Abstract
A tandemly repeated C-terminal domain (CTD) of the largest subunit of RNA polymerase II is functionally essential and strongly conserved in many organisms, including animal, yeast and plant models. Although present in simple, ancestral red algae, CTD tandem repeats have undergone extensive modifications and degeneration during the evolutionary transition to developmentally complex rhodophytes. In contrast, CTD repeats are conserved in both green algae and their more complex land plant relatives. Understanding the mechanistic differences that underlie these variant patterns of CTD evolution requires knowledge of CTD-associated proteins in these 2 lineages. To provide an initial baseline comparison, we bound potential phospho-CTD associated proteins (PCAPs) to artificially synthesized and phosphorylated CTD repeats from the unicellular red alga Cyanidioschyzon merolae and green alga Chlamydomonas reinhardtii. Our results indicate that red and green algae share a number of PCAPs, including kinases and proteins involved in mRNA export. There also are important taxon-specific differences, including mRNA splicing-related PCAPs recovered from Chlamydomonas but not Cyanidioschyzon, consistent with the relative intron densities in green and red algae. Our results also offer the first experimental indication that different proteins bind 2 distinct types of repeats in Cyanidioschyzon, suggesting a division of function between the proximal and distal CTD, similar to patterns identified in more developmentally complex model organisms.
Keywords: C-terminal domain, green algae, phosphorylation, red algae, RNA polymerase II
Abbreviations
- CTD
Carboxyl-terminal domain
- PCAPs
Phospho-CTD associated proteins
- CDK
Cyclin-dependent kinase
Introduction
RNA polymerase II is a large complex containing 12 subunits; the largest (RPB1) has a unique carboxyl-terminal domain (CTD) that has attracted the interest of many scientists since it was discovered in the 1980s.1,2 In model systems where most functional studies of the CTD have been carried out, the domain is composed of a varied number of tandemly repeated heptapeptides (yeast 26, human 52, Arabidopsis 34) with the consensus sequence Y1S2P3T4S5P6S7. Initial functional studies of the CTD employed truncation mutants in yeast and human cells3; they showed that the domain is essential for viability and there is functional redundancy among CTD repeats.4-6 Genetic substitution screens in yeast revealed that Y1 residues and the 2 SP pairs are essential, consistent with their stronger evolutionary conservation than the T4 and S7 positions.7,8 Further, insertions between individual heptapeptides proved to be lethal in fission and budding yeast, whereas insertions between paired repeats were not, indicating that the smallest CTD functional unit lies within pairs of heptapeptides.9,10 Further studies narrowed the smallest functional CTD unit in budding yeast to 2 Y1 residues surrounded by 3 SP pairs; that is, YSPxSPxYSP or SPxYSPxSPxY (x represents any amino acid).11 Consistent with genetic analyses, cumulative structural studies indicate that most CTD interactions with binding partners involve motifs between one and 2 heptapeptides in length, and usually not starting from a Y1 residue.12 Although great insights into the functional significance of CTD residues have been gained from experimental analyses, primarily in yeast and animals, comprehensive evolutionary investigations have shown that CTD sequence diversity precludes broader generalization of these results to many other organisms.13 This has been borne out by functional studies, for example, the demonstration that the CTD is indispensable in Trypanosoma brucei despite the absence of any of the essential motifs or repetitive structures required in yeast and animal models.14 Parallel with studies of the CTD sequence itself, further investigations have implicated the domain’s role in a wide variety of metabolic pathways in yeasts, animals and Arabidopsis, including transcription initiation and elongation, pre-mRNA processing, mRNA transport, and chromatin modification among others.15
The main way that the CTD performs these functions is by recruiting other proteins involved in the various pathways to create transcription/processing factories. Different modifications of heptapeptide residues provide a code that allows for the widely varied interactions between the CTD and many target proteins.16,17 Among the possible residue modifications, phosphorylations of S2 and S5 are the most common, and mainly relate to co-transcriptional functions like mRNA 5′ capping, mRNA 3′ end processing and pre-mRNA splicing.15 Interestingly, these core mRNA processing functions are broadly conserved across the eukaryotic domain, as are CTD-directed kinases responsible for these modifications.18
Very little empirical evidence exists for CTD functions in most eukaryotic groups. To our knowledge, there has been no previous direct experimental work reported in red or green algae. Interestingly, the CTDs of these groups have evolved in very different ways. Comparable to what has been found in animals,13 simple forms of green algae have CTDs consisting of canonical tandem repeats, whereas developmentally complex land plants display both tandemly repeated proximal regions and more modified distal regions (Fig. 1). Tandem repeats also are present in unicellular red algae; however, multicellular rhodophytes have highly modified CTDs without retention of any tandem repeats (Fig. 1). Why the CTD has adopted such different evolution trajectories in green plants and red algae is unknown, but it undoubtedly relates to underlying differences in the types and numbers of protein partners in the 2 lineages. Given the limited genetic tools available for investigating CTD function in rhodophytes, we undertook a biochemical comparison of baseline CTD-protein interactions in unicellular red and green algae as a reasonable first step toward elucidating comparative CTD function in the 2 groups.
Figure 1.
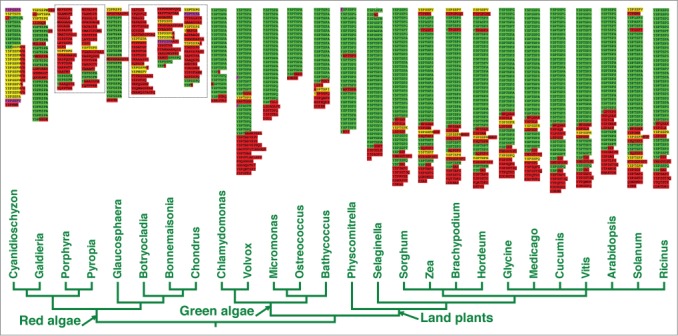
The CTD in green plants and red algae. The tree reflecting the relationships among green algae/plants and red algae was constructed based on the Tree of Life Web Project. Annotated CTDs for each genus are shown above the taxa included in the tree (CTD N-termini are at the top of each sequence). Sequences from multicellular red algae are shown in boxes; they have highly modified CTDs with no discernable repetitive structures that are present in unicellular (ancestral) forms. Green indicates regions with at least 2 continuous canonical (YSPxSPx) heptapeptides; yellow indicates the presence of isolated heptads, not in tandem with another canonical repeat; purple indicates the presence of the non-canonical motif “FSPTSPS;" red regions are without any canonical heptapeptides whatsoever. For more detail on these annotations, see our previous publication.13
Chlamydomonas reinhardtii is a well-studied green alga with a CTD comprising 20 tandem heptapeptides with the consensus YSPTSPA. The red unicell, Cyanidioschyzon merolae, has a CTD with 7 proximal tandem heptapeptides (YSPTSPA) and, surprisingly, 11 distal tandem nonapeptides (YSPSSPNVA). This latter structure is unique among all CTD sequences known.13 Complete genomes are available for both of these algae, permitting identification of proteins through mass spectrometry. Applying methods used previously to identify PCAPs from both yeast and mammalian cells,19,20 we isolated proteins that bind to bi-phosphorylated (S2 and S5), tri-heptapeptide CTD repeats from both algae, and tri-nonapeptide CTD repeats from C. merolae. We aimed to 1) identify proteins that bind differentially to the 2 different CTD regions in C. merolae, and 2) provide a first view of CTD-protein interactions that were in place before the CTD was modified differently in multicellular green plants and rhodophytes.
Results
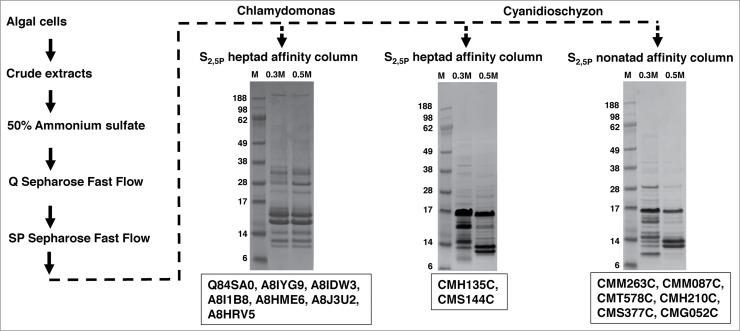
We isolated 154 total proteins from C. reinhardtii that bound the phospho-CTD, and 133 from C. merolae, yields that are very similar to those reported from yeast using comparable methods.20 Through careful screening and annotation, we identified 7 proteins from C. reinhardtii (Table 1) and 8 from C. merolae (Table 2) that we consider to be likely PCAPs. Six of the 8 red algal proteins were eluted from the nonapeptide affinity column and 2 from the heptapeptide column. The fact that this group of proteins bound only to the heptapeptide or nonapeptide repeats, but not to both, suggests they have specific CTD-motif affinities and are not simple, opposite charge attractions to phospho-serines. Other reasonable PCAP candidates were recovered (full lists provided in Tables S1 and S2, and see further discussion below), including a number that were specific to only one type of CTD repeat. In this report, however, we focus primarily on proteins for which there is prior experimental evidence of a CTD-interaction in model organisms, and on cases that highlight potential variation in CTD functions in the anciently diverged Rhodophyta.
Table 1.
The 7 potential Chlamydomonas PCAPs identified. 0.3M and 0.5M represent the NaCl elution concentrations. The numbers of the MS/MS identified matching peptides from the elutions are shown under each salt concentration. The annotations are based on predicted homologs in yeast, human and Arabidopsis, with e-values shown for the best Blast matches. The conventions are used in Table 2 and supplementary tables
| Heptapeptide column |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein names | 0.3M | 0.5M | Annotations | Best hit in yeast and the e-value | Best hit in human and the e-value | Best hit in Arabidopsis and e-value | |||
| Q84SA0 | 19 | 16 | Casein kinase I | HRR25 | e-139 | P48730 | e-165 | AT4G26100 | e-171 |
| A8IYG9 | 8 | 4 | Casein kinase II subunit α | CKA2 | e-111 | E7EU96 | e-118 | AT2G23070 | e-129 |
| A8J3U2 | 2 | Component of U2 snRNP | LEA1 | 1.00E-08 | P09661 | 3.00E-55 | AT1G09760 | 2.00E-59 | |
| A8IDW3 | 1 | 2 | Histone H2A deubiquitinase | RPN11 | 2.00E-06 | Q5VVJ2 | 3.00E-14 | AT3G09600 | 4.00E-08 |
| A8HME6 | 2 | Polyadenylate-binding protein | PAB1 | e-127 | P11940 | e-135 | AT1G49760 | e-127 | |
| A8I1B8 | 3 | RNA export factor | PAB1 | 7.00E-07 | Q86V81 | 4.00E-21 | AT5G59950 | 1.00E-24 | |
| A8HRV5 | 1 | U2B component of U2 snRNP | MSL1 | 7.00E-10 | P08579 | 1.00E-75 | AT1G06960 | 6.00E-88 | |
Table 2.
The 8 potential Cyanidioschyzon PCAPs identified
| Protein names | Heptapeptide column |
Nonapeptide column |
Annotations | Best hit in yeast and the e-value | Best hit in human and the e-value | Best hit in Arabidopsis and the e-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.3M | 0.5M | 0.3M | 0.5M | ||||||||
| CMM263C | 24 | TOP1 | TOP1 | e-155 | P11387 | e-152 | AT5G55300 | e-180 | |||
| CMM087C | 1 | SWIB/MDM2 domain containing protein | TRI1 | 1.00E-12 | F8VUB0 | 7.00E-06 | AT3G19080 | 7.00E-20 | |||
| CMT578C | 5 | Similar to methylated-DNA–protein-cysteine methyltransferase | MGT1 | 2.00E-09 | |||||||
| CMH210C | 7 | peptidyl-prolyl cis-trans isomerase activity | ESS1 | 5.00E-24 | Q13526 | 8.00E-18 | AT2G18040 | 7.00E-23 | |||
| CMS377C | 13 | 6 | Casein kinase I isoform | HRR25 | e-142 | B0QY34 | e-160 | AT4G26100 | e-156 | ||
| CMG052C | 8 | Myb-related transcription factor | BAS1 | 6.00E-13 | E9PJ96 | 6.00E-24 | AT3G18100 | 2.00E-33 | |||
| CMH135C | 4 | 2 | mRNA export | YRA1 | 2.40E-05 | E9PB61 | 8.00E-09 | AT5G59950 | 5.00E-10 | ||
| CMS144C | 1 | TBP-associated factor TAF12 | TAF12 | 1.00E-14 | Q16514 | 5.00E-24 | AT3G10070 | 8.00E-14 | |||
Potential PCAPs with functions shared in both algae
The proteins shared between C. reinhardtii and C. merolae fall into 2 functional groups, and co-purification of these proteins from both organisms further implies that they are biologically relevant PCAPs. One shared functional group contains 3 casein kinases, serine/threonine-targeting enzymes, Q84SA0 and A8IYG9 from C. reinhardtii and CMS377C from C. merolae. Q84SA0 and CMS377C show significant similarity to casein kinase I (CK1) and A8IYG9 to casein kinase II (CK2). Considering that CMS377C is most similar (1e-152) to Q84SA0 in reciprocal Blast searches, the 2 appear to be homologous. Inferred homologs of both of these algal proteins in yeast (Hrr25), human and Arabidopsis are annotated as CK1 isoforms.
The catalytic domain of CK1 lies in its N-terminus, with variable domains in the C-terminus that confer substrate specificity for protein-protein interactions or subcellular localization.21 Budding yeast contains 4 CK1 isoforms, and Hrr25 is the only one that is localized to the nucleus.21 Hrr25 is involved in transcriptional response to DNA damage through physical interactions with the transcription factor Swi6, a component of cell cycle regulatory complex SBF.22 Notably, comparable affinity column assays in yeast also recovered Hrr25 as a PCAP.20 The fact that homologs from yeast, and now both C. reinhardtii and C. merolae, all bind to phospho-CTD repeats, strongly implicates this protein as a conserved functional CTD partner. The third protein in this group, C. reinhardtii A8IYG9, is most likely the α subunit of CK2, which has been reported to phosphorylate the most C-terminal serine of the mammalian CTD,23 although its association with CTD heptapeptides has not been reported previously. Moreover, CK2 has been implicated as the main kinase that phosphorylates FCP1 in Xenopus, a CTD phosphatase that binds transcription factor IIF.24 Thus, prior evidence indicates at least indirect associations between CK2 and the CTD, and our results suggest that CK2 could serve as a CTD-dependent kinase in Chlamydomonas.
The second shared functional group includes A8I1B8 and A8HME6 from C. reinhardtii and CMH135C from C. merolae. All contain RNA recognition motifs and appear to be related to mRNA export based on similarity scores in reciprocal Blast searches that recovered putative human and Arabidopsis homologs. The human homolog of CMH135C is ALY/REF, an mRNA export factor that shuttles between the nucleus and cytoplasm. Previous studies showed that metazoan ALY/REF couples pre-mRNA splicing and mRNA export by associating with spliced mRNPs, and also that ALY/REF co-localizes with splicing factors.25 The apparent yeast homolog of both CMH135C and ALY/REF is Yral, also an mRNA export factor, and it is perhaps the more likely functional model given the relative paucity of introns in both yeast and red algae. Interestingly, Yra1 is another of the proteins that was recovered from comparable binding experiments with bi-phosphorylated heptapeptides in yeast.20 Further, experiments substituting negatively charged glutamates for phospho-serines indicated the interaction between Yra1 and phospho-CTD is specific rather than a binding artifact.26 In addition, structural analysis revealed that both the RNA binding and CTD interaction domains of Yra1 are located in its N-terminus, and partial N-terminal truncations resulted in a severe decrease of Yra1 recruitment to elongating genes.26 Mutations resulting in deficient RNA binding or CTD interactions both negatively impact mRNA export,26 indicating that Yra1 is likely recruited to transcriptionally elongating genes by the phospho-CTD.
The closest match from yeast to both Chlamydomonas sequences A8HME6 and A8I1B8 is Pab1, a poly(A) binding protein that also functions in mRNA export; however, based on similarity scores, Pab1 is more closely related to A8HME6 (they are reciprocal best hits). Although A8I1B8 does not share significant similarity with Yra1 from yeast (E-value = 0.078), it is the reciprocal match to CMH135C, the Yra1 homolog (see above) from Cyanidioschyzon (3e-04). A8HME6 is not only identified as the homolog of Pab1 from yeast, but also from human and Arabidopsis. Previous studies have shown that Pab1 binds the poly(A) tail of pre-mRNA and could be involved in final trimming of the tail, mRNA release from transcription sites, and its transport to the cytoplasm.27 To our knowledge, Pab1 has never been shown to interact with the CTD; however, given the confirmed relationship between other mRNA export factors and the CTD, for example, Npl3 in yeast,28 the proteins in this functional class from Chlamydomonas and Cyanidioschyzon are reasonable candidates for further experimental validation as bona fide PCAPs. Taken together, our results suggest that the coupling of mRNA processing and export to the phospho-CTD, previously characterized in animals and yeast, also is conserved in both red and green algae.
Potential PCAPs found only in C. reinhardtii
Two of the proteins isolated only from Chlamydomonas appear to be related to pre-mRNA splicing. A8J3U2 and A8HRV5 both are most similar to components of the U2 snRNP complex, which combines with pre-mRNAs and other snRNPs to form spliceosomes. The homologs of A8J3U2 in yeast, human and Arabidopsis are U2A components, and those of A8HRV5 are U2B components. Previous studies have shown a strong functional link between the CTD and pre-mRNA splicing, including several splicing factors that physically interact with the phospho-CTD; for example, Prp40 in yeast, a component of the U1 snRNP.29 Moreover, a recent study reported that the auxiliary factor 65-kDa subunit (U2AF65) of the U2 snRNP and Prp19 complex is recruited by the CTD to promote splicing activation, and that U2AF65 interacts directly with the CTD.30 Although there is no evidence for direct interactions between the CTD and U2 snRNP complex components, considering the importance of the CTD in pre-mRNA splicing, along with established CTD/spliceosome interactions, broader or even slightly different direct interactions between the CTD and spliceosome components is reasonable.
Another putative PCAP identified in C. reinhardtii is A8IDW3, which contains both SANT and MPN domains. The human homolog of A8IDW3 is histone H2A deubiquitinase MYSM1, a chromatin regulator. Domain analysis showed that A8IDW3 shares the SANT and MPN domains with its human counterpart and, therefore, is likely to function as a histone H2A deubiquitinase in C. reinhardtii. Human histone H2A deubiquitinase regulates transcriptional activation and elongation of many genes (hormone related genes, for example) by deubiquitination of H2A, which enhances the dissociation of linker histone H1 from the nucleosome.31 Previous studies reported that several proteins related to chromatin modifications are associated with the phospho-CTD, including histone methyltransferases Set1 and Set2.3 Such interactions are consistent with our recovery of a green algal H2A deubiquitinase as a putative PCAP; if demonstrated in vivo, this would identify a new function of the CTD in chromatin modification.
Potential PCAPs only in C. merolae
In addition to the 2 proteins discussed above (CMS377C bound the nonapeptide and CMH135C the heptapeptide columns, respectively), there are another 6 likely PCAPs identified only from C. merolae; 5 (CMH210C, CMT578C, CMM263C, CMM087C and CMG052C) bound to nonapeptides and one (CMS144C) to heptapeptides.
CMH210C is a nonapeptide-associated PCAP that is likely to be a peptidyl-prolyl cis/trans isomerase (PPIase), based on its strong similarity to homologs from yeast, human and Arabidopsis. The yeast homolog Ess1 and the human homolog Pin1 both have been confirmed experimentally to interact with the phospho-CTD; their putative function is to help Ssu72 dephosphorylate S5 on CTD repeats by making the S5P-P6 bond take on a cis conformation.32 Ess1 and Pin1 interact with the CTD through their WW domains.3 Although CMH210C does not have a recognizable WW domain, it does contain a SurA domain with predicted PPIase function as in yeast and human. Instead of a WW domain, however, CMH210C has a FHA domain at its N-terminus, which also is a phospho-peptide (mostly phospho-threonine) interacting domain present in many regulatory proteins.33 CMH210C was eluted only from the nonapeptide column, suggesting this protein does not interact strongly with phosphorylated heptapeptides in the CTD of C. merolae. This certainly could be explained by the presence of a FHA instead of a WW domain; in both yeast and human homologs, the latter interacts only with phosphorylated heptapeptides.
CMT578C, another nonapeptide-associated PCAP, is homologous with Mgt1 from yeast. No reciprocal homolog was found in human, but the nearest match (e-value exceeded our threshold) was MGMT, homologous to yeast Mgt1. Although the similarity between CMT587C and MGMT is not significant based on our a priori cutoff, the significant relationships between Mgt1 and MGMT, and between Mgt1 and CMT587C, make it likely that all 3 are homologs. Both Mgt1 and MGMT are 6-O-methylguanine-DNA methyltransferases that use cysteine residues to interact with alkyl groups, which are transferred from toxic lesions of alkylated guanine in DNA.34,35 If CMT578C has the same function in C. merolae, it is the first time this methyltransferase has been implicated as interacting with the RNAP II CTD.
The nonapeptide-associated PCAP CMM263C is likely to be a Topoisomerase I, based on its strong similarity to yeast, human and Arabidopsis Top I genes. During transcription, Top I relaxes superhelical stress in unwinding DNA. Early analyses indicated that the N-terminal domain of Drosophila Top I could associate with RNA polymerase II,34 and later work revealed that both human and yeast Top I physically bind the phospho-CTD.20 A more recent study demonstrated that both Drosophila and human Top I use the proximal half of the N-terminal domain for this interaction.36 Therefore, our identification of Top I as a PCAP in C. merolae is consistent with established Top I interactions with the phospho-CTD.
Another nonapeptide-associated protein, CMM087C, contains a SWIB/MDM2 domain found in both SWI/SNF complex B and in MDM2, a regulator of the p53 tumor suppressor gene. SWI/SNF components, first characterized in chromatin remodeling complexes in yeast,37 are widely conserved in eukaryotes. Cumulative studies indicated that they remove nucleosome blocks on interactions between DNA and regulatory proteins like transcription factors.38 In doing so, SWI/SNF complexes regulate many biological processes, including RNAP II transcription initiation, elongation and associated DNA repair.39 To our knowledge, no direct interaction between SWI/SNF complex subunits and phospho-CTD has been established. Nevertheless, given the phospho-CTD’s apparent recruitment of histone acetyltransferase (HAT) complexes and deacetylase complexes (HDACs) that remodel nucleosomes around the elongating RNAP II,40 it is reasonable that SWI/SNF components, which also accompany RNAP II transcription factory, could have evolved direct CTD interactions in some organisms. Thus, our recovery of a SWI/SNF-like subunit acting as a PCAP in C. merolae could be the first evidence of a more broadly important CTD protein interaction.
Another nonapeptide-associated protein from Cyanidioschyzon, CMG052C, is inferred to be homologous with yeast Bas1, a MYB-related transcription factor required for transcriptional regulation of a number of genes related to the biosynthesis of purine, pyrimidine and several amino acids; for example, the ADE3 gene encoding the purine and glycine biosynthetic enzyme tetrahydrofolate synthase.41 The most similar sequence to CMG052C in Arabidopsis is an R2R3 transcription factor, which belongs to a MYB-protein subfamily in plants. Cumulative research on plant R2R3-type MYB factors suggests they are involved in controlling development, determination of cell fate, and transcriptional activation.42 To date, there is no experimental evidence for a Bas1/CTD interaction in yeast, or any reports of a CTD association with MYB-related transcription factor in plants and animals. Moreover, if the CTD in C. merolae is hyperphosphorylated during transcript elongation rather than initiation, as is true in all CTD model organisms,17 then a relevant biological interaction between an MYB factor and the phospho-CTD is not immediately apparent.
The same can be said for a potential PCAP, CMS144C, which bound to phospho-heptapeptides. All yeast, human and Arabidopsis homologs are identified as TFIID subunit 12 (Taf12), a TATA-binding protein associated factor. Previous studies in yeast have shown an association between the CTD and the TFIID complex43,44; however, the specific component(s) of TFIID that is/are the target(s) for this interaction remain(s) unclear. Interestingly, a recent study revealed that another TFIID subunit, Taf15, can interact with the unphosphorylated CTD in vitro through its polymerized Low Complexity (LC) domain, and that this interaction is deterred by phosphorylation of the CTD.45 This suggests that recruitment of RNAP II during transcription initiation is facilitated by interactions between the unphosphorylated CTD and Taf15, and that its release from the transcription initiation complex is promoted by CTD phosphorylation.45 No clear homolog of Taf15 is present in yeast, however, and we likewise found no Taf15 homolog in C. merolae through extensive Blast searches using human Taf15 as the query. Taf12 does not contain a LC domain, suggesting it might not interact with the unphosphorylated CTD in Cyanidioschyzon. Thus, if the interaction of Taf12 with the phospho-CTD in C. merolae is biologically relevant, it suggests a more complicated relationship between the TFIID complex and the CTD, at least in red algae.
Other proteins that bound phospho-CTD peptides
In addition to the 15 likely PCAPs we singled out for in-depth comparative analyses, many other proteins bound to our phospho-CTD affinity columns. A number have putative functions that are relevant to the CTD’s established roles in transcription and mRNA processing, while many others have no recognizable homologs that allow a prediction of function or cellular localization. Like the 8 red algal PCAPs discussed above, many of these proteins from Cyanidioschyzon bind to either the heptapeptide or nonapeptide column, but not to both (Table S2). Thus, it is reasonable that a number of other proteins we isolated are biological relevant CTD partners.
Interestingly, most of the proteins that can be annotated do not function in the nucleus based on inferred yeast, human and Arabidopsis homologs. The largest fractions are ribosomal proteins, consistent with prior results from yeast where numerous ribosomal proteins bound to bi-phosphorylated CTD affinity columns.20 This is not surprising, given that these proteins generally interact with uniform, negative phosphate charges on rRNAs within the ribosome. Although individual ribosomal proteins are imported into the nucleus, where major ribosome components are assembled before transport to the cytoplasm, the physical separation between the nucleolus (site of ribosome synthesis) and RNAP II transcription factories suggests that their direct contact with the phospho-CTD (present only where RNAP is actively elongating mRNA transcripts) as individual proteins is unlikely.
We also found a similar result using the E. coli proteome, an additional negative control for assessing non-specific binding. Given that the CTD is present only on RNAP II in eukaryotes, there has been no selective pressure on E. coli proteins to avoid binding inappropriately to negative charges on a phospho-CTD. Sixty-five percent E. coli proteins that bound phospho-CTD peptides were ribosomal proteins (Table S3). Therefore, it appears that their recovery represents the major issue with non-specific protein binding to phospho-CTD peptides.
Despite potential binding artifacts, it has been demonstrated that the methodologies employed here are effective in recovering numerous bona fide PCAPs from both yeast and human cells.19,20,47,47 We believe this is because inside the nucleus, where elaborate and intricate regulation of transcription and mRNA processing is carefully orchestrated, there must be strong selection for more highly specific interactions between the phospho-CTD and its binding partners. In other words, transcription-related nuclear proteins are likely to be under strong selection to avoid simple opposite surface charge attractions, whereas cytoplasmic proteins that do not encounter the CTD will have experienced weaker or no selection to avoid non-specific interactions. Because our results are comparable to those reported previously, we thought it important to test this hypothesis by examining the proportions of nuclear and cytoplasmic proteins that bound our CTD affinity columns.
Of the 116 C. reinhardtii proteins recovered with identifiable homologs, 26 (22.4%) are putatively related to processes occurring in the nucleus; for C. merolae, 23 of the 113 (20.4%) are nucleus-related. Because of the large numbers of genes without known homologs in their genomes, clear ratios of nuclear to total proteins are difficult to estimate for these 2 algal species. In more thoroughly characterized budding yeast and Arabidopsis genomes, however, the ratios are 35.2% (2070/5887) and 32.4% (9356/28912) respectively, according to GO annotations. We therefore set a conservative estimate of 30% as the fraction of the nuclear localized proteins for both algae, and ran binomial tests to determine whether, as predicted, there is evidence for reduced non-specific binding of artificial phospho-CTD peptides for nuclear proteins. For the purposes of this analysis, we used the highly unlikely and conservative assumption that none of the nuclear proteins isolated were true PCAPs, and that all proteins had an equal probability of binding the phospho-CTD. For C. reinhardtii, the 26 (22.4%) nuclear proteins recovered are significantly fewer than 30% (P = 0.044, one-tailed), which also was true for the 23 (20.4%) C. merolae proteins (P = 0.014, one-tailed). Thus, even assuming that no legitimate PCAPs were recovered from either alga, our results are consistent with natural selection having diminished non-specific CTD-protein interactions within the nucleus. Clearly, if even some of the nuclear proteins isolated are legitimate PCAPs, the differences are that much greater in non-specific binding between nuclear and cytoplasmic proteins. The support we find for selection favoring CTD binding specificity in nuclear proteins also offers further validation of PCAPs inferred in previous studies using comparable methods.
Discussion
Although we are unaware of experimental investigations showing the CTD is phosphorylated at S2 and S5 in red or green algae, our phylogenetic analyses revealed that, except for the absence of CDK8 from the 2 unicellular red algae Cyanidioschyzon and Galdieria, members of all CDK subfamilies are conserved in both the red and green lineages (Fig. S1). Therefore, the presence of homologs of the CDK7 and CDK9/12/13 subfamilies, those mainly responsible for S2 and S5 CTD phosphorylations in human and yeast,18 suggests that this phosphorylation pattern also is conserved in Chlamydomonas and Cyanidioschyzon, and is predicted to be present during transcription elongation. Thus, using S2P and S5P CTD peptides as bait for PCAPs appears reasonable for both algae.
Our proteomics analyses provide the first experimental evidence of CTD-protein interactions in red and green algae. Although the potential for nonspecific binding to artificial CTD repeats dictates caution when interpreting results from this sort of assay, a number of factors suggest we have identified viable PCAP candidates in both algal species. First, our data are consistent with natural selection favoring reduced non-specific CTD binding by proteins that function in the nucleus, where they could encounter the CTD by chance. We think this result is important, in itself, given that similar non-specific binding has been reported in prior studies, and is always a concern in any assay of protein binding to a highly charged peptide like the CTD.
Second, a number of the homologs of known PCAPs were recovered from both algal taxa, which is unlikely to be coincidental given the small fractions of the proteomes involved. Third, a number of the proteins we recovered are inferred homologs of yeast and human proteins that have been shown to bind comparable phospho-CTD affinity columns for those organisms,19,20 and for which there is additional corroborating evidence of a CTD interaction. Perhaps more compelling, however, is the level of differential binding of nuclear proteins to heptapeptides and nonapeptides from Cyanidioschyzon. Of the 23 proteins with nuclear annotations, only 5 bound to both peptide affinity columns. In contrast, over half (48 of 90) cytoplasmic proteins bound to both versions of the phospho-CTD. This demonstrates a significant (P = 0.01, binomial test, one tailed) tendency for nuclear proteins to bind specifically to one or the other type of CTD repeats present in Cyanidioschyzon, as would be expected if CTD-protein interactions are spacio-temporally arranged as in model systems.12
Finally, despite focusing on only the 15 proteins for which CTD interactions can be argued from prior research, differences in PCAP functional categories recovered relate to a clear and important biological difference between the 2 algae; PCAPs associated with pre-mRNA splicing were recovered from Chlamydomonas, but not from Cyanidioschyzon. Only 27 introns (in 26 genes) have been identified in C. merolae and several spliceosome-related proteins appear to be missing from the complete genome.48 In Chlamydomonas, by contrast, over 90% of protein-encoding genes contain introns, with 8.3 exons per gene on average.49 Thus, it is unlikely that spliceosomal proteins that could interact with the CTD are expressed as highly in Cyanidioschyzon as in Chlamydomonas. Comparable methods applied in S. cerevisiae recovered splicing-related proteins,20 despite the relatively paucity of yeast introns50 compared to Chlamydomonas, suggesting the possibility that fewer or no splicing factors interact with the CTD in C. merolae. Given the likelihood that splicing is an ancient CTD function,13 it will be interesting to determine whether the CTD remains involved in co-transcriptional splicing in other eukaryotes that, like red algae, are thought to have lost most of their ancestral introns.51
Such differences highlight the importance of further experimental investigations of CTD function in red algae and other diverse eukaryotes. When considering our results, it is important to note that red algae have been evolving independently from other eukaryotes for well over a billion years,52 and relatively few gene functions have been determined experimentally. Moreover, patterns of CTD evolution among eukaryotic taxa are far more diverse than was suggested by early comparative studies.13 Thus, our recovery of 2 different proteins implicated in transcription initiation among our potential PCAPs, could be a first suggestion that patterns of CTD hypo- and hyper-phosphorylation in at least some red algae differ from those established in model systems.17 Interestingly, we found neither CDK8 (Fig. S1), nor most components of mediator, in the Cyanidioschyzon genome; this is in line with potential differences in CTD phosphorylation during transcription initiation.
Given the great evolutionary distances between major eukaryotic lineages, the single CTD phosphorylation pattern we examined, the small fractions of the algal proteomes recovered and even smaller fractions that have identifiable homologs, it is interesting that we uncovered as many shared putative homologs and functional categories as we did. This suggests there could be functional conservation of a number of core CTD-protein interactions across broad eukaryotic diversity. In addition to the cases we highlight, other nuclear proteins were recovered from both species (Tables S1 and S2). Some have inferred functions that are biologically relevant to the CTD’s established roles, whereas others have no identifiable homologs to provide predictions of function and cellular localization. Many could prove to be interesting PCAP candidates as validated or at least predicted functions become available.
In conclusion, our study provides the first experimental evidence of CTD-protein interactions in simple, undifferentiated unicellular red and green algae. They permit an initial comparison of potential PCAPs with those recovered in comparable previous investigations in yeast and mammals. The PCAPs shared among all these groups indicate that a number of CTD-protein interactions are widely conserved, at least among eukaryotic groups that ultimately evolved multicellularity. In contrast, differential PCAP binding to heptapeptides and nonapeptides in the red alga further highlights the importance of lineage-specific modifications, which have punctuated CTD evolution during the diversification of major eukaryotic phyla.13 Indeed, the large number of unclassified proteins that bind specifically to nonapeptide repeats from Cyanidioschyzon (Table S2) suggests the presence of a variety of new, taxon-specific CTD-protein interactions. This variation likely reflects differences in how CTD-protein interactions have elaborated and diversified, providing what Zachary Burton53 has called the “New Testament” in the Genesis of organismal complexity through elaborations of CTD-based mechanisms for controlling gene expression. Our investigation provides a first glimpse into the chapters of that book on red and green algae.
Materials and Methods
Cell culture and lysis
C. reinhardtii (CC-503 cw92 mt+) was cultured in TAP medium54 at room temperature and 24 hrs light, and C. merolae (N-1804) was cultured in Allen Culture medium55 at 42°C and 24 hrs light. Escherichia coli (DH5α) was cultured in LB medium at 37°C overnight. Harvested algal and E. coli cells were suspended in cold BY-AS400 buffer (25 mM HEPES, pH 7.6; 1 mM EDTA; 1 mM PMSF; 400 mM AmSO4; protease inhibitor cocktail for plants 1:100 dilution) using 2-3 ml buffer per gram of cells. A French press (12,000 psi) was used twice to break suspended cells and obtain crude protein extracts. The crude extracts were centrifuged in a SS34 rotor at 20,000ċg for 45 minutes at 4°C, and the supernatant was collected. A flowchart of the protein purification methodology is shown in Fig. 2.
Figure 2.
PCAP purification process. The PCAP purification process is shown step by step as indicated by the direction of the arrows. The elution from each affinity column was subjected to SDS-PAGE followed by staining with Coomassie blue. The gels run on elutions are shown for each affinity column. M represents molecular weight (KDa) marker, 0.3 M and 0.5 M indicates elution with those concentrations of NaCl in BH buffer. The putative PCAPs from each elution highlighted in our results section are shown under the respective gels.
Ammonium sulfate precipitation
The detergent NP-40 was added to the SS34-supernatant to a final concentration of 1%, and (NH4)2SO4 was gradually added to a final concentration of 50% (~313g/l) while stirring at 4°C. The ammonium sulfate suspension was then centrifuged again in a SS34 rotor at 30,000ċg for 45 minutes at 4°C. The (NH4)2SO4 pellet was collected and suspended with enough cold BH buffer (25 mM HEPES, pH 7.6; 1 mM EDTA; 1 mM DTT; 1 mM PMSF; 8% glycerol) to bring conductivity in the suspension approximately equal to 0.15 M NaCl.
Ion exchange chromatography
In order to increase concentration of proteins with positive surface charges that could bind CTD phospho-serines, we employed 2 steps of ion-exchange chromatography modified from the protocol of Greenleaf and colleagues.20 This both enriched potential PCAPs, and removed remaining cell debris and undesired proteins (e.g., chromoproteins) that were not eliminated by initial centrifugations.
The BH-suspension was passed through a ~21 ml (1.5 cm ċ 12 cm) anion exchange column (Q Sepharose Fast Flow, GE Healthcare) at a flow rate of ~1.4 ml/min, and the column was washed with 4 column volumes of BH buffer + 0.15 M NaCl. The flow through from the column was collected and loaded on a same size cation exchange column (SP Sepharose Fast Flow, GE Healthcare) with the same flow rate. The column also was washed with 4 column volumes of BH buffer + 0.15 M NaCl, and eluted with BH buffer + 1 M NaCl. The elution from cation exchange column was collected and desalted by dilution and ultrafiltration.
Affinity chromatography
One ml CTD affinity columns were constructed using NeutrAvidin Agarose Resin (Thermo Scientific) bound to biotin-labeled, synthetic CTD tri-heptapeptides (Biotin-YSpPTSpPAYSpPTSpPAYSpPTSpPA) or tri-nonapeptides (Biotin-YSpPSSpPNVAYSpPSSpPNVAYSpPSSpPNVA), which were constructed at Eton Bioscience Inc., each containing 3 repeats phosphorylated at all S2 and S5 residues. Because these peptides are very similar in sequence, and in phosphorylation patterns, each represents an excellent negative control for non-specific binding to the other. That is, if a protein cannot bind to one of these very similar phospho-peptides, it is strong evidence of a specific affinity for the other. A 1 ml control column also was made containing only the NeutrAvidin Agarose Resin. We chose this phosphorylation pattern to allow direct comparison to PCAPs isolated previously from the far more thoroughly characterized Saccharomyces cerevisiae genome.20
We added a PhosSTOP phosphatase inhibitor cocktail tablet to each cation-elution pool (~4 mg of protein) to avoid de-phosphorylation of the CTD peptides and then passed the pool through the appropriate heptapeptide or nonapeptide affinity column. All columns were washed with 16 column volumes of BH + 0.1 M NaCl. Bound proteins were eluted sequentially with increasing salt concentrations (1ml BH buffer + 0.3, 0.5, 1.0 M NaCl), with each elution step collected in 4 250-μl aliquots. To assay the presence and quality of eluted proteins, 25 μl of each aliquot was examined using SDS-PAGE (4-20% Tris HCl gradient gels from Bio-Rad) stained with Coomassie blue (gels not shown). The control column (resin with no CTD peptides) followed the same procedure as above, and showed no indication of protein binding (gels not shown). The middle 2 250-µl aliquots from each elution concentration were pooled, desalted and concentrated. 10 μg of proteins from each elution pool were subjected to SDS-PAGE, followed by Coomassie blue stain (gels shown in Fig. 2); the rest were submitted to Duke University Proteomics Center for mass spectrometry (LC/ESI/MS/MS) identification.
Protein annotations
Because functions assigned to genes in both the C. reinhardtii and C. merolae genome are based primarily upon sequence similarity to genes from more well-developed models, we relied on annotated functions of apparent homologs from yeast, human and Arabidopsis to identify potential CTD-binding partners in both algae. Homologs were identified through reciprocal Blast searches between the C. reinhardtii or C. merolae and each of the 3 reference genomes (E-value cutoff of 1e-04). Reciprocal best hits were considered to be homologous sequences. Protein domain analyses were based on the National Center for Biotechnology Information (NCBI) structure online service http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
Phylogenetic analyses of CDKs
We performed phylogenetic analyses of putative cyclin-dependent kinases (CDKs) from both algae to verify that appropriate homologs are present to expect the pattern of CTD-phosphorylation analyzed in this study. According to previous investigations, human CDKs can be divided into well-defined subfamilies.56,57 Therefore, we applied reciprocal Blast searches to identify the homologs of each CDK subfamily from yeast, Arabidopsis, Chlamydomonas, Cyanidioschyzon and 2 additional complete red algal genomes (Chondrus crispus and Galdieria sulphuraria). For each organism, the putative CDK homolog with the highest similarity score to each subfamily was chosen for phylogenetic analyses together with the representative human CDKs. A multiple sequence alignment was performed in MUSCLE58 (online service: http://www.ebi.ac.uk/Tools/msa/muscle/) and Gblocks 0.91b59 (http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type =gblocks) was used to select the conserved blocks appropriate for tree-building. Phylogenetic analysis were performed in MrBayes using a WAG + invgamma model60 as determined through maximum-likelihood model estimation in MEGA 5.2.2.61 Relative support for the presence of CTK1/CDK9 homologs was inferred from Bayesian posterior probabilities estimated from all trees (106 generations) sampled after the average standard deviation of split frequencies had converged on a value < 0.01.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank J.W Thompson, A. Mosley and L. Dubois from the Duke University Proteomics Facility for technical support and help with analysis of MS/MS protein identification.
Funding
This study was supported by National Science Foundation Grant 0849586.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 1985; 42:599-610; PMID:3896517; http://dx.doi.org/ 10.1016/0092-8674(85)90117-5 [DOI] [PubMed] [Google Scholar]
- 2. Corden JL, Cadena DL, Ahearn JM, Dahmus ME. A Unique Structure at the Carboxyl Terminus of the Largest Subunit of Eukaryotic Rna Polymerase-Ii. Proc Natl Acad Sci U S A 1985; 82:7934-8; PMID:2999785; http://dx.doi.org/ 10.1073/pnas.82.23.7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corden JL. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem Rev 2013; 113:8423-55; PMID:24040939; http://dx.doi.org/ 10.1021/cr400158h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 1995; 140:1223-33; PMID:7498765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 1987; 50:909-15; PMID:3304659; http://dx.doi.org/ 10.1016/0092-8674(87)90517-4 [DOI] [PubMed] [Google Scholar]
- 6. Bartolomei MS, Halden NF, Cullen CR, Corden JL. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol 1988; 8:330-9; PMID:3275873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu P, Greenleaf AL, Stiller JW. The essential sequence elements required for RNAP II carboxyl-terminal domain function in yeast and their evolutionary conservation. Mol Biol Evol 2008; 25:719-27; PMID:18209193; http://dx.doi.org/ 10.1093/molbev/msn017 [DOI] [PubMed] [Google Scholar]
- 8. Schwer B, Shuman S. Deciphering the RNA Polymerase II CTD Code in Fission Yeast. Mol Cell 2011; 43:311-8; PMID:21684186; http://dx.doi.org/ 10.1016/j.molcel.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stiller JW, Cook MS. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot Cell 2004; 3:735-40; PMID:15189994; http://dx.doi.org/ 10.1128/EC.3.3.735-740.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwer B, Sanchez AM, Shuman S. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci U S A 2012; 109:18024-9; PMID:23071310; http://dx.doi.org/ 10.1073/pnas.1208995109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol 2010; 27:2628-41; PMID:20558594; http://dx.doi.org/ 10.1093/molbev/msq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA; 2013; 4:1-16. [DOI] [PubMed] [Google Scholar]
- 13. Yang C, Stiller JW. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci U S A 2014; 111:5920-5; PMID:24711388; http://dx.doi.org/ 10.1073/pnas.1323616111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das A, Bellofatto V. The Non-Canonical CTD of RNAP-II Is Essential for Productive RNA Synthesis in Trypanosoma brucei. PLoS One 2009; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 2013; 113:8456-90; PMID:23952966; http://dx.doi.org/ 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- 16. Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging Views on the CTD Code. Genet Res Int 2012; 2012:347214; PMID:22567385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet 2008; 24:280-8; PMID:18457900; http://dx.doi.org/ 10.1016/j.tig.2008.03.008 [DOI] [PubMed] [Google Scholar]
- 18. Bartkowiak B, Greenleaf AL. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription 2011; 2:115-9; PMID:21826281; http://dx.doi.org/ 10.4161/trns.2.3.15004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics 2002; 1:598-610; PMID:12376575; http://dx.doi.org/ 10.1074/mcp.M200029-MCP200 [DOI] [PubMed] [Google Scholar]
- 20. Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 2004; 43:15702-19; PMID:15595826; http://dx.doi.org/ 10.1021/bi048364h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee JY. Versatile casein kinase 1: multiple locations and functions. Plant Signal Behav 2009; 4:652-4; PMID:19820321; http://dx.doi.org/ 10.4161/psb.4.7.8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1997; 94:581-6; PMID:9012827; http://dx.doi.org/ 10.1073/pnas.94.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Payne JM, Laybourn PJ, Dahmus ME. The Transition of Rna Polymerase-Ii from Initiation to Elongation Is Associated with Phosphorylation of the Carboxyl-Terminal Domain of Subunit-Iia. J Biol Chem 1989; 264:19621-9; PMID:2584185 [PubMed] [Google Scholar]
- 24. Palancade B, Dubois MF, Bensaude O. FCP1 phosphorylation by casein kinase 2 enhances binding to TFIIF and RNA polymerase II carboxyl-terminal domain phosphatase activity. J Biol Chem 2002; 277:36061-7; PMID:12138108; http://dx.doi.org/ 10.1074/jbc.M205192200 [DOI] [PubMed] [Google Scholar]
- 25. Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 2000; 407:401-5; PMID:11014198; http://dx.doi.org/ 10.1038/35030160 [DOI] [PubMed] [Google Scholar]
- 26. MacKellar AL, Greenleaf AL. Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. J Biol Chem 2011; 286:36385-95; PMID:21856751; http://dx.doi.org/ 10.1074/jbc.M111.268144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 2003; 4:223; PMID:12844354; http://dx.doi.org/ 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One 2008; 3:e3273; PMID:18818768; http://dx.doi.org/ 10.1371/journal.pone.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 2000; 275:39935-43; PMID:10978320; http://dx.doi.org/ 10.1074/jbc.M004118200 [DOI] [PubMed] [Google Scholar]
- 30. David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev 2011; 25:972-83; PMID:21536736; http://dx.doi.org/ 10.1101/gad.2038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 2007; 27:609-21; PMID:17707232; http://dx.doi.org/ 10.1016/j.molcel.2007.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 2011; 286:5717-26; PMID:21159777; http://dx.doi.org/ 10.1074/jbc.M110.197129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Durocher D, Jackson SP. The FHA domain. FEBS Lett 2002; 513:58-66; PMID:11911881; http://dx.doi.org/ 10.1016/S0014-5793(01)03294-X [DOI] [PubMed] [Google Scholar]
- 34. Shaiu WL, Hsieh TS. Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I. Mol Cell Biol 1998; 18:4358-67; PMID:9632819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007; 6:429-42; PMID:17112791 [DOI] [PubMed] [Google Scholar]
- 36. Wu J, Phatnani HP, Hsieh TS, Greenleaf AL. The phosphoCTD-interacting domain of Topoisomerase I. Biochem Biophys Res Commun 2010; 397:117-9; PMID:20493173; http://dx.doi.org/ 10.1016/j.bbrc.2010.05.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 1992; 8:387-91; PMID:1332230; http://dx.doi.org/ 10.1016/0168-9525(92)90300-S [DOI] [PubMed] [Google Scholar]
- 38. Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 2007; 27:6987-95; PMID:17709398; http://dx.doi.org/ 10.1128/MCB.00717-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem 2012; 287:30897-905; PMID:22952240; http://dx.doi.org/ 10.1074/jbc.R111.309302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spain MM, Govind CK. A role for phosphorylated Pol II CTD in modulating transcription coupled histone dynamics. Transcription 2011; 2:78-81; PMID:21468233; http://dx.doi.org/ 10.4161/trns.2.2.14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joo YJ, Kim JA, Baek JH, Seong KM, Han KD, Song JM, Choi JY, Kim J. Cooperative regulation of ADE3 transcription by Gcn4p and Bas1p in Saccharomyces cerevisiae. Eukaryot Cell 2009; 8:1268-77; PMID:19525417; http://dx.doi.org/ 10.1128/EC.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 2001; 4:447-56; PMID:11597504; http://dx.doi.org/ 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 43. Conaway RC, Bradsher JN, Conaway JW. Mechanism of Assembly of the Rna Polymerase-Ii Preinitiation Complex - Evidence for a Functional Interaction between the Carboxyl-Terminal Domain of the Largest Subunit of Rna Polymerase-Ii and a High Molecular Mass Form of the Tata Factor. J Biol Chem 1992; 267:8464-7; PMID:1569096 [PubMed] [Google Scholar]
- 44. Koleske AJ, Buratowski S, Nonet M, Young RA. A Novel Transcription Factor Reveals a Functional Link between the Rna Polymerase-Ii Ctd and Tfiid. Cell 1992; 69:883-94; PMID:1591782; http://dx.doi.org/ 10.1016/0092-8674(92)90298-Q [DOI] [PubMed] [Google Scholar]
- 45. Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013; 155:1049-60; PMID:24267890; http://dx.doi.org/ 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3'-End formation. J Biol Chem 1999; 274:31583-7; PMID:10531363; http://dx.doi.org/ 10.1074/jbc.274.44.31583 [DOI] [PubMed] [Google Scholar]
- 47. Carty SM, Goldstrohm AC, Sune C, Garcia-Blanco MA, Greenleaf AL. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci U S A 2000; 97:9015-20; PMID:10908677; http://dx.doi.org/ 10.1073/pnas.160266597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Yoshida Y, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2004; 428:653-7; PMID:15071595; http://dx.doi.org/ 10.1038/nature02398 [DOI] [PubMed] [Google Scholar]
- 49. Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007; 318:245-50; PMID:17932292; http://dx.doi.org/ 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spingola M, Grate L, Haussler D, Ares M. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. Rna-a Publication of the Rna Society 1999; 5:221-34; http://dx.doi.org/ 10.1017/S1355838299981682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Csuros M, Rogozin IB, Koonin EV. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol 2011; 7:e1002150; PMID:21935348; http://dx.doi.org/ 10.1371/journal.pcbi.1002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 2000; 26:386-404; http://dx.doi.org/ 10.1666/0094-8373(2000)026%3c0386:BPNGNS%3e2.0.CO;2 [DOI] [Google Scholar]
- 53. Burton ZF. The Old and New Testaments of gene regulation: Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD. Transcription 2014; 5:e28764. http://dx.doi.org/ 10.4161/trns.28674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A 1965; 54:1665-9; PMID:4379719; http://dx.doi.org/ 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol 2004; 45:667-71; PMID:15215501; http://dx.doi.org/ 10.1093/pcp/pch087 [DOI] [PubMed] [Google Scholar]
- 56. Cao L, Chen F, Yang X, Xu W, Xie J, Yu L. Phylogenetic analysis of CDK and cyclin proteins in premetazoan lineages. BMC Evol Biol 2014; 14:10; PMID:24433236; http://dx.doi.org/ 10.1186/1471-2148-14-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo Z, Stiller JW. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 2004; 5:69; PMID:15380029; http://dx.doi.org/ 10.1186/1471-2164-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-7; PMID:15034147; http://dx.doi.org/ 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-52; PMID:10742046; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 60. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001; 17:754-5; PMID:11524383; http://dx.doi.org/ 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 61. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-9; PMID:21546353; http://dx.doi.org/ 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.