Abstract
Structural comparisons of initiating RNA polymerase complexes and structure-based amino acid sequence alignments of general transcription initiation factors (eukaryotic TFIIB, archaeal TFB and bacterial σ factors) show that these proteins are homologs. TFIIB and TFB each have two-five-helix cyclin-like repeats (CLRs) that include a C-terminal helix-turn-helix (HTH) motif (CLR/HTH domains). Four homologous HTH motifs are present in bacterial σ factors that are relics of CLR/HTH domains. Sequence similarities clarify models for σ factor and TFB/TFIIB evolution and function and suggest models for promoter evolution. Commitment to alternate modes for transcription initiation appears to be a major driver of the divergence of bacteria and archaea.
Abbreviations:
- BRE
TFB/TFIIB recognition element
- CLR/HTH
cyclin-like repeat/helix-turn-helix domain
- DPBB
double psi beta barrel
- DDRP
DNA-dependent RNA polymerase
- GTF
general transcription factor
- LECA
last eukaryotic common ancestor
- LUCA
last universal common ancestor
- Ms
Methanocaldococcus sp. FS406-22,
- PIF
primordial initiation factor
- RDRP
RNA-dependent RNA polymerase
- RNAP
RNA polymerase
- Sc
Saccharomyces cerevisiae
- TFB
transcription factor B
- TFIIB
transcription factor for RNAP II
- factor B
Tt, Thermus thermophilus
Introduction
Bacteria and archaea appear to have diverged about the time of LUCA–the last universal common ancestor and one of the first DNA-based organisms. Eukaryotes emerged at LECA–the last eukaryotic common ancestor. Eukaryotes are a chimeric fusion of multiple bacteria and at least one archaea.1 Multi-subunit RNA polymerases (RNAPs) are DNA-dependent RNAPs (DDRPs) conserved in bacteria, archaea and eukaryotes. These dynamic molecular motors that accurately translocate stepwise on DNA are of the 2-double psi beta barrel (2-DPBB) type.2-4 Because some eukaryotes have interfering RNA-dependent RNAPs (RDRPs) of the 2-DPBB type, multi-subunit RNAPs appear rooted in the distant RNA-protein world.3 Bacterial σ factors bind to RNAP “core” (α2ββ’ω) to form RNAP “holoenzyme” (α2ββ’ωσ), which is capable of accurate initiation from a promoter DNA sequence.5,6 Archaeal core RNAP is more ornate (i.e., 14 subunits), similar to eukaryotic RNAPs I, II and III.7 Archaeal RNAPs utilize TFB-TBP (transcription factor B and TATA-binding protein) rather than σ factors for initiation.7
Despite conservation of 2-DPBB RNAPs within the three domains of life, the evolutionary relationship between bacterial σ factors and eukaryotic RNAP II general transcription factors (GTFs) has remained enigmatic.6,8 Recently, however, several reports have indicated the possible functional analogy and/or evolutionary relatedness of bacterial σ factors and eukaryotic TFIIB, which is one of a collection of GTFs for accurate initiation by RNAP II.4,9,10 TFIIB is a paralog of yeast Rrn7 (human TAF1B) for RNAP I and Brf-1 for RNAP III.11-14 In archaea, TFB is the homolog of eukaryotic TFIIB family proteins.7
In RNAP complexes with an open transcription bubble, σ factors and TFIIB both closely approach catalytic sites indicating direct and similar roles in initiation. Furthermore, σ factors and TFIIB each have multiple DNA binding helix-turn-helix (HTH) motifs, which typically include three crossing helices and two turns: H1-T1-H2-T2-H3. H3 is referred to as the “recognition helix” because sequences within T2 and toward the N-terminal end of H3 are most important for sequence recognition within the DNA major groove. TFIIB has two HTH motifs, each at the C-terminal end of a larger five-helix bundle termed a cyclin-like repeat (CLR or here CLR/HTH to emphasize that these are DNA binding domains and not cyclins). By contrast, group 1 σ factors include four HTH motifs.15 Aravind and co-workers, therefore, raised the issue of the evolutionary relatedness of σ factors and TFIIB based on apparent conservation of two C-terminal HTH motifs,4 but they did not extend their analysis to sequence comparisons or detailed structural models. Cramer and Kornberg and their colleagues have also indicated the possible relatedness or analogy of σ factors and TFIIB.9,10,16 Here, 2-HTH motifs of bacterial σ factors and eukaryotic TFIIB are shown to occupy homologous environments within initiating RNAP and RNAP II complexes.9,10,17 Based on extensive apparent structural homology, amino acid sequence alignments were generated, supporting the conclusion that σ factors, TFB and TFIIB are homologs. Furthermore, the 4-HTH domains in σ appear to be derived from larger CLR/HTH domains similar to those retained as recognizable repeats in TFB/TFIIB. This view of σ factors, TFB and TFIIB generates predictions for conserved mechanisms of transcription initiation within the three domains of extant organisms.2
Homology of σ factors and TFB/TFIIB, furthermore, suggests a simple model for promoter evolution. A typical promoter sequence in Escherichia coli includes a -35 region (consensus -35TTGACA-30) and a -10 region (consensus -12TATAAT-7).17-19 Spacing between -35 and -10 regions is typically 16-18 bp, with a spacing of 17 bp generally preferred. A promoter may include an extended -10 region (i.e., -15TGXTATAAT-7) and/or a discriminator region (i.e., -6GGG-4).20 In archaea/eukaryotes, promoters may include TFB/TFIIB recognition elements (BREdown and BREup) surrounding a TATAAAAG element to bind TFB/TFIIB downstream and upstream of TBP. Assuming a primordial initiation factor (PIF) had 4-CLR/HTH domains similar to a bacterial σ factor, primordial promoter structures and their co-evolution with GTFs can be modeled.
Results and Discussion
A structural comparison of bacterial RNAP and eukaryotic RNAP II initiating complexes is shown in Figure 1. σ factors include 4-HTH motifs, and TFIIB includes 2-HTH motifs within larger CLR/HTH domains. In Figure 1, HTH motifs are color-coded: H1 (yellow), H2 (red), and H3 (green) using brighter shades for σ than for TFIIB. Because the more C-terminal TFIIB CLR/HTH2 domain was missing from the original 4BBS structure, it was placed by modeling. The positioning is approximate, but the double-stranded (ds) BREup, to which TFIIB CLR/HTH2 binds, must be located near where CLR/HTH2 was placed. The two C-terminal σ and TFIIB HTH motifs appear to occupy homologous positions in the structures. The quality of the alignment can be judged from coincidence of homologous secondary structures in the overlay.
Figure 1.
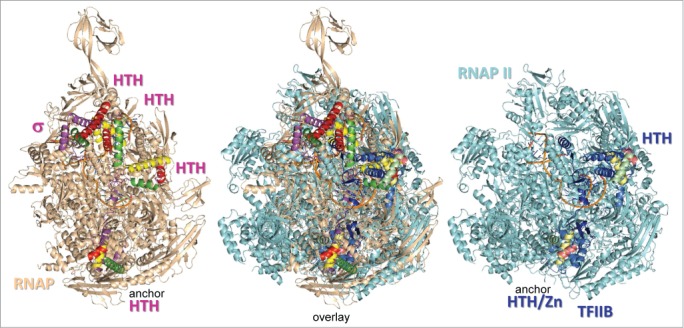
Structural alignment of initiating bacterial RNAP (PDB 4G7O) and yeast RNAP II (PDB 4BBS) complexes shows that σ factors and TFIIB occupy homologous positions.9,17 HTH motifs are colored yellow (H1), red (H2) and green (H3). Brighter colors are used for σ and duller shades for TFIIB. TFIIB CLR/HTH2 was placed by modeling, based on its predicted position bound to the ds BREup DNA anchor, which is missing in the structure.
In order to emphasize the similarities of bacterial σ factors and eukaryotic TFIIB, a simplified view was produced by removing RNAP and RNAP II from the image (Fig. 2). The quality of the alignment (and the alignment in Fig. 1) can be judged from the overlay of Mg and nucleic acids. The overlapping image of TFIIB CLR/HTH1 results from the overlay of two structures in creating the TFIIB model (PDBs 4BBS and 1AIS). Remarkably, σ CLR/HTH3.0-3.1 and TFIIB CLR/HTH1 occupy homologous positions, and σ CLR/HTH4.1-4.2 and TFIIB CLR/HTH2 also appear to occupy homologous positions. Again, the location of TFIIB CLR/HTH2 is not certain because it results from a model, not a structure, but the domain is placed with some confidence according to the expected location of ds BREup, which remains double-stranded during transcription bubble opening and to which CLR/HTH2 binds. Additionally, according to the model, TFIIB CLR/HTH2 appears to closely approach or interact with the N-terminal TFIIB Zn finger, providing a testable prediction of the model.
Figure 2.
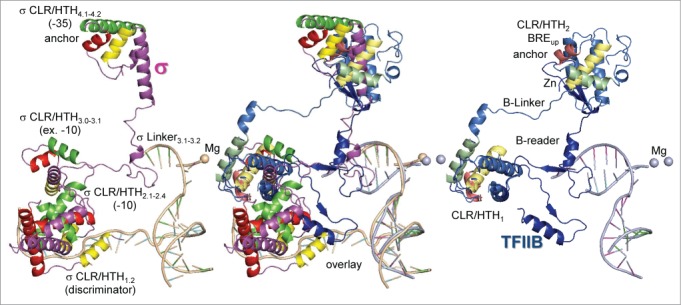
Relative positions of σ and TFIIB in initiating complexes. HTH motifs are colored yellow (H1), red (H2) and green (H3). Brighter colors are used for σ and duller shades for TFIIB. TFIIB CLR/HTH2 was placed by modeling, based on its predicted position bound to the ds BREup DNA anchor. Placement of TFIIB CLR/HTH2 indicates that the N-terminal Zn ribbon and the C-terminal CLR/HTH2 are likely to interact. The quality of the alignment is indicated by the overlay of the Mg atoms and nucleic acids.
To further the argument for homology of σ factors and TFB/TFIIB, a structural comparison of ds DNA promoter binding is shown (Fig. 3). Bacterial -35 promoter regions and archaeal/eukaryotic BREup are considered here to be “anchor” DNA sequences. Binding of C-terminal CLR/HTH domains to ds upstream anchor DNA is posited to allow directional downstream promoter opening by more mobile N-terminal CLR/HTH domains, a concept in promoter evolution and GTF structure and function discussed in further detail below. In Figure 3, a structure of bacterial σ CLR/HTH4.1-4.2 bound to ds -35 region promoter DNA is aligned with archaeal CLR/HTH2 in a co-crystal of ds BRE and TATAAAAG box DNA with GTFs TFB and TBP. Despite distortion of ds DNA by TBP binding in the minor groove, the overlay of σ CLR/HTH4.1-4.2 and TFB CLR/HTH2 is surprisingly precise and supports the structural homology of σ CLR/HTH4.1-4.2 and TFB CLR/HTH2. From this comparison, furthermore, bacterial -35 regions and archaeal/eukaryotic BREup DNA anchor sequences appear to serve a similar anchoring function in the evolution of promoters. Very interestingly, TFIIB CLR/HTH1 and CLR/HTH2 bound to ds BREdown and BREup are clustered on either side of the TBP “stirrup” (Fig. 3), but spread apart in the structural model for initiation (Figs. 1–2). As BREdown becomes single-stranded, CLR/HTH1 must move in the downstream direction for transcription initiation, but CLR/HTH2 is expected to remain in place, bound to the ds BREup anchor. A similar model for spreading of CLR/HTH domains from the ds -35 anchor DNA during bubble opening is considered likely for bacterial σ factors (see below).
Figure 3.
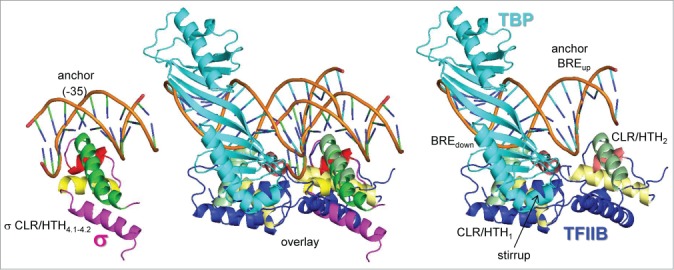
Homologous binding of σ CLR/HTH4.1-4.2 to the -35 region (PDB 1KU7) and archaeal TFB CLR/HTH2 to BREup ds anchor DNA (PDB 1AIS).26,33 Anchor DNA-binding CLR/HTH domains are expected to remain in place as the bubble opens, but σ CLR/HTH3.0-3.1 and TFB CLR/HTH1 are expected to move in a downstream direction. In the overlay, note the structural homology of H1, T1, H2, T2 and H3 of σ and TFB. Colors are as shown in Figures 1–2.
Based on structural homology, protein sequence alignments of Saccharomyces cerevisiae (Sc) TFIIB, Methanocaldococcus sp. FS406-22 (Ms) TFB and Thermus thermophilus (Tt) σ factor were generated (Fig. 4). Remarkably, with structure as a guide, a reasonable σ to TFB and σ to TFIIB homology model was constructed. Similarities between the Tt σ factor and Ms/Sc TFB/TFIIB are most notable in T1, H2, T2 and H3. Because of sequence identity and similarity, when aligning according to turns and helices, bacterial σ factors are concluded to be homologs of archaeal/eukaryotic TFB/TFIIB. To reinforce this conclusion, multiple archaeal TFB and bacterial σ factor sequences are aligned in Figures S1-S4. Consistent with structural alignments (Figs. 1–3), multiple sequence alignments of bacterial σ CLR/HTH3.0-3.1 and CLR/HTH4.1-4.2 show remarkable in-phase similarity to archaeal TFB, particularly across T1, H2, T2 and H3 (Figs. S3-S4). Consistent with the homology model, within σ CLR/HTH4.1-4.2 H2, some alternate σ factors show significantly higher identity and similarity to TFB CLR/HTH2 compared to the Tt σ factor used in the original alignment (Fig. 4; Fig. S4). Despite the ancient time of divergence for σ factors and TFB/TFIIB, alignments of linear sequence are convincing (Fig. 4; Figs. S1–S4), and structural comparisons are compelling (Figs. 1–3) for homology among these GTFs.
Figure 4.

Sc TFIIB, Ms TFB and Tt σ factors are homologs. σ factors have 4-CLR/HTH motifs (regions 1.2, 2.1-2.4, 3.0-3.1 and 4.1-4.2). Grey shading indicates helical regions. Amino acids that are identical between either Sc TFIIB or Ms TFB and Tt σ are red; amino acids that are similar are in black bold type. Greatest similarity is within T1, H2, T2 and H3.
From structural and sequence similarities, a model for σ and TFB/TFIIB homology is proposed (Fig. 5). σ factor regions (extending from N→C 1, 2, 3 and 4) were defined historically based on homology among σ factors.21,22 The model in Figure 5 posits that σ homology regions 1-4 are each degenerated from one of 4-CLR/HTH domains, similar to the 2-CLR/HTH domains found in TFB/TFIIB. To distinguish C-terminal CLR/HTH domains from the more mobile N-terminal CLR/HTH domains, C-terminal CLR/HTH domains that bind anchor DNA sequences are colored red. σ CLR/HTH4.1-4.2 binds ds -35 anchor DNA, which remains double-stranded during bubble opening and initiation (Figure 3). σ CLR/HTH domains 1-3 are posited to spread from the ds anchor DNA sequence in the downstream direction as the bubble opens, as observed in Figures 1–2. Similarly, TFB/TFIIB has two CLR/HTH domains that are clustered for ds promoter recognition (Fig. 3). CLR/HTH2 remains bound to ds BREup anchor DNA, and CLR/HTH1 must spread from CLR/HTH2 and its bound ds DNA anchor in the downstream direction during bubble opening and initiation (Figs. 1–3). The -35 region and BREup DNA anchors, conserved in the three domains of life on earth, therefore, support the directionality of bubble opening and transcription. The initiating RNAP structure (Figs. 1–2) shows σ in its opened conformation. Currently, there is no RNAP-σ structure available that only contains ds promoter DNA.
Figure 5.
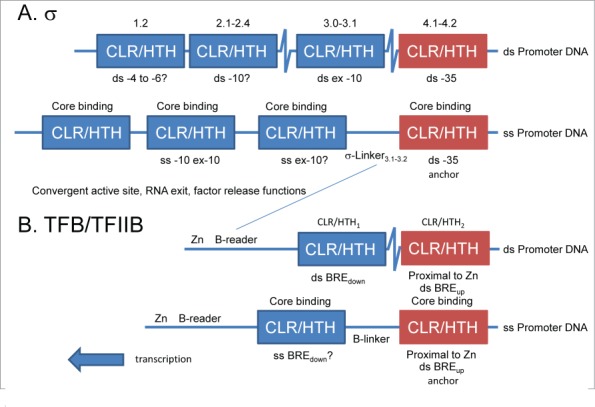
(A) A model for σ and (B) a model for TFB/TFIIB in initial ds promoter recognition and in initiating RNAP and RNAP II complexes with an open bubble. σ and TFIIB are proposed to cluster CLR/HTH domains for initiation. Contact to ds anchor DNA maintains the position of the most C-terminal CLR/HTH domain (red). For bubble opening and initiation, more N-terminal CLR/HTH domains (blue) unpack in the downstream direction.
The 4-CLR/HTH domains of σ factors and their roles in transcription are described in Figure 5A. σ CLR/HTH1.2, which may bind the -4 to -6 “discriminator”,6 appears to be the most degenerated from a CLR/HTH (Fig. 4; Fig. S1). With five bunched helices, σ CLR/HTH2.1-2.4 appears similar in overall secondary structure to a TFB/TFIIB CLR/HTH domain, but CLR/HTH2.1-2.4 is highly specialized in evolution for promoter -10 region opening through a base flipping mechanism (Figs. 2 and 4; Fig. S5) (PDB 3UGO, 3UGP, 4LUP, 47GO).17-19,23,24 Although evolution has fused two helices to eliminate a turn, σ CLR/HTH3.0-3.1 also shows sequence similarity to a CLR/HTH motif (Fig. 4; Fig. S3). σ HTH3.0-3.1 appears to contact the extended -10 region of promoters as ds DNA, but may also have a role in DNA strand separation.25 σ CLR/HTH4.1-4.2 appears to have lost or fused the most N-terminal CLR helix but maintains similarity to a CLR outside the HTH4.2 motif (Fig. 4). Upstream of the melted promoter region, σ CLR/HTH4.1-4.2 contacts the anchor ds -35 region (PDB 1KU7, 2H27) (Fig. 3 and 5).26,27 In σ factors, CLR/HTH domains are highly specialized in evolution for recognition of promoter DNA sequences (regions 2 and 4), for opening the transcription bubble (region 2) and for RNAP core recruitment (regions 1-4) (Fig. 5A). Separating σ HTH3.0-3.1 and CLR/HTH4.1-4.2 is the flexible σ-Linker3.1-3.2 that approaches the RNAP active site in initiating complexes and makes contacts with the early transcript (~6-13 nts) that are important to dissociate σ during RNA chain elongation and perhaps for initially dissociating RNA from the template DNA strand.17 Because σ CLR/HTH4.1-4.2 (-35 recognition), HTH3.0-3.1 (extended -10 recognition), and CLR/HTH2.1-2.4 (-10 recognition) are predicted to be packed together within the major groove of the ds promoter DNA and then to spread apart in the initiating complex with an open bubble, the distances between σ HTH regions, are expected to increase as the bubble expands (PDB 4G7O) (Figs. 1–2).17 Because the -35 anchor region of the promoter remains as ds DNA bound to σ CLR/HTH4.1-4.2, σ regions 1-3 are expected to spread from region 4 in the direction of transcription (upstream→downstream) as indicated in Figure 5A.
In Figure 5B, a schematic of TFB/TFIIB regions is shown. Near the N-terminus, TFIIB has a Zn ribbon. Next, is the B-reader region consisting of the B-reader helix, the B-reader loop and the B-reader strand.9 The B-reader region approaches the RNAP II active site and, although not homologous by orientation (N→C) or sequence to σ-Linker3.1-3.2, appears to have convergent functions in initiation and promoter escape.9,10 TFIIB includes two CLR/HTH domains (CLR/HTH1 and CLR/HTH2) separated by the B-linker (Figure 5B). As indicated in Figures 4, S3 and S4, σ HTH3.0-3.1 is most similar to TFB/TFIIB CLR/HTH1, and σ CLR/HTH4.1-4.2 is most similar to TFB/TFIIB CLR/HTH2. In initiation from a TATA box-containing promoter, TBP binds within the DNA minor groove and induces a ~90 degree bend in the DNA. The TFIIB 2-CLR/HTH domains cluster on either side of TBP touching the TBP “stirrup” (Fig. 3). TFIIB CLR/HTH2 inserts into the DNA major groove upstream of the TATA box and can recognize a BREup through a typical HTH-DNA interaction (Fig. 3).28 TFB/TFIIB CLR/HTH2 binding to BREup anchors the initiating complex on ds DNA and establishes the direction of transcription analogously to the anchoring of σ CLR/HTH4.1-4.2 binding to the ds -35 region of the bacterial promoter (Figs. 3 and 5). TFB/TFIIB CLR/HTH1 contacts the DNA major groove at BREdown.29,30 The B-linker separating TFB/TFIIB CLR/HTH1 and CLR/HTH2 is compressed in the pre-initiation complex on ds DNA (Fig. 3), indicating that TFIIB CLR/HTH1 and CLR/HTH2 must unpack and separate during bubble opening for initiation (Figures 1–2), as proposed for σ CLR/HTH1.2, CLR/HTH2.1-2.4, CLR/HTH3.0-3.1 and the upstream anchoring CLR/HTH4.1-4.2 (Fig. 5A).25 The TFB/TFIIB Zn ribbon may provide a bulky group to help maintain the conformation of the B-reader region to support initiation functions, and the Zn ribbon is predicted from modeling to interact with TFIIB CLR/HTH2 (Fig. 2). Such an interaction is expected to stabilize B-reader threading through RNAP II by helping to position the Zn ribbon. In σ factors, the evolutionarily convergent region is σ-Linker3.1-3.2, which is bounded and positioned by bulky domains σ CLR/HTH3.0-3.1 and σ CLR/HTH4.1-4.2. Similar to σ factors, TFB/TFIIB is predicted to cluster its CLR/HTH domains for formation of a pre-initiation complex (as in PDB 1VOL, 1C9B, 1AIS, and 1D3U) (Figs. 3 and 5B) and unpack and separate CLR/HTH domains as the bubble opens (Figs. 1, 2, and 5B).
Promoter co-evolution assuming a 4-CLR/HTH PIF
Homology among bacterial σ factors and archaeal/eukaryotic TFB/TFIIB suggests a simple model for a primordial initiation factor (PIF) and for promoter evolution based on a 4-CLR/HTH PIF at about the time of LUCA (Fig. 6). The 4-CLR/HTH PIF is posited to have arisen by duplication of a single CLR/HTH domain followed by subsequent duplication of the 2-CLR/HTH domains to form the 4-CLR/HTH factor. Because of the evolutionary path, σ CLR/HTH1.2 and CLR/HTH3.0-3.1 are most similar to TFB CLR/HTH1 (odd numbered repeats are most similar among σ and TFB), and σ CLR/HTH2.1-2.4 and CLR/HTH4.1-4.2 are most similar to TFB CLR/HTH2 (even numbered repeats are most similar among σ and TFB). In Figure 6A, a model is shown for a 4-CLR/HTH domain PIF supporting bi-directional initiation on an AT-rich negatively supercoiled primordial promoter. A 4-CLR/HTH domain PIF is hypothesized because it is difficult to imagine a 2-CLR/HTH domain PIF, resembling TFB/TFIIB, opening even negatively supercoiled AT-rich DNA without cooperation of additional GTFs. Because of powerful selection for promoter directionality, anchor DNA sequences are posited to have rapidly evolved near the upstream edge of the AT-rich segment (Fig. 6B). Bacterial -35 regions and archaeal/eukaryotic BREup are posited to be relics of primordial anchor DNA sequences (Figs. 3, 5 and 6B). The AT-rich primordial promoter is posited to have been compressed in evolution into the bacterial Pribnow box (TATAAT) and the archaeal/eukaryotic TATAAAAG box through co-evolution with RNAP and GTFs. According to this simple working model, promoters have not changed very much since the advent of DNA genomes at about the time of LUCA.
Figure 6.
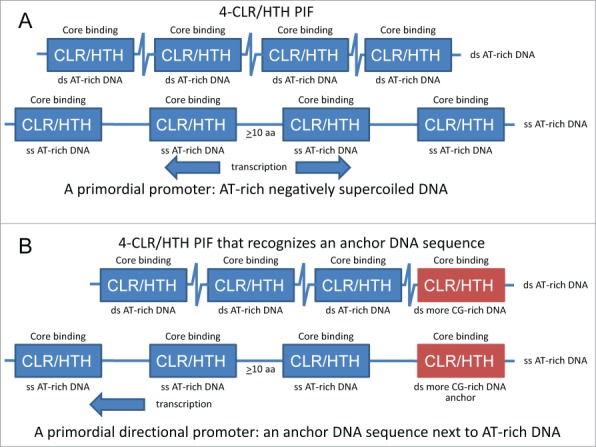
A model for early evolution of promoters based on a 4-CLR/HTH PIF. A) a 4-CLR/HTH PIF for initiation on a bidirectional primordial promoter. B) a 4-CLR/HTH PIF for initiation on a unidirectional primordial promoter with an anchor DNA sequence. The C-terminal CLR/HTH domain is shaded red to indicate that it binds anchor DNA. Bacterial -35 and archaeal/eukaryotic BREup DNA elements are posited to be relics of primordial anchor DNA sequences. TATAAT (Pribnow box) and TATAAAAG boxes are posited to be derived from AT-rich primordial promoter sequences. CLR/HTH domains are posited to separate from the upstream anchor in the downstream direction as the bubble opens.
Ancient Evolution
Previously, a working model was proposed for genesis of life on earth that concentrated on the likely central roles of 2-DPBB type multi-subunit RNAPs found in bacteria, archaea and eukaryotes.2 The model gave insight into the RNA-protein world, LUCA, LECA and the role of the RNAP II CTD and its extensive interactome in evolution of eukaryote complexity. An updated version of the model that incorporates σ homology to TFB/TFIIB, promoter anchor DNA sequences and promoter evolution is shown in Figure 7. The concept of a 4-CLR/HTH PIF at LUCA enhances the model for radiation of bacteria with 4-CLR/HTH σ factors with 4 degenerate CLR/HTH repeats and archaea with 2-CLR/HTH TFB with 2 highly conserved CLR/HTH repeats. Bacterial σ factors, therefore, are posited to be most similar to a 4-CLR/HTH PIF in overall domain structure, and conserved archaeal TFB CLR/HTH repeats are posited to be most similar to PIF CLR/HTH domains in sequence. It is posited that bacteria and archaea are early radiations from LUCA that diverged largely because of their fundamental differences in GTFs and, therefore, their alternate approaches to genome interpretation. Bacteria became committed to σ factors for promoter recognition, and RNAP recruitment is tightly coupled to σ factor binding to core RNAP in bacteria. Co-evolution of bacterial σ factors and RNAP, therefore, is proposed to have driven streamlining to a simpler RNAP subunit structure (α2ββ’ω). Evolution of the bacterial promoters is posited to be driven by co-evolution with interacting σ CLR/HTH domains. Archaeal TFB is proposed to have lost 2-CLR/HTH domains from a 4-CLR/HTH PIF and gained a N-terminal Zn ribbon and B-reader region. In archaea, cooperation of TFB with TBP is posited to have allowed for the losses of 2-CLR/HTH domains from the PIF and the gain of the N-terminal Zn ribbon and B-reader. In initiation mechanisms, TFB-TBP GTFs appear less strongly coupled to archaeal RNAP than bacterial σ factors and bacterial RNAP. This difference in GTFs may have allowed archaea to retain a more ornate RNAP subunit structure than observed in bacteria.
Figure 7.
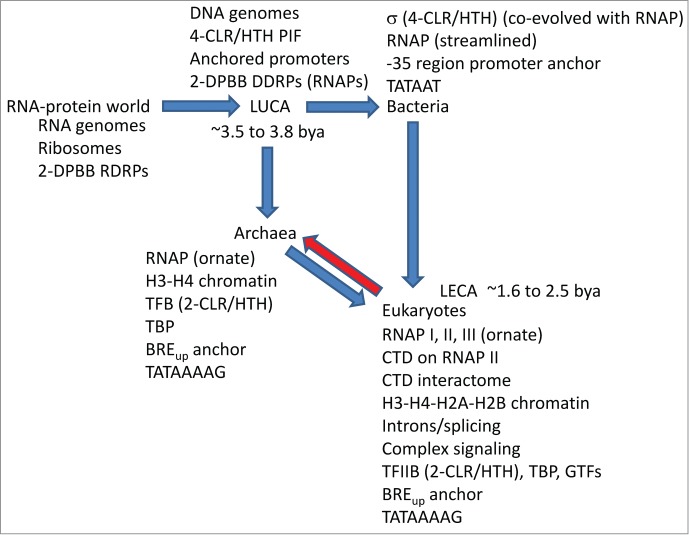
A model for the genesis of life on earth focusing on multi-subunit RNAPs, GTFs and promoters. The red arrow indicates strong competition by eukaryotes suppressing mesophilic archaea. See the text for details.2
Eukaryotes are a chimeric fusion of at least one generalist archaea and several bacteria, for instance, resulting from multiple endosymbioses and genome fusions (Figure 7).1 The archaeal species proposed to have given rise to eukaryotes at LECA appears now to be extinct but may be represented by gene clusters now dispersed to a number of surviving archaeal species. The red arrow in Figure 7 indicates competition won by eukaryotes over mesophilic archaea, driving archaea into more extreme niches and complex symbioses and forcing genetic losses and archaeal extinctions. Because archaeal TFB is such a close homolog of eukaryotic TFIIB (Fig. 4), the current work gives most insight into bacterial and archaeal divergence at LUCA rather than eukaryote generation at LECA.
Methods
PDBs 4G7O (initiating Tt σ-RNAP-DNA-RNA)17 and 4BBS (Sc TFIIB-RNAP II-DNA-RNA)9 were aligned for the two largest RNAP subunits using Pymol (Schrodinger; www.pymol.org). Remarkably, σ HTH3.0-3.1 and TFIIB CLR/HTH1 were observed to occupy comparable positions in respective RNAP initiating complexes. Because TFIIB CLR/HTH2 was missing from PDB 4BBS, CLR/HTH2 was placed by modeling. Approximate placement of TFIIB CLR/HTH2 was done based on comparison of TFB-TBP-DNA complexes and based on the likely positioning of ds BREup. Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id = index)31 was used for homology threading to adapt the archaeal TFB-TBP-DNA structure (PDB 1AIS) to Sc TFIIB sequences and structures. After placing CLR/HTH2, the B-linker was adjusted using the program Modeller (https://salilab.org/modeller/9.12/manual/)32 to connect the 2-CLR/HTH domains. Structural figures were prepared using Pymol.
Because of extensive apparent structural homology comparing bacterial σ factors and eukaryotic TFIIB, a structure-based linear amino acid sequence alignment was generated, aligning Tt σ and Sc TFIIB according to secondary structure elements. Because archaea are more closely related to bacteria than are eukaryotes, Ms TFB was included in the comparison. Although bacteria and archaea must have diverged about the time of LUCA, only these sequences were necessary to generate the alignment. To reinforce the conclusion of homology, multiple sequence alignments were done to determine whether additional sequence similarities would be detected comparing more sequences and/or alternate σ factors (Fig. S1-S4). This prediction of the original alignment model was justified (i.e., see Fig. S4; H2). Sequences for archaeal TFB multiple alignments were collected from the Uniprot database (http://www.uniprot.org/). Sequences for σ multiple alignments were collected from the NCBI database (http://www.ncbi.nlm.nih.gov/) and aligned using the SDSC Biology Workbench tools (http://workbench.sdsc.edu/). Based on structural alignments and sequences compatible with turns, some minor adjustments to some alignments were made.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Koonin EV, Yutin N. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harbor Perspect biol 2014; 6:a016188; PMID:24691961; http://dx.doi.org/ 10.1101/cshperspect.a016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton ZF. The old and New testaments of gene regulation: evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD. Transcription 2014; 5; PMID:24802897; http://dx.doi.org/ 10.4161/trns.28674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol 2003; 3:1;PMID:12553882; http://dx.doi.org/ 10.1186/1472-6807-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer LM, Aravind L. Insights from the architecture of the bacterial transcription apparatus. J Struct Biol 2012; 179:299–319; PMID:22210308; http://dx.doi.org/ 10.1016/j.jsb.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feklistov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 2014;68:357–76; PMID:25002089; http://dx.doi.org/ 10.1146/annurev-micro-092412-155737 [DOI] [PubMed] [Google Scholar]
- 6.Decker KB, Hinton DM. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Ann Rev Microbiol 2013; 67:113–39; PMID:23768203; http://dx.doi.org/ 10.1146/annurev-micro-092412-155756 [DOI] [PubMed] [Google Scholar]
- 7.Werner F. Molecular mechanisms of transcription elongation in archaea. Chem Rev 2013; 113:8331–49; PMID:24024741; http://dx.doi.org/ 10.1021/cr4002325 [DOI] [PubMed] [Google Scholar]
- 8.Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder RG. Sequence of general transcription factor TFIIB and relationships to other initiation factors. Proc Natl Acad Sci U S A 1991; 88:9553–7; PMID:1946368; http://dx.doi.org/ 10.1073/pnas.88.21.9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 2013; 493:437–40; PMID:23151482; http://dx.doi.org/ 10.1038/nature11715 [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 2010; 327:206–9; PMID:19965383; http://dx.doi.org/ 10.1126/science.1182015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Gene Deve 1992; 6:1940–9; PMID:1398071; http://dx.doi.org/ 10.1101/gad.6.10.1940 [DOI] [PubMed] [Google Scholar]
- 12.Hahn S, Roberts S. The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Gene Deve 2000; 14:719–30; PMID:10733531 [PMC free article] [PubMed] [Google Scholar]
- 13.Knutson BA, Hahn S. TFIIB-related factors in RNA polymerase I transcription. Biochim Biophys Acta 2013; 1829:265–73; PMID:22960599; http://dx.doi.org/ 10.1016/j.bbagrm.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutson BA, Hahn S. Yeast rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Sci 2011; 333:1637–40; PMID:21921198; http://dx.doi.org/ 10.1126/science.1207699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 2002; 417:712–9; PMID:12000971; http://dx.doi.org/ 10.1038/nature752 [DOI] [PubMed] [Google Scholar]
- 16.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Sci 2004; 303:983–8; PMID:14963322; http://dx.doi.org/ 10.1126/science.1090838 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Sci 2012; 338:1076–80; PMID:23086998; http://dx.doi.org/ 10.1126/science.1227786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feklistov A, Darst SA. Crystallographic analysis of an RNA polymerase sigma-subunit fragment complexed with -10 promoter element ssDNA: quadruplex formation as a possible tool for engineering crystal contacts in protein-ssDNA complexes. Acta crystallogr Sect F, Struct Biol Cryst Commun 2013; 69:950–5; PMID:23989139; http://dx.doi.org/ 10.1107/S1744309113020368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 2011; 147:1257–69; PMID:22136875; http://dx.doi.org/ 10.1016/j.cell.2011.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the 'extended-10' motif at promoters. EMBO J 1997; 16:4034–40; PMID:9233812; http://dx.doi.org/ 10.1093/emboj/16.13.4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gribskov M, Burgess RR. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res 1986; 14:6745–63; PMID:3092189; http://dx.doi.org/ 10.1093/nar/14.16.6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 1992; 174:3843–9; PMID:1597408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darst SA, Feklistov A, Gross CA. Promoter melting by an alternative sigma, one base at a time. Nature Struct Mol Biol 2014; 21:350–1; PMID:24699085; http://dx.doi.org/ 10.1038/nsmb.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campagne S, Marsh ME, Capitani G, Vorholt JA, Allain FH. Structural basis for -10 promoter element melting by environmentally induced sigma factors. Nat Struct Mol Biol 2014; 21:269–76; PMID:24531660; http://dx.doi.org/ 10.1038/nsmb.2777 [DOI] [PubMed] [Google Scholar]
- 25.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 2003; 13:31–9; PMID:12581657; http://dx.doi.org/ 10.1016/S0959-440X(02)00005-2 [DOI] [PubMed] [Google Scholar]
- 26.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell 2002; 9:527–39; PMID:11931761; http://dx.doi.org/ 10.1016/S1097-2765(02)00470-7 [DOI] [PubMed] [Google Scholar]
- 27.Lane WJ, Darst SA. The structural basis for promoter -35 element recognition by the group IV sigma factors. PLoS Biol 2006; 4:e269; PMID:16903784; http://dx.doi.org/ 10.1371/journal.pbio.0040269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Gene Dev 1998; 12:34–44; PMID:9420329; http://dx.doi.org/ 10.1101/gad.12.1.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 1995; 377:119–28; PMID:7675079; http://dx.doi.org/ 10.1038/377119a0 [DOI] [PubMed] [Google Scholar]
- 30.Tsai FT, Sigler PB. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J 2000; 19:25–36; PMID:10619841; http://dx.doi.org/ 10.1093/emboj/19.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Sternberg MJ. Protein structure prediction on the web: a case study using the phyre server. Nat Protoc 2009; 4:363–71; PMID:19247286; http://dx.doi.org/ 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 32.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics / editoral board, Andreas D Baxevanis ; et al] 2006; Chapter 5:Unit 5.6; PMID:18428767; http://dx.doi.org/ 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosa PF, Ghosh G, DeDecker BS, Sigler PB. The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci U S A 1997; 94:6042–7; PMID:9177165; http://dx.doi.org/ 10.1073/pnas.94.12.6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


