Abstract
Pervasive, or genome-wide, transcription has been reported in all domains of life. In bacteria, most pervasive transcription occurs antisense to protein-coding transcripts, although recently a new class of pervasive RNAs was identified that originates from within annotated genes. Initially considered to be non-functional transcriptional noise, pervasive transcription is increasingly being recognized as important in regulating gene expression. The function of pervasive transcription is an extensively debated question in the field of transcriptomics and regulatory RNA biology. Here, we highlight the most recent contributions addressing the purpose of pervasive transcription in bacteria and discuss their implications.
Keywords: antisense transcription, asRNA, double-stranded RNA, Hfq, pervasive transcription, promoter, RNA, RNase III, spurious transcription, sigma factor, transcriptome
Abbreviations
- asRNA
antisense RNA
- dsRNA
double-stranded RNA
- intraRNA
intragenic RNA
- ncRNA
non-coding RNA
Introduction
Pervasive transcription refers to the idea that the vast majority of the genome is transcribed. In humans, about 75–85% of the genome is transcribed, although only 1.5–2% is protein-coding DNA.1 Similarly, transcripts have been detected across the genomes of most bacteria.2 Unlike the human genome, bacterial genomes are compact, with around 90% consisting of protein-coding genes. Consequently, most newly identified transcripts in bacteria are antisense RNAs (encoded on the DNA strand opposite to protein-coding genes). Therefore, the terms pervasive transcription and antisense transcription are used interchangeably in bacteria, but neither term indicates the function of the transcript. Regulatory antisense transcripts were first observed on plasmids, opposite to transposon, phage and toxin genes.3,4 Antisense transcripts regulate gene expression via several different mechanisms: transcription interference, transcription attenuation, translation stimulation or inhibition, and RNA stability. Antisense transcripts can be independent bona fide cis-RNAs (not associated with any annotated RNA), as well as overlapping 5′- or 3′UTRs of mRNAs from divergent or convergent gene pairs, respectively.2,5 trans-encoded sRNAs are often described as regulating their targets via an antisense mechanism, however these are not asRNAs. Instead, trans-encoded sRNAs are transcribed from separate genomic loci than their targets and bind with only partial complementarity. Furthermore, transcripts originating from within annotated genes, termed intraRNAs, have been recently reported and represent another aspect of genome-wide transcription.6,7 Some of the newly identified transcripts, found using next generation sequencing, are regulatory non-coding RNAs (ncRNAs) that control gene expression, but the majority remain functionally uncharacterized. Both antisense (as-) and intragenic (intra-) RNAs can be non-functional products of spurious transcription events, byproducts and/or precursors of functional RNAs, or they can be functional ncRNAs themselves. The utility of pervasive transcription has been extensively debated in the field of RNA biology. Here we examine the evidence and arguments for pervasive transcription having a function and discuss strategies for identifying functional RNAs in bacteria.
Evidence for Spurious Antisense Transcription
One could argue that most asRNAs are functional and contribute to bacterial fitness because producing RNA is costly. Alternatively, genome-wide asRNA expression might represent non-adaptive transcriptional noise. Examining the degree of conservation among homologous sequences is an effective method to differentiate functional sequences from non-functional sequences. Using this approach, Raghavan et al.8 recently compared asRNAs originating within protein-coding genes in Escherichia coli and Salmonella enterica Typhimurium, grown under similar conditions (exponential phase, LB medium). Although around 1200 genes contained asRNAs in each bacterium, only 343 genes had asRNAs in both species, and only eight genes had highly expressed asRNAs in both. These data show that the vast majority of asRNAs are not conserved between E. coli and Salmonella. However, more than 70% of asRNAs in both bacteria were associated with an identifiable −10 promoter element, indicating that most asRNAs were expressed under the control of σ70, the primary sigma factor during exponential phase growth.
Even if the observed lack of overlap between E. coli and Salmonella asRNA repertoires was due to experimental variations, one would expect asRNA promoter sequences to be conserved in both bacteria if they are functional. Sequences of functional importance experience purifying selection; that is to say, most new mutations are deleterious and are therefore eliminated from the population. As a result, functional sequences show lower rates of sequence evolution than non-functional sequences. Promoters for mRNAs exhibited reduced nucleotide divergence between E. coli and Salmonella, especially around the –35 and –10 elements; however, there was no evidence of purifying selection on asRNA promoter regions in E. coli and Salmonella. Furthermore, similar results were obtained when comparing E. coli with Escherichia fergusonii, and, at the intraspecific level, by analyzing 41 strains of E. coli and Shigella. The lack of conservation in asRNAs between E. coli and Salmonella might indicate that asRNAs function largely in a species-specific manner. However, because there is no evidence of conservation or functional constraint acting within the genus Escherichia or even among different strains of E. coli, the alternative interpretation is that the majority of asRNAs in bacterial genomes is non-functional. A recent examination of the Bacillus subtilis transcriptome also came to a similar conclusion because many asRNAs were found to originate from evolutionarily less conserved promoter sequences.9
Promoter-like sequences can arise spontaneously by point mutations in any locus of a bacterial genome.10 However, promoter-like sequences are underrepresented within coding regions compared to other genomic regions, indicating that selection acts to purge spurious promoters.11,12 Nevertheless, the average intensity of selection against such elements is weak, and, consequently, many spurious promoter-like sequences persist within populations.13 Because uncontrolled transcription from genome-wide promoter-like sequences are potentially dangerous, bacteria have several systems in place to control the generation of spurious transcripts: (i) The histone-like nucleoid structuring protein (H-NS) suppresses transcription initiation from intragenic promoters,14 (ii) the termination factor Rho and its cofactor NusG function in the termination of asRNA transcription,15,16 and (iii) multiple RNases degrade aberrant RNAs.17
A lack of asRNA conservation among closely related bacteria might not necessarily indicate lack of function because, as shown recently in Drosophila, functional genes can arise rapidly in a lineage-specific manner.18 Additionally, ncRNAs evolve rapidly in eukaryotes, with the rate of evolutionary turnover similar to other regulatory sequences.19 Even if most asRNAs do not have a clear function, there are undoubtedly functional asRNAs in bacteria. Previous studies have described functional asRNAs in both E. coli and Salmonella5 and an analysis of the transcriptomes of a number of Gram-positive bacteria suggests a role for asRNAs in genome-wide mRNA processing.20 The promoter conservation analysis by Raghavan et al.8 identified 17 putatively functional asRNAs in E. coli, and, in concordance, 7 of them were detected by a recent study that described the double-stranded transcriptome of E. coli.21 In addition, Lybecker et al.21 detected 13 of the 80 non-conserved asRNAs, suggesting a cellular role for some of the species-specific asRNAs.
Evidence for Functional asRNAs
There are several recent reports demonstrating that many asRNAs in various bacteria are likely functional. Different features have been used to characterize asRNAs as functional including: regulated expression, binding to regulatory proteins, binding to target RNA, and regulating expression of the corresponding sense gene. In both Listeria monocytogenes and Staphylococcus aureus a subset of asRNAs are dependent on the alternative sigma factor SigB, suggesting these transcripts are regulated and functional.20,22 In a recent report, 67 bona fide asRNAs were co-immunoprecipitated with the RNA chaperone Hfq in E. coli.7 Hfq is often required for the function of trans-encoded sRNAs in Gram-negative bacteria, but its role in gene regulation via cis-encoded asRNAs was not previously reported. The association of these asRNAs with Hfq in vivo suggests they are functional. In addition, a new model of antisense-mediated gene regulation, termed the excludon, was characterized in L. monocytogenes.22,23 Excludon regulation occurs at divergently transcribed genes, with a long asRNA contributing to the transcription of one gene, while inhibiting the other through an antisense mechanism. RNase III, a well-conserved double-stranded RNA specific endoribonuclease, has been shown to be an important player in asRNA-dependent gene regulation.20,21,24 Lasa et al.20 demonstrated that S. aureus has an RNase III-dependent genome-wide gene regulation via asRNAs. Moreover, an RNase III co-immunopreciptiation assay in S. aureus identified asRNAs and overlapping transcripts bound to RNase III.24 Recently, a set of functional asRNAs was identified in E. coli by isolating and deep sequencing asRNAs found duplexed with their sense counterparts.21 The majority of dsRNAs identified in this study were RNase III-dependent, further demonstrating the important role of RNase III in antisense-mediated gene regulation in bacteria. The dsRNAs identified were only a small subset of the potential dsRNA-forming regions in E. coli because not all overlapping transcripts form dsRNA. In contrast, Lasa et al.20 report that most (75%) of the mRNAs expressed in S. aureus have overlapping transcripts associated with them and these potential dsRNA regions have processing products generated by RNase III, suggesting that dsRNA formation and subsequent RNase III digestion is occurring at nearly all sites of overlapping transcription. The identification of asRNAs in the absence of an RNA degradation factor, such as RNase III, is reminiscent of what was observed in yeast: novel non-coding transcripts (originally categorized as CUTs, SUTs and XUTs) were found as a consequence of depleting several components of RNA degradation pathways.25
All known mechanisms of asRNA-mediated regulation, except transcription interference, require that an asRNA interacts with the complementary sense RNA (forming double-stranded RNA). Most asRNA-mediated gene regulation mechanisms requiring an RNA/RNA interaction affect the stability and/or translation efficiency or attenuate transcription of the mRNA. RNase III can cleave dsRNA resulting in either the destabilization or stabilization of one or both transcripts. In this mechanism, RNase III plays a direct role in the regulation of gene expression via dsRNAs, as proposed for several Gram-positive bacteria.20,24 Alternatively, the formation of the dsRNA itself may regulate gene expression and the dsRNA (subsequently degraded by RNase III) would be a byproduct of the regulation. In this mechanism, gene regulation is independent of RNase III, but the resulting dsRNA levels are RNase III-dependent (Fig. 1). Specifically, an asRNA that overlaps the ribosome-binding site (RBS) of its cognate mRNA could prevent the ribosome from binding and inhibit translation; subsequently the dsRNA would be degraded by RNase III, but the translational regulation would not be RNase III-dependent. Translation could also be stimulated by dsRNA formation by releasing the RBS for ribosome binding. Finally, dsRNA formation could cause transcriptional attenuation and termination, also resulting in a dsRNA byproduct, which would be degraded by RNase III. The dsRNA-mediated gene regulation mechanism is supported by the observation in E. coli that the regions of RNAs that are double-stranded are the most stable fragments.21 There are many factors that may influence the pairing of 2 transcripts, including transcript abundance, RNA structure, and the presence of ribosomes or proteins on the transcripts. An RNA chaperone likely aids in the restructuring and annealing of the complementary RNAs. One candidate is the RNA chaperone Hfq. Notably, forty-eight of the transcripts found in dsRNA duplexes were also co-immunoprecipitated with Hfq.7,21 These data suggest that Hfq may play a role in the annealing of antisense and sense RNAs in the cell.
Figure 1.
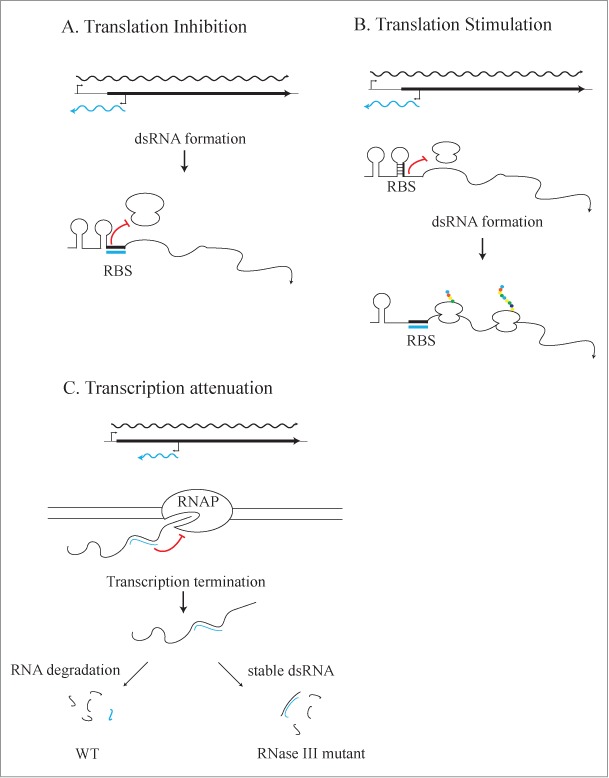
Mechanisms of dsRNA-mediated gene regulation. (A) dsRNA-mediated translation inhibition. An asRNA that overlaps the RBS of its cognate mRNA could prevent the ribosome from binding the RBS and inhibit translation; the dsRNA would then be degraded by RNase III, but the translational regulation would not be dependent on RNase III. (B) dsRNA-mediated translation stimulation. Translation could also be stimulated by dsRNA formation by releasing the RBS for ribosome binding. (C) dsRNA-mediated transcription attenuation. dsRNA formation could cause transcriptional attenuation and termination, also resulting in a dsRNA byproduct, which would be degraded by RNase III.
HN-S, Rho and NusG have been implicated in controlling the transcription initiation or termination of pervasive transcription in bacteria (as discussed above). Specifically, HN-S binds DNA within protein-coding genes and inhibits transcription from promoter-like elements found in genes.14 However, HN-S also binds promoter regions and represses many mRNAs, acting as a transcriptional repressor. Taking into account the role of H-NS in regulation of canonical transcripts, the repression of intragenic transcripts indicates that intraRNAs are indeed regulated, and should not be considered non-functional simply because they are not characterized. Interestingly, in E. coli 25 intraRNAs were co-immunoprecipitated with Hfq.7 These Hfq-binding intraRNAs may have their own promoters within the mRNA or be stable processing products. Similarly, Rho and NusG have been implicated in terminating pervasive transcription. However, over 50% of the overlapping transcripts that are identified are formed from 3′-overlapping UTRs of mRNAs16, suggesting that Rho is involved in the termination of these mRNAs and that non-functional overlapping transcripts are likely produced in the absence of functional Rho. Notably, only a few dsRNA regions were identified at overlapping 3′ UTRs,21 suggesting that these Rho-dependent overlapping transcripts are not functional and are artifacts of read-through transcription.
Conclusions and Future Directions
In addition to the already discussed mechanisms of asRNA-mediated gene regulation, we hypothesize that pervasive transcripts may function as RNA scaffolds for nucleoid structure, similar to what is observed in eukaryotes. RNAs in eukaryotes specifically interact with protein effectors to mediate long-range chromatin interactions and architecture.25 Pettijohn and Hecht in 1974 first suggested that RNA plays a role in maintaining the nucleoid structure in E. coli.26 Recently, Macvanin et al.27 reported that 2 novel non-coding RNAs bind to the architectural DNA-binding protein HU and affect the nucleoid structure, implying that RNA molecules play an important role in genome organization. Moreover, we expect that some of the asRNAs and intraRNAs may code for small peptides, further increasing the protein-coding potential of genomes. Pervasive transcripts in bacteria could also function as sponges for proteins or other ncRNAs. Long non-coding RNAs and circular RNAs in eukaryotes have been shown to regulate gene expression by binding regulatory RNAs or proteins and sequestering them from their regulatory targets.28,29
Mechanistic and functional studies of asRNA and intra-RNAs remain scarce due to the technical difficulty in studying these transcripts. A traditional loss-of-function assay is challenging to perform without disturbing the corresponding coding region. Precise characterization and mutation of asRNA and intraRNA promoters will be necessary to begin elucidating their functions. As- and intra-RNAs can be overexpressed in trans on plasmids, but an asRNA, if acting in cis, will not be transcribed in its endogenous context in close proximity to the sense RNA, expression of which might influence the activity of asRNA. In addition, the presence of a non-physiologically abundant RNA is likely to yield artifactual results. An overexpressed asRNA or intra-RNA may bind to Hfq or another regulator and sequester it away from its normal substrates, creating a phenotype that is not specific to the studied RNA. High-throughput transcriptome analyses examine a bacterial population rather than a single bacterium, producing a composite genome-wide transcription picture, so the number of antisense and intragenic transcripts detected by these approaches that are produced in each bacterial cell is not known. Advances in single-cell transcriptomic technologies are needed to understand the scale of pervasive transcription at the cellular level. Each bacterium may only produce a few spurious non-functional transcripts, and RNases, HN-S, Rho and NusG may be capable of neutralizing their potential negative consequence. In addition, these non-functional transcripts might not only be “junk,” but may also serve as a reservoir for evolutionary innovation.
Recent work aimed at distinguishing likely functional RNAs from non-functional transcription, by identifying RNAs that either display functionally specific features (such as forming a duplex with its sense counterpart or binding a major mediator of ncRNA-regulation) or are conserved in related species, have identified almost 400 putative functional RNAs in E. coli. Extending these studies to different bacteria and environmental conditions should identify more novel transcripts and reveal the regulatory potential of pervasive transcription. Detailed and careful analyses of specific asRNAs need to be performed to further address the question of the function of pervasive asRNA transcription in bacteria.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Scott Samuels, Renée Schroeder and the Schroeder laboratory for thoughtful and critical readings of the manuscript.
Funding
ML and IB are supported by the Austrian Science Fund (Grants FWF I538-B12 and F4301) and the University of Vienna. Research in Raghavan lab is supported by Portland State University, and by grants from Collins Medical Trust and American Heart Association.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. . Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/ 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011; 75:286-300; PMID:21646430; http://dx.doi.org/ 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J 1994; 13:3599-607; PMID:7520390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 1994; 48:713-42; PMID:7826024; http://dx.doi.org/ 10.1146/annurev.mi.48.100194.003433 [DOI] [PubMed] [Google Scholar]
- 5.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 2010; 44:167-88; PMID:20707673; http://dx.doi.org/ 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 2012; 31:4005-19; PMID: 22922465; http://dx.doi.org/ 10.1038/emboj.2012.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilusic I, Popitsch N, Rescheneder P, Schroeder R, Lybecker M. Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol 2014; 11:e00156-12; PMID:24922322; http://dx.doi.org/ 10.4161/rna.29299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio 2012; 3; PMID:22872780; http://dx.doi.org/ 10.1128/mBio.00156-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. . Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 2012; 335:1103-6; PMID: 22383849; http://dx.doi.org/ 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- 10.Stone JR, Wray GA. Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol 2001; 18:1764-70; PMID:11504856; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a003964 [DOI] [PubMed] [Google Scholar]
- 11.Froula JL, Francino MP. Selection against spurious promoter motifs correlates with translational efficiency across bacteria. PLoS One 2007; 2:e745; PMID:17710145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huerta AM, Francino MP, Morett E, Collado-Vides J. Selection for unequal densities of sigma70 promoter-like signals in different regions of large bacterial genomes. PLoS Genet 2006; 2:e185; PMID:17096598; http://dx.doi.org/ 10.1371/journal.pgen.0020185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn MW, Stajich JE, Wray GA. The effects of selection against spurious transcription factor binding sites. Mol Biol Evol 2003; 20:901-6; PMID: 12716998; http://dx.doi.org/ 10.1093/molbev/msg096 [DOI] [PubMed] [Google Scholar]
- 14.Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev 2014; 28:214-9; PMID:24449106; http://dx.doi.org/ 10.1101/gad.234336.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leela JK, Syeda AH, Anupama K, Gowrishankar J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 2013; 110:258-63; PMID:23251031; http://dx.doi.org/ 10.1073/pnas.1213123110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 2012; 26:2621-33; PMID:23207917; http://dx.doi.org/ 10.1101/gad.196741.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. . The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 2010; 34:883-923; PMID:20659169 [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Saelao P, Jones CD, Begun DJ. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 2014; 343:769-72; PMID: 24457212; http://dx.doi.org/ 10.1126/science.1248286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet 2012; 8:e1002841; PMID:22844254; http://dx.doi.org/ 10.1371/journal.pgen.1002841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, et al. . Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA 2011; 108:20172-7; PMID:22123973; http://dx.doi.org/ 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci USA 2014; 111:3134-9; PMID:24453212; http://dx.doi.org/ 10.1073/pnas.1315974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 2012; 8:583; PMID:22617957; http://dx.doi.org/ 10.1038/msb.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol 2013; 11:75-82; PMID:23268228; http://dx.doi.org/ 10.1038/nrmicro2934 [DOI] [PubMed] [Google Scholar]
- 24.Lioliou E, Sharma CM, Caldelari I, Helfer AC, Fechter P, Vandenesch F, Vogel J, Romby P. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 2012; 8:e1002782; PMID:22761586; http://dx.doi.org/ 10.1371/journal.pgen.1002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell 2013; 52:473-84; PMID:24267449; http://dx.doi.org/ 10.1016/j.molcel.2013.10.032 [DOI] [PubMed] [Google Scholar]
- 26.Pettijohn DE, Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol 1974; 38:31-41; PMID:4598638; http://dx.doi.org/ 10.1101/SQB.1974.038.01.006 [DOI] [PubMed] [Google Scholar]
- 27.Macvanin M, Edgar R, Cui F, Trostel A, Zhurkin V, Adhya S. Noncoding RNAs binding to the nucleoid protein HU in Escherichia coli. J Bacteriol 2012; 194:6046-55; PMID:22942248; http://dx.doi.org/ 10.1128/JB.00961-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; http://dx.doi.org/ 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013; 14:880-93; PMID:24217315; http://dx.doi.org/ 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]


