick, and held true across all species.1 In the last 2 decades, however, exceptions have been emerging to the concept of proteins as the sole effectors of the genetic code in organisms. With the release of the human genome sequence, it became clear that only a small fraction of DNA encodes proteins.2 However, a large proportion of the non-protein-coding genome is transcribed temporally and spatially in a well-regulated manner.3 The transcribed pool of this non-coding RNA has given rise to a variety of new classes of regulatory RNA molecules, which appear to have numerous functions in cellular differentiation, development, and disease (Table 1).4-7
Table 1.
Prominent lncRNAs and their functions in development and disease
| lncRNAs | Organisms | Functions | Phenotypes/disease | References |
|---|---|---|---|---|
| ANRIL/CDKN2B-AS1 |
Human |
Transcriptional regulation |
Prostate cancer, leukemia |
5 |
| Airn, Kcnq1ot1 |
Mouse |
Epigenetic regulation; Embryonic gene activation |
Growth defects; breast, colon carcinoma |
57,58 |
| Malat1 |
Mouse, Human |
Splicing, gene regulation |
Tumor; myoblast differentiaiton |
5,34,35,38,71 |
| HOTAIR |
Mouse, Human |
Hox gene regulation; Recruitment of PRC2 and LSD1 |
Tumor formation; cancer metastasis |
76-80 |
| Hottip |
Chicken |
HoxA gene regulation |
Defect in limb formation |
25 |
| Xist, Tsix |
Mouse |
Dosage compensation |
Loss of function causes embryonic lethality |
17-21 |
| Fendrr, Braveheart |
Mouse, Human |
Heart development |
Loss of function causes embryonic lethality |
73-75 |
| Miat, Six3os1, Tug1, Vax2os |
Mouse |
Retinal development |
Defects in retinal specification; photoreceptor differentiation |
49,50,53,54,56 |
| Dlx1os, Dlx6os1 |
Mouse |
Brain development |
Neurological deficit |
51,52 |
| Megamind |
Zebrafish |
Brain and eye development |
Defects in brain and eye development |
55 |
| H19 |
Mouse, Human |
Posttranscriptional regulation by producing microRNAs |
Skeletal muscle differentiation, regeneration, cancer |
65,67,81 |
| SRA |
Mouse, Human |
Transcriptional activity of MyoD and p53 |
Skeletal muscle differentiation; breast, uterus, ovary tumor |
30-33 |
| Linc-MD1 |
Mouse, Human |
Sequestration of microRNAs |
Myogenic differentiation, Duchenne Muscular Dystrophy |
42 |
| SINE-containing lncRNAs |
Mouse |
Staufen-mediated mRNA degradation |
Myogenic differentiation |
72 |
| bII NAT |
Mouse |
Suppress MHC IIb transcription |
Skeletal muscle development |
70 |
| CE, DRR lncRNAs |
Mouse |
Transcriptional regulation of MyoD |
Skeletal muscle differentiation |
62 |
| YAMS | Mouse | Transcriptional regulation | Myogenic differentiation | 69 |
In retrospect, the concept of functional RNA should hardly be surprising; ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) were discovered in the 1950s as the most basic and essential components of cellular machinery of protein synthesis in all organisms.8 The discovery of small-interfering RNAs and microRNAs established non-coding RNAs as powerful regulators of development that could alter the expression of hundreds of targets, and hold equal footing with transcription factors as powerful controllers of gene expression.9-12 Still, the functional categories of the majority of the transcribed non-coding RNAs are difficult to predict and they have poor evolutionary sequence conservation, indicating either that a high level of transcriptional noise is present in the cell, or that numerous uncharacterized, species-specific classes of non-coding RNA exist.
Long non-coding RNAs (lncRNAs) are >200 bases long with low or no protein coding potential. These RNAs often regulate epigenetic silencing through chromatin remodeling. They are also now known to regulate splicing, recruit transcription factors, and regulate mRNA stability.13 Intersections of ChIP-seq and RNA-seq studies have found thousands of lncRNAs in both mice and humans.14-16 Since sequence conservation across species is poor, predicting the function of these molecules is difficult, but contemporary experimental approaches allow their genetic manipulation in vitro and in vivo and the discovery of protein- and genomic-binding partners. Such studies have established lncRNA molecules as important regulators of diverse biological functions. This review provides an update on the roles lncRNAs play during cellular differentiation, development, and disease.
Various Modes of Action of Long Non-coding RNA in Regulating Gene Expression
Many long non-coding RNAs are encoded in regions proximal to the promoters of known coding genes, or as antisense transcripts to coding genes. lncRNAs are regulated independently of adjoining genes and have their own specific histone modifications and splicing signals; in some cases, lncRNAs are located in genomic regions distant from known protein coding genes.13 Genomic maps of long intergenic non-coding RNAs now include thousands of these RNAs in mice and humans. These non-coding genes contain histone H3K4me3 marking transcriptional start sites and H3K36me3 indicating actively transcribed regions. RNA sequencing reveals that many of these genes produce transcripts encoded by exons that are separated by introns, which are spliced out using conventional splicing signals. However, as mentioned above, sequence conservation of lncRNAs between species is much lower than that of protein-coding sequences; some of the lncRNA sequences show higher evolutionary conservation than that of non-expressed genomic sequences. In addition, many lncRNAs are regulated by a number of important developmental and homeostatic transcription factors.14,15 We describe below several examples of how these lncRNAs regulate gene expression, highlight their diverse modes of action, and their significance in development and disease. A number of these examples are shown in Table 1 and Figure 1.
Figure 1.

lncRNAs regulate gene expression using diverse modes of action.
X-chromosome-inactivating long non-coding RNAs
Xist is one of the earliest examples of lncRNA with a prominent role in regulating X-chromosome inactivation (Fig. 1A). In mammalian females, the majority of genes contained on one of the 2 X-chromosomes in each cell are silenced, accounting for the similar level of expression of these genes between females and males. During female development, the non-coding RNA Xist is transcribed from the X-chromosome that is destined to become inactivated in each cell.17 Xist associates with the regions of the chromosome that are to be silenced, resulting in the formation of “Xist clouds” and recruitment of Polycomb Group Repressive complex 2 (PRC2).18,19 PRC2 is comprised of Suz12, Eed/Ezh1/Ezh2 (H3K27 methyltransferase) and RbAp48, and represses promoters by trimethylation of H3K27. Another lncRNA, Tsix, is produced from the Xist gene in the antisense direction on the active X chromosome. Tsix antagonizes Xist to prevent inactivation of the active X chromosome.20 Interestingly, Xist was recently used as an approach for therapeutic intervention for Down syndrome, a genetic disorder caused by trisomy of chromosome 21.21 In this work, the authors introduced an inducible Xist transgene into the DYRK1A locus on chromosome 21 of induced pluripotent stem (iPS) cells derived from Down syndrome patients and successfully inactivated the extra chromosome 21 by stable heterochromatin formation.21 This finding raises hope that lncRNAs will have therapeutic applications, especially when considered in tandem with regenerative medicine.
Cis-acting long non-coding RNAs
lncRNAs have important roles in regulating transcription of protein coding genes. lncRNAs transcribed from enhancer regions of protein coding genes, called e-RNAs, often regulate the expression of adjoining protein coding genes in cis through the recruitment of transcription factors.22-24 For example, Hottip lncRNA is a well-studied cis-acting lncRNA expressed from the HOXA cluster.25 It activates transcription of nearby genes by binding the MLL-WDR5 complex and facilitating the addition of activating histone marks (H3K4me3) to the gene promoter (Fig. 1C). The expression of the bHLH transcription factor Neurogenin1 is dependent on the expression of an e-RNA, utNgn1, encoded 7 kb upstream of the Neurogenin1 transcriptional start site. Polycomb group proteins suppress the expression of both utNgn1 and Neurogenin1, and knockdown of utNgn1 results in reduction in Neurogenin1, suggesting that the expression of Neurogenin1 is positively regulated by the expression of the utNgn1 e-RNA.26
A group of these cis-acting e-RNAs, termed non-coding RNA activating (ncRNAa), acts through recruitment of the transcriptional co-activator Mediator (Fig. 1D). Mediator physically interacts with a number of these lncRNAs, and depletion of these lncRNAs or of Mediator decreases expression of adjacent target genes. This interaction was found to facilitate chromatin looping of the adjacent genes, leading to their transcriptional activation and expression.27 These examples suggest that many enhancer-encoded lncRNAs in the mammalian genome are essential for the cis-activating function of their corresponding enhancers, at least in part, by facilitating DNA looping so as to bring their target genes in proximity to protein factors required for transcription.
Trans-acting long non-coding RNAs
In addition to cis-acting lncRNAs, there are interesting examples of gene expression regulation by lncRNAs in trans, e.g., 7SK and B2 lncRNAs.28,29 These lncRNAs impact global transcription by negatively regulating RNA polymerase II (RNAP II) activity. 7SK lncRNA negatively regulates transcription elongation factor PTEFβ, while B2 lncRNA represses RNAP II activity by binding RNAP II C-terminal domain (CTD) and inhibiting its phosphorylation.28,29 Both of these lncRNAs are upregulated in response to stress signals and, thus, shut down global transcription, most likely for cellular protection.
A fascinating case of an lncRNA regulating transcription in trans is that of the steroid receptor activator (SRA) gene, which encodes both a protein (SRAP) and a functional lncRNA (SRA) that act as co-regulators of nuclear receptor transcriptional activity (Fig. 1E).30 In addition to nuclear receptors, SRA facilitates the transcriptional activity of p5331 and MyoD32 in different cellular and developmental contexts. Overexpression of SRA RNA, but not SRA protein (SRAP), along with MyoD, facilitates trans-differentiation of mouse fibroblasts to skeletal muscle. SRAP, on the other hand, inhibits muscle differentiation by binding SRA RNA and preventing it from activating MyoD.33 This is a remarkable example of a gene encoding 2 products with opposing effects on transcription: a stimulatory lncRNA and an inhibitory protein.
Long non-coding RNAs in alternative splicing of pre-mRNAs
Alternative splicing of pre-mRNAs is an important event in the regulation and diversification of gene function, with a majority of multi-exonic human transcripts known to undergo alternative splicing. Several lncRNAs regulate alternative splicing. The lncRNA Metastasis Associated Lung Adenocarcinoma Transcript 1 (Malat1), also known as Nuclear-Enriched Abundant Transcript 2 (Neat2) is an example of such a lncRNA (Fig. 1F).34 Malat1 was originally discovered as a prognostic factor for metastasis in several human cancers, including lung cancer, but its role in cellular physiology remained elusive for many years.35 Alternative splicing is regulated by several trans-acting protein factors including a well-characterized class of RNA-binding proteins called the Serine Arginine (SR) family proteins.36,37 Malat1 is now known to predominantly localize to nuclear speckles, the sites where other splicing factors are often located, and believed to regulate the alternative splicing of a large set of genes by recruiting SR splicing factors to these nuclear speckles. Malat1 interacts with SR splicing factors and both Malat1 and the interacting SR splicing factors are conserved among species.34 Despite this conservation, however, alternative splicing is still seen in Malat1 knockout mice or human knockout cell lines, suggesting the presence of unidentified molecules or pathways that can substitute for Malat1 in alternative splicing.38
Competing endogenous long non-coding RNAs
microRNAs bind to the 3’UTR of their target genes and negatively regulate gene expression either by repressing translation or by promoting mRNA decay in P bodies.39 microRNA sponging was developed as an experimental strategy in which a designed exogenous RNA with multiple target sites for a particular microRNA is expressed to "sponge up" the cellular microRNA and inhibit the repression of other cellular targets by the microRNA.40 Interestingly, recent studies report naturally occurring microRNA sponges, termed competing endogenous RNAs (ceRNAs) (Fig. 1G).41 These ceRNAs consist of a variety of RNA species that include protein-coding mRNAs, pseudogenes, lncRNAs, and circular RNAs. These ceRNAs also cross-talk and co-regulate each other by competing for binding of microRNAs for which they share common target sites.41
linc-MD1 is a ceRNA with a role in skeletal muscle development and disease. linc-MD1 plays a critical role in skeletal muscle differentiation by titrating away miR-133 and miR-135 from their targets, the MAML1 and MEF2C mRNAs.42 MAML1 and MEF2C mRNAs encode transcription factors that are induced during muscle differentiation and are required for proper development. Excess linc-MD1 titrates away the microRNAs, de-represses MAML1 and MEF2C and thus promotes differentiation. Conversely, differentiation is delayed upon knockdown of linc-MD1. The importance of linc-MD1 in myoblast differentiation is underlined by its decrease in Duchenne muscular dystrophy (DMD), a devastating muscle degenerative disease. Myoblasts obtained from DMD patients show delay and defects in differentiation.42 Restoration of linc-MD1 to DMD myoblasts restored differentiation and, in particular, expression of MAML1 and MEF2C proteins.
Inhibition of microRNAs by titration by lncRNAs has also been shown to be important in embryonic stem (ES) cell renewal. OCT4, SOX2 and NANOG are essential transcription factors required for ES cell self-renewal. linc-RoR is abundantly expressed in human ES cells, sequesters miR-145, and thus protects OCT4, SOX2 and NANOG from miR-145-mediated repression.43,44 Introduction of mutations in miR-145-binding sites in linc-RoR lncRNA abolished its ability to repress miR-145. Recently, a large number of natural circular transcripts, termed circRNAs, containing multiple target sites of the same microRNA have been reported, suggesting that circRNAs can sequester highly abundant microRNAs.45,46 One such circRNA, circular RNA sponge for miR-7 (ciRS-7), contains multiple putative miR-7 target sites and is expressed in the human and mouse brain.45,46 Overexpression of ciRS-7 impaired brain development in zebrafish, a similar phenotype to that seen in miR-7 knockdown.45,46 The same research group has shown that the testis-specific circRNA sex-determining region Y (Sry) sponges miR-138, indicating that the phenomenon of microRNA sponging is not an isolated example.45 Together, these findings suggest that ceRNAs are important regulators of diverse biological functions, and that unraveling the cross-talk between these lncRNAs will provide valuable insights into a number of developmental and pathological processes.45
Long Non-coding RNAs in Cellular Differentiation and Development
A large number of lncRNAs are involved in cellular differentiation, maintenance of stem cell pluripotency, and development of tissues or organs. Several lncRNAs, such as lncRNA-ES1, lncRNA-ES2, and Linc-ROR, are associated with the maintenance of pluripotency of embryonic stem cells or iPS cells.43,44,47 A number of lncRNAs are involved in organ development, such as brain and eye,48-56 and growth.57,58 We will now focus on the role of lncRNAs in skeletal and cardiac muscle development as examples of the importance of these molecules in differentiation and development.
The role of long non-coding RNAs in skeletal muscle development
Skeletal muscle differentiation and development are well coordinated and tightly regulated processes. A well characterized family of transcription factors known as Myogenic Regulatory Factors (MRFs), comprised of MyoD, Myf5, Myogenin, MRF4, and the Myocyte Enhancer Factor-2 (MEF2A-D), are known to play key roles in these processes.59,60 Recent studies have identified a number of lncRNAs upregulated during muscle differentiation that play important roles in regulating several of these important transcription factors, including MyoD expression and activity.
lncRNAs overlap with a number of MyoD binding sites across the genome, and are transcribed in a MyoD-dependent manner.61 Such lncRNAs are enriched in the enhancer regions of MyoD target genes and appear to play a role in myogenesis. In a recent study, 2 eRNAs, referred to as CE and DRR lncRNAs, generated from the upstream regulatory regions of MyoD, were shown to regulate MyoD and myogenin expression by altering chromatin accessibility and recruitment of RNAP II.62 SRA lncRNA is another example of a lncRNA that regulates myogenesis. As described earlier, SRA and its protein isoform SRAP have opposite roles in facilitating MyoD activity. The ratio of SRA to SRAP increases during myogenesis, which rescues SRA from repression by SRAP and allows SRA to act as a co-activator of MyoD.33 MyoD has also been shown to regulate the lncRNA H19, which is located at the Igf2 imprinted locus and expressed only from the maternal allele. H19 expression represses transcription of the adjoining gene Igf2. Igf2 protein interacts with MyoD in vitro and indirectly inhibits MyoD expression.63 Thus, MyoD de-represses its own expression by inducing H19 and thus repressing Igf2 RNA and protein.63
H19 was previously called MyoH when identified in the same screen for inducers of myogenic differentiation that identified MyoD.64 H19 is abundantly expressed during embryonic development but strongly repressed in all adult tissues, except skeletal muscle. We have recently demonstrated that H19 has a direct role in skeletal muscle differentiation and regeneration (Fig. 2).65
Figure 2.
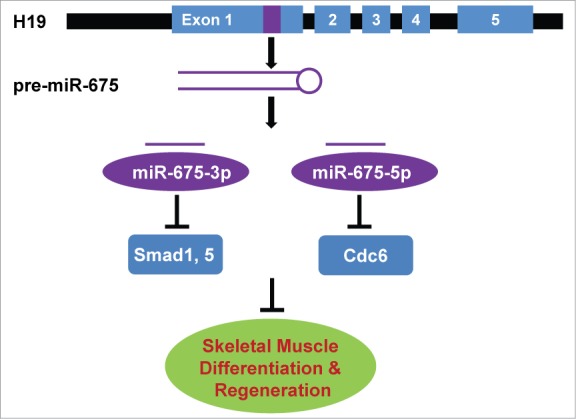
H19 lncRNA generates miR-675-3p and -5p and promotes skeletal muscle differentiation and regeneration by inhibiting repressors of myogenesis.
H19 encodes 2 conserved microRNAs, miR-675-3p and -5p. We showed that the biological function of H19 is mediated through miR-675-3p and -5p in both muscle differentiation in vitro and muscle regeneration in vivo.65 miR-675-3p represses the anti-myogenic bone morphogenetic protein (BMP) pathway by directly targeting the transcription factors Smad1 and Smad5. Thus, H19 lncRNA promotes myogenesis by generating a microRNA to inhibit this negative regulator of muscle differentiation. The other microRNA, miR-675-5p, directly targets and represses the DNA replication initiation factor Cdc6. Cdc6 was shown to be activated by MyoD during the myoblast stage,66 but the mechanism by which it is downregulated during myogenesis was previously unknown.
Paradoxically, a recent study reports that H19 sponges let-7 in the 293T kidney cell line and suggests that H19 inhibits C2C12 myoblast differentiation by sponging let-7.67 However, our data strongly support a role of H19 as a pro-myogenic factor both for myoblast cells differentiation in vitro and muscle regeneration in vivo.65 In contrast to their finding, we did not observe a marked upregulation of let-7 in our differentiation system. These results indicate that sponging of let-7 by H19 may not be physiologically relevant in skeletal muscle differentiation and regeneration,65 but it does not rule out the possibility that H19 may function by other mechanisms in different tissue types and developmental contexts. It is also possible that, in other contexts, H19 can act as a lncRNA independently of the creation or sponging of microRNAs.
Our findings provide a new insight into how lncRNAs function through production of embedded microRNAs. Consistent with this, a genome-wide study predicted that a large number of lncRNAs encode microRNAs.68 It will be interesting to identify how many in the rapidly expanding compendium of lncRNAS act through microRNA pathways to regulate gene expression in different cellular contexts.
Other lncRNAs important for myogenesis are YAM 1-4 (YY1-associated muscle 1-4), Malat1, and bII NAT.69-71 YAMs 1-4 are regulated by the myogenic transcription factor YY1. YY1 ChIP-seq data showed that YY1 binds to the regulatory elements of these lncRNAs, facilitating their expression.69 Interestingly, Yam-2 and Yam -3 were shown to promote myogenic differentiation whereas Yam-1 and Yam-4 inhibited differentiation. Malat1 lncRNA was originally discovered to be involved in cancer metastasis, but was later found to play a role in skeletal muscle differentiation.71 Malat1 expression is upregulated during C2C12 myoblast differentiation, and siRNA-mediated knockdown of this lncRNA arrests the cell cycle in G0/G1, suggesting a role in differentiation. Malat1 is suppressed by myostatin, a well known inhibitor of myogenesis. As discussed earlier, Malat1 is believed to be a regulator of alternative splicing, but a more detailed mechanism of the pro-myogenic function of Malat1 remains to be elucidated. The myosin heavy chain (MHC) proteins found in skeletal muscle have multiple isoforms encoded at several locations on the genome. A natural antisense RNA (termed bII NAT), encoded at the MHC IIb locus, was found to suppress MHC IIb transcription, playing a role in determining which MHC isoforms are expressed during postnatal development.70 Recently, a study illustrated the role of short interspersed element (SINE)-containing lncRNAs in regulating myogenic differentiation.72 Briefly, a lncRNA called m1/2-sbsRNA2(B2) contains a B2 element that pairs with the B2 element present in the 3' UTR of mTRAF6 and promotes the degradation of mTRAF6, by Staufen-mediated degradation. mTRAF6 is a pro-myogenic factor, making m1/2sbsRNA2(B2) an anti-differentiation factor. Together, these studies establish that lncRNAs are important contributors to the regulation of skeletal muscle development.
The role of long non-coding RNAs in cardiac muscle development
A large number of lncRNAs are expressed during cardiomyocyte differentiation.73-75 Two such lnRNAs, braveheart (Bvht) and a gene that is adjacent to Foxf1a called Fetal-lethal non-coding developmental regulatory RNA (Fendrr), were recently shown to be required in the development of the mammalian heart, and strongly highlighted the importance of lncRNAs in organ development.73,74 siRNA-mediated knockdown of Bvht in mouse neonatal cardiomyocytes altered cardiac-specific gene expression and blocked their differentiation into mature cardiomyocytes. In cardiac progenitor cells, Bvht promotes expression of mesoderm progenitor 1 (Mesp1), a critical transcription factor in the network of genes that has to be activated for cardiac differentiation, by sequestering PRC2 (a writer of the repressive histone modification H3K27me3). However, Bvht is not conserved among species and whether the functional role of Bvht in humans is played by an equivalent molecule still remains to be determined.
Fendrr was also identified as a regulator of heart development.73 Loss of function of Fendrr caused embryonic lethality due to defective heart morphology and function. Loss of Fendrr was associated with increased expression of a subset of cardiac transcription factors, including NKx2.5 and Gata6. The increase in the level of these transcripts was accompanied by a concomitant increase in H3K4me3 level in their target promoters. Fendrr is believed to regulate cardiac genes both in cis and in trans. Consistent with its cis regulatory function, Fendrr regulates its neighboring gene Foxf1a by interacting with and recruiting the PRC2 complex to the Foxf1a promoter. In addition, Fendrr interacts with the TrxG/MLL activating complex (a writer of the activating histone mark H3K4me3) and is important for activating cardiac-specific genes in trans. Thus, Fendrr is required for the maintenance of a fine balance between repressive and activating marks at promoters of various genes during cardiogenesis. In contrast to Bvht, Fendrr is conserved in humans, associates with PRC2, and likely plays an important role in human heart development.
Long Non-coding RNAs in Human Diseases
The list of lncRNAs implicated in human diseases is growing very fast. As lncRNAs are being discovered in key biological and developmental processes, it is likely that misregulation of lncRNAs will lead to disease. A large number of studies have implicated lncRNA expression in the pathophysiology of various cancers.5 HOTAIR, a lncRNA encoded within the HOX gene cluster, promotes metastasis in breast, hepatocellular, nasopharyngeal, colorectal, pancreatic, and ovarian cancers.76-80 HOTAIR lncRNA is overexpressed in these metastatic cancers, and alters the occupancy of PRC2 across the genome, rearranging the landscape of repressive H3K27me3 in cells (Fig. 1B). In particular, HOTAIR acts in trans to alter the target specificity of PRC2 and thus repress a number of anti-metastatic genes.77-80
The pro-myogenic role of H19 is consistent with previous studies in which inactivation of H19 was linked to the development of rhabdomyosarcoma (RMS).81 RMS is a childhood tumor that arises from defective skeletal muscle differentiation. As miR-675-3p and -5p can promote myogenic differentiation, these 2 microRNAs might have a therapeutic potential for treatment of RMS. Since H19 inactivation has also been linked to the development of Wilms' tumor,81 it is worth investigating whether the tumor suppressor activity of H19 works, in this context, via the production of the same 2 microRNAs. Several other lncRNAs involved in the pathophysiology of various cancers have been reviewed elsewhere.5
Apart from cancers, lncRNAs are also involved in other diseases, particularly those in which defects in differentiation are observed. For example, a few lncRNAs show altered expression in various forms of muscular dystrophies, including DMD and facioscapulohumeral muscular dystrophy (FSHD).42,82 The expression of linc-MD1 is reduced in myoblasts isolated from DMD patients. The abnormal kinetics of differentiation in myoblasts isolated from DMD patients was partly corrected by reintroducing exogenous linc-MD1, suggesting its important role in DMD.42 As we have demonstrated a critical role of H19 lncRNA in skeletal muscle differentiation and regeneration, we are interested in investigating whether H19 expression, or processing into microRNAs, is altered in differentiation-defective myoblasts from DMD patients. A third example is the lncRNA encoded by the D4Z4‑binding element (DBE‑T), which is selectively expressed in FSHD patient samples.82 DBE‑T increases H3K36me2 by recruiting the MLL1 complex and results in excessive transcriptional activation of the FSHMD1A locus in patients with FSHD.82
Conclusions and Future Perspectives
With the advent of ultra-high-throughput sequencing, the universe of non-coding RNAs is getting bigger every day. It is becoming clear that a significant part of the non-protein-coding mammalian genome could be essential for development and physiology through the expression of the various classes of lncRNAs. lncRNAs play a critical role in various aspects of biology, and many lncRNAs have now been found to direct differentiation and development, while leading to disease when misregulated. However, compared to coding genes, there are significant gaps in the current understanding of lncRNA regulation and mechanism of action. This is partly because lncRNA sequences are not evolutionarily conserved as well as protein-coding sequences are. It remains difficult to classify lncRNAs into categories beyond their genomic locations and expression patterns. Therefore, careful study of loss- or gain-of-function mutants of lncRNAs in cell lines and in appropriate animal models is essential to discern their function. However, since lncRNAs are less conserved between humans and mice than protein-coding genes are, it may not be possible to apply the findings generated with animal models to humans. Indeed, this variability in lncRNAs could explain many of the phenotypic differences in higher eukaryotes. In that scenario, advances will need to be made in computational algorithms predicting lncRNAs secondary structures, domains, and protein interactions in order to determine the best in vitro models and developmental contexts to study a given lncRNA. Importantly, lncRNAs are dysregulated in numerous biological processes and are becoming rapidly linked to numerous human diseases. The lncRNA field is still very young, but new mechanistic insights into lncRNA function are bound to emerge, and will lead to a greater understanding of many complex and devastating disorders.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Crick F. Central dogma of molecular biology. Nature 1970;227:561-3; PMID:4913914; http://dx.doi.org/ 10.1038/227561a0 [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA., Holt RA, et al. The sequence of the human genome. Science 2001; 291:1304-51; PMID:11181995; http://dx.doi.org/ 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- 3.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447:799-816; PMID:17571346; http://dx.doi.org/ 10.1038/nature05874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21:354-61; PMID:21550244; http://dx.doi.org/ 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Bhan A, Mandal SS. Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease. ChemMedChem 2014. (pre-print); PMID:24677606 [DOI] [PubMed] [Google Scholar]
- 6.Dey BK, Mueller AC, Dutta A. Non-micro-short RNAs: the new kids on the block. Mol Biol Cell 2012; 23:4664-7; PMID:23239791; http://dx.doi.org/ 10.1091/mbc.E12-10-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagan J, Dey BK, Dutta A. MicroRNAs regulate and provide robustness to the myogenic transcriptional network. Curr Opin Pharmacol 2012; 12:383-8; PMID:22386695; http://dx.doi.org/ 10.1016/j.coph.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920-30; PMID:10937990; http://dx.doi.org/ 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- 9.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013; 14:475-88; PMID:23800994; http://dx.doi.org/ 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- 10.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 2011; 31:203-14; PMID:21041476; http://dx.doi.org/ 10.1128/MCB.01009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 2012; 26:2180-91; PMID:23028144; http://dx.doi.org/ 10.1101/gad.198085.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522-31; PMID:15211354; http://dx.doi.org/ 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 13.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013; 14:699-712; PMID:24105322; http://dx.doi.org/ 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 2010; 28:503-10; PMID:20436462; http://dx.doi.org/ 10.1038/nbt.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223-7; PMID:19182780; http://dx.doi.org/ 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011; 25:1915-27; PMID:21890647; http://dx.doi.org/ 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992; 71:527-42; PMID:1423611; http://dx.doi.org/ 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- 18.Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 2004; 167:1025-35; PMID:15596546; http://dx.doi.org/ 10.1083/jcb.200409026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 2009; 19:1361-73; PMID:19581487; http://dx.doi.org/ 10.1101/gr.092643.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 1999; 21:400-4; PMID:10192391; http://dx.doi.org/ 10.1038/7734 [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature 2013; 500:296-300; PMID:23863942; http://dx.doi.org/ 10.1038/nature12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010; 143:46-58; PMID:20887892; http://dx.doi.org/ 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, [gamma]-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 2010; 29:1434-45; PMID:20224553; http://dx.doi.org/ 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010; 8:e1000384; PMID: 20485488; http://dx.doi.org/ 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120-4; PMID:21423168; http://dx.doi.org/ 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci U S A 2012; 109:16939-44; PMID:23027973; http://dx.doi.org/ 10.1073/pnas.1202956109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013; 494:497-501; PMID: 23417068; http://dx.doi.org/ 10.1038/nature11884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 2007; 13:583-96; PMID:17307818; http://dx.doi.org/ 10.1261/rna.310307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001; 414:317-22; PMID:11713532; http://dx.doi.org/ 10.1038/35104575 [DOI] [PubMed] [Google Scholar]
- 30.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999; 97:17-27; PMID:10199399; http://dx.doi.org/ 10.1016/S0092-8674(00)80711-4 [DOI] [PubMed] [Google Scholar]
- 31.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J 2005; 24:543-53; PMID:15660129; http://dx.doi.org/ 10.1038/sj.emboj.7600550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 2006; 11:547-60; PMID:17011493; http://dx.doi.org/ 10.1016/j.devcel.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 33.Hube F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res 2011; 39:513-25; PMID:20855289; http://dx.doi.org/ 10.1093/nar/gkq833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet 2013; 9:e1003368; PMID:23555285; http://dx.doi.org/ 10.1371/journal.pgen.1003368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73:1180-9; PMID:23243023; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell 2006; 126:37-47; PMID:16839875; http://dx.doi.org/ 10.1016/j.cell.2006.06.023 [DOI] [PubMed] [Google Scholar]
- 37.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 2010; 11:75-87; PMID:20019688; http://dx.doi.org/ 10.1038/nrg2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012; 18:1487-99; PMID:22718948; http://dx.doi.org/ 10.1261/rna.033217.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 40.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4:721-6; PMID:17694064; http://dx.doi.org/ 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505:344-52; PMID:24429633; http://dx.doi.org/ 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011; 147:358-69; PMID:22000014; http://dx.doi.org/ 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009; 137:647-58; PMID:19409607; http://dx.doi.org/ 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev Cell 2013; 25:69-80; PMID:23541921; http://dx.doi.org/ 10.1016/j.devcel.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 45.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 46.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 47.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 2012; 31:522-33; PMID:22193719; http://dx.doi.org/ 10.1038/emboj.2011.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ, Pillai B. Non-coding RNA interact to regulate neuronal development and function. Front Cell Neurosci 2014; 8:47; PMID:24605084; http://dx.doi.org/ 10.3389/fncel.2014.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA 2012; 18:111-23; PMID:22128341; http://dx.doi.org/ 10.1261/rna.029454.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mustafi D, Kevany BM, Bai X, Maeda T, Sears JE, Khalil AM, Palczewski K. Evolutionarily conserved long intergenic non-coding RNAs in the eye. Hum Mol Genet 2013; 22:2992-3002; PMID:23562822; http://dx.doi.org/ 10.1093/hmg/ddt156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 2006; 20:1470-84; PMID:16705037; http://dx.doi.org/ 10.1101/gad.1416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraus P, Sivakamasundari V, Lim SL, Xing X, Lipovich L, Lufkin T. Making sense of Dlx1 antisense RNA. Dev Biol 2013; 376:224-35; PMID:23415800; http://dx.doi.org/ 10.1016/j.ydbio.2013.01.035 [DOI] [PubMed] [Google Scholar]
- 53.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 2010; 10:49; PMID:20459797; http://dx.doi.org/ 10.1186/1471-213X-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev 2011; 6:32; PMID:21936910; http://dx.doi.org/ 10.1186/1749-8104-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011; 147:1537-50; PMID:22196729; http://dx.doi.org/ 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005; 15:501-12; PMID:15797018; http://dx.doi.org/ 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 57.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012; 338:1469-72; PMID:23239737; http://dx.doi.org/ 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- 58.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 2006; 20:1268-82; PMID:16702402; http://dx.doi.org/ 10.1101/gad.1416906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 2005; 16:585-95; PMID:16099183; http://dx.doi.org/ 10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 60.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development 2007; 134:4131-40; PMID:17959722; http://dx.doi.org/ 10.1242/dev.008367 [DOI] [PubMed] [Google Scholar]
- 61.Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev 2012; 26:2763-79; PMID:23249738; http://dx.doi.org/ 10.1101/gad.200113.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 2013; 51:606-17; PMID:23993744; http://dx.doi.org/ 10.1016/j.molcel.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borensztein M, Monnier P, Court F, Louault Y, Ripoche MA, Tiret L, Yao Z, Tapscott SJ, Forne T, Montarras D, et al. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development 2013; 140:1231-9; PMID:23406902; http://dx.doi.org/ 10.1242/dev.084665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987; 51:987-1000; PMID:3690668; http://dx.doi.org/ 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- 65.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014; 28:491-501; PMID:24532688; http://dx.doi.org/ 10.1101/gad.234419.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang K, Sha J, Harter ML. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol 2010; 188:39-48; PMID:20048262; http://dx.doi.org/ 10.1083/jcb.200904144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013; 52:101-12; PMID:24055342; http://dx.doi.org/ 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He S, Su H, Liu C, Skogerbo G, He H, He D, Zhu X, Liu T, Zhao Y, Chen R. MicroRNA-encoding long non-coding RNAs. BMC Genomics 2008; 9:236; PMID:18492288; http://dx.doi.org/ 10.1186/1471-2164-9-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu L, Sun K, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J 2013; 32:2575-88; PMID:23942234; http://dx.doi.org/ 10.1038/emboj.2013.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandorf CE, Jiang W, Qin AX, Bodell PW, Baldwin KM, Haddad F. Regulation of an antisense RNA with the transition of neonatal to IIb myosin heavy chain during postnatal development and hypothyroidism in rat skeletal muscle. Am J Physiol Regul Integr Comp Physiol 2012; 302:R854-67; PMID:22262309; http://dx.doi.org/ 10.1152/ajpregu.00591.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watts R, Johnsen VL, Shearer J, Hittel DS. The myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol 2013; 304:C995-1001; PMID:23485710; http://dx.doi.org/ 10.1152/ajpcell.00392.2012 [DOI] [PubMed] [Google Scholar]
- 72.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev 2013; 27:793-804; PMID:23558772; http://dx.doi.org/ 10.1101/gad.212639.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013; 24:206-14; PMID:23369715; http://dx.doi.org/ 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013; 152:570-83; PMID:23352431; http://dx.doi.org/ 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 2012; 151:206-20; PMID:22981692; http://dx.doi.org/ 10.1016/j.cell.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol 2013; 425:3707-22; PMID:23375982; http://dx.doi.org/ 10.1016/j.jmb.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; http://dx.doi.org/ 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep 2013; 29:946-50; PMID:23292722; http://dx.doi.org/ 10.3892/or.2012.2219 [DOI] [PubMed] [Google Scholar]
- 79.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013; 104:458-64; PMID:23281836; http://dx.doi.org/ 10.1111/cas.12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011; 18:1243-50; PMID:21327457; http://dx.doi.org/ 10.1245/s10434-011-1581-y [DOI] [PubMed] [Google Scholar]
- 81.Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, Hess I, Dullin C, Kimmina S, Pirngruber J, Johnsen SA, et al. Antitumor effects of a combined 5-aza-2'deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res 2009; 69:887-95; PMID:19155313; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0946 [DOI] [PubMed] [Google Scholar]
- 82.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 2012; 149:819-31; PMID:22541069; http://dx.doi.org/ 10.1016/j.cell.2012.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]


