Abstract
In mature gametes and during the oocyte-to-embryo transition, transcription is generally silenced and gene expression is post-transcriptionally regulated. However, we recently discovered that major transcription can occur immediately after fertilization, prior to pronuclear fusion, and in the first cell division of the oocyte-to-embryo transition in the nematode Ascaris suum. We postulate that the balance between transcriptional and post-transcriptional regulation during the oocyte-to-embryo transition may largely be determined by cell cycle length and thus the time available for the genome to be transcribed.
Keywords: early development, maternal deposition, oocyte-to-embryo transition, post-transcriptional regulation, zygotic transcription
Maternal Deposition and Post-transcriptional Regulation During Development
During late gametogenesis, the oocyte-to-embryo transition, and early embryogenesis, transcription is thought to be largely quiescent, yet differential gene expression is thought to be necessary for these processes. How then could development proceed without transcription? During animal oogenesis, significant transcription occurs in the diplotene stage of meiosis before the genome is silenced, and large amounts of RNAs and proteins are produced and accumulate in the mature oocyte. A variety of adaptations in animals ensure that the mature oocyte has the necessary RNAs, proteins and nutritional components to develop and navigate through the oocyte-to-embryo transition and early embryogenesis, before zygotic transcription is re-activated for subsequent development. For example, lampbrush chromosomes are often formed to allow massive transcription during meiosis; excess germ cells in C. elegans1 or nurse cells in Drosophila2 contribute RNAs and proteins to the maturing oocyte; and rRNA genes are amplified up to ∼1,000-fold in Xenopus3 to enable massive rRNA transcription and accumulation.
The deposited RNAs and proteins are differentially and coordinately used to drive developmental processes. This is mediated by a variety of post-transcriptional regulatory mechanisms including translational repression and activation, mRNA clearance, post-translational modifications, and protein degradation.4,5 Translational repression of mRNAs is often achieved through interaction of RNA-binding proteins with the 3’-UTR of messages.6 For example, in a conserved mechanism first discovered in Xenopus7 oocytes, maternal mRNAs are translationally repressed through binding of RNA-binding proteins, such as Cytoplasmic Polyadenylation Element Binding proteins (CPEBs), to sequence motifs known as Cytoplasmic Polyadenylation Elements (CPEs) in their 3’-UTRs. During the oocyte-to-embryo transition, phosphorylation of CPEBs activates mRNA translation by promoting polyadenylation.8 In addition to repression, degradation of maternally contributed mRNAs can also be achieved through the interaction of RNA-binding proteins with 3’-UTR cis-elements. RNA-binding proteins that elicit decay of maternal mRNAs include SMAUG in Drosophila9 and PolyC-Binding Proteins (PCBPs) in C. elegans.10 There is also evidence that miRNAs can promote degradation of maternal transcripts, as seen in zebrafish embryos where miR-430 targets the 3’-UTR of mRNAs destined for decay.11 Proteins are also targeted for degradation during early development through the ubiquitin-proteasome pathway and macroautophagy (see reviews12,13).
Zygotic Transcriptional Landscapes in Model Organisms
A prevailing view in developmental biology is that after fertilization, the zygotic genome remains transcriptionally silenced until the onset of the maternal-to-zygotic transition, a developmental period where maternally contributed RNAs are degraded and new transcription is required for development to proceed.14 Studies in multiple organisms have revealed that the activation of transcription during early embryo development occurs in 2 waves. The first and minor wave transcribes only a few dozen to hundreds of genes. The second and major wave transcribes thousands of genes.14 The timing of these transcription waves varies between organisms and has been described using multiple methods. Early studies used radioactive or bromouridine labeling of RNA to define the onset and amount of transcribed RNAs. Northern blots, quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR), and in situ hybridization were used later to trace individual mRNA changes during development. More recently, high-throughput approaches such as microarray and RNA-seq combined with advanced sampling technologies were used to systematically examine gene transcription and RNA changes in early development (Table 1). These genome-wide studies not only revealed the dynamic nature of transcription and RNA turnover during early development, but have also provided valuable resources for evolutionary comparisons of early development in various organisms.23,30–32 Interestingly, these newer studies observed that transcription occurs in much earlier developmental stages than previously thought in several organisms, including Drosophila,19 zebrafish,23 and mammals.29 This is likely due to the use of more sensitive methods that can identify the minor wave of early transcription.
Table 1.
Genome-wide studies on early developmental transcriptomes in model organisms
| Organism | Embryo Samples | Genomic Approaches | Reference |
|---|---|---|---|
| S. purpuratus | Staged embryos | Microarray | Wei et al, 200615 |
| Staged embryos | Whole-genome tiling array | Samanta et al, 200616 | |
| C. elegans | Handpicked embryos | Microarray | Baugh et al, 200317 |
| Picked or cell sorting | RNA-seq | Stoeckius et al, 201410 | |
| D. melanogaster | Chromosomal ablation | Microarray | De Renzis et al, 200718 |
| Genetic cross | RNA-seq & SNP analysis | Ali-Murthy et al, 201319 | |
| D. rerio | Staged embryos | RNA-seq (SOLiD) | Aanes et al, 201120 |
| Staged embryos | RNA-seq and lncRNA analysis | Pauli et al, 201221 | |
| Genetic cross | RNA-seq & SNP analysis | Harvey et al, 201322 | |
| Metabolic RNA labeling | RNA-seq | Heyn et al, 201423 | |
| X. tropicalis | Staged embryos | Microarray | Yanai et al, 201124 |
| Staged embryos | RNA-seq | Tan et al, 201325 | |
| Staged embryos | RNA-seq (poly-A and ribo-zero) | Paranjpe et al, 201326 | |
| M. musculus | Staged embryos | Microarray | Hamatani et al, 200427 |
| Staged embryos | Microarray | Xie et al, 201028 | |
| Staged embryos | Single-cell RNA-seq & SNP analysis | Xue et al 201329 |
Although transcription is now known to occur very early in development in many organisms (Table 1), its specific contributions to early development are largely unknown. Furthermore, in many cases where early transcription occurs, transcriptional inhibition using α-amanitin does not immediately lead to inhibition of development (see review14). Thus, additional studies are needed to unveil the functions of early transcription during embryogenesis.19,33
Variation in Gene Regulation Programs During Early Development
Variations in the onset of major transcription in different organisms are observed ranging from the 2-cell stage in mouse to > 4000-cell stage in frog and fruit fly.14 In addition, large differences in the overall contributions of post-transcriptional versus transcriptional gene regulation driving early development can occur in related species. For example, significant differences in the maternal contribution and requirements for early transcription have been observed in nematodes.34 C. elegans embryos can develop to 100–150 cells when zygotic transcription is blocked by α-amanitin34 or a mutant RNA polymerase.35 In contrast, Acrobeloides nanus embryos are arrested at the 5-cell stage when transcription is blocked by α-amanitin.34 C. elegans develops very rapidly with an average cell cycle length of 20–25 min, while A. nanus develops 4–5 times slower. The differences in requirements for transcription in these nematode embryos may be due to differences in cell cycle length and thus the speed of early development, as fast development and progression through mitosis can cause abortion of nascent transcripts.36 Although differences in embryonic pattern formation and cell-specification may also account for these differences,34 the slower developing A. nanus may be more dependent on new transcription due to smaller contributions from maternal deposition. We recently compared the developmental transcriptome dynamics of C. elegans with an extremely slow-developing nematode, Ascaris suum.32 A. suum and C. elegans appear to have identical early cleavage and developmental patterns, but the early cell cycle lengths of A. suum (1,200 min) are ~50-fold longer than those in C. elegans (20–25 min) (Fig. 1). In C. elegans, the minor, initial wave of transcription is thought to begin at the 4-cell stage and the major wave does not start until the ~100-cell stage. In contrast, A. suum major transcription initiates immediately following fertilization prior to pronuclear fusion and appears to drive early development (Fig. 1). Developmental polysome profiles revealed little translational regulation in A. suum, further supporting the notion that transcriptional regulation drives A. suum early development.32
Figure 1.
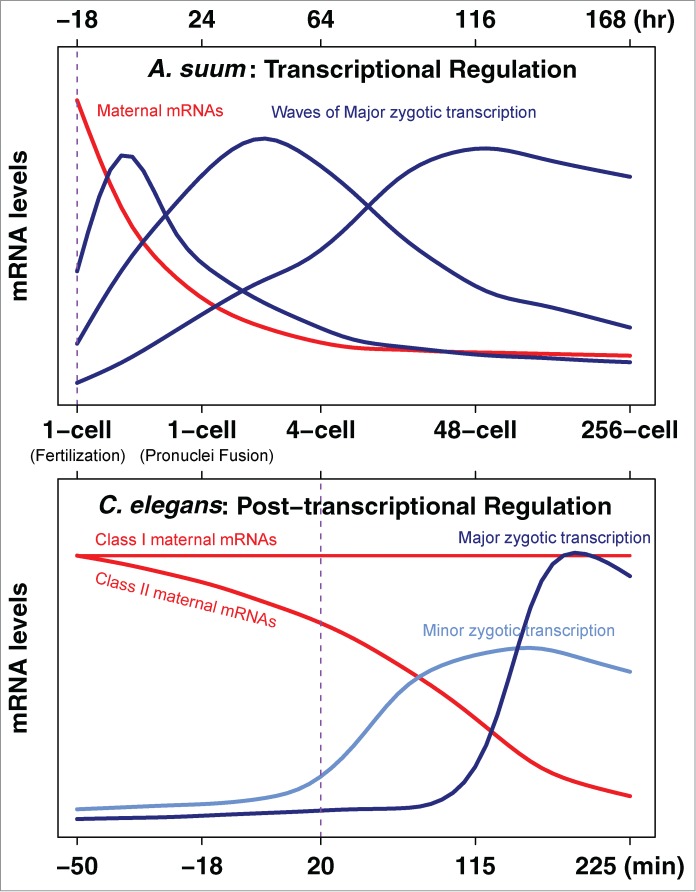
Rewiring of nematode gene expression program during early development. In A. suum, maternal mRNA degradation and zygotic transcription initiates immediately after fertilization and continues prior to pronuclear fusion. Thus, the maternal-to-zygotic transition in A. suum occurs right after fertilization (vertical purple line). Very little maternal RNA remains after the 4-cell stage suggesting that de novo transcription drives A. suum early development. In contrast, the C. elegans maternal-to-zygotic transition starts at ~4-cell stage (vertical purple line) and major transcription does not occur until ~100 cells. Maternal mRNA levels remain high during C. elegans early development and differential gene expression is post-transcriptionally regulated.
Zygotic Transcription May Be Dependent on the Cell Cycle Length
The cell cycle length and thus the time available for the genome to be transcribed may be a key determinant for the contributions of transcriptional vs. post-transcriptional regulation of gene expression in early embryogenesis.36,37 We speculate that the predominant use of newly transcribed genes in A. suum may have been enabled by its unusually long cell cycles and protracted early development. Indeed, in organisms with long cell cycle times during early cell divisions, such as mouse and human, significant numbers of genes are transcribed during the 1–4 cell stages.27,29 A comparison of organisms with fast or slow cell cycles during early development suggests that the time when the genome is transcriptionally activated correlates well with the length of cell cycle during early cleavages (Fig. 2). As many model organisms have a short life cycle and are fast-developing (except mammals), features that make them favorable for research, many studies have found that these fast-developing animals use a gene regulation program that primarily depends on post-transcriptional control during early development.
Figure 2.
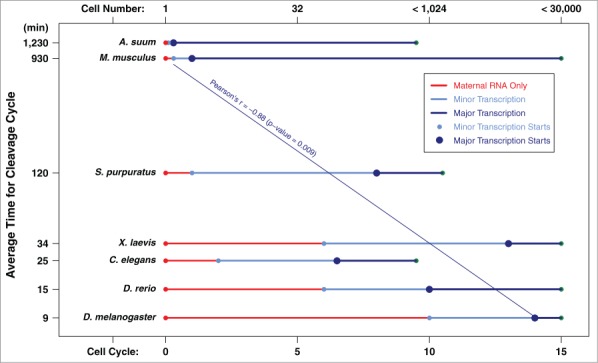
Zygotic genome activation is correlated with early development cell cycle length. The average cell-cycle length for each organism’s early cell divisions was calculated from 1 to 128 cells. The cell cycle (labeled at the bottom) and cell number (labeled at the top) for minor and major transcription in model organisms were derived from Tadros and Lipshitz.14 Data for A. suum was from our recent study.32 Note the y-axis is in log scale.
The lack of transcriptional activity during early development has been explained by several non-mutually exclusive mechanisms (see reviews14,38), including: (1). There is an excess of a repressor that maintains the repressed chromatin status. This repressor is diluted out during developmental cleavages to re-activate transcription (the “excess repressor model”); (2). The transcription machinery is incomplete and the missing components need to be expressed in development to assemble the functional transcription complex (the “limited machinery model”); (3). There is a maternal clock that is triggered by egg activation or fertilization that is independent of cell cycle and nucleo-cytoplasmic ratio but dependent on the absolute time of development (the “maternal clock model”); (4). The chromatin from early cell stages is not competent or ready for transcription (the “incompetent chromatin model”); and (5). Rapid cell cycle and DNA replication without G1 and G2 during embryo cleavage leads to transcript abortion (the “rapid cell cycle model”). Since major transcription is seen before pronuclear fusion and in 1- to 4-cell embryos in A. suum and mammals, this argues against the excess repressor model, the limited machinery model, and the incompetent chromatin model in these organisms. While a maternal clock cannot be ruled out, the correlation between zygotic genome activation and cell cycle length (Fig. 2) strongly suggests organisms with longer cell cycles transcribe significantly more genes during early development. Consistent with this, significant transcription occurs in fast-developing animals when the cell cycle length increases during late cleavage and gastrulation. In addition, there appears to be a bias for transcription of relatively short genes (such as intron poor or intronless genes) in these organisms.23 Thus, one determining factor for the overall level of transcription in early development may be related to the time available for the genome to be accessed by transcription machinery, supporting the rapid cell cycle model.
The Balance Between Maternal Deposition, Transcriptional and Post-transcriptional Regulation During Development
The amount of maternal deposition and the relative contributions of transcriptional versus post-transcriptional regulation to early development are ultimately determined by the life cycles and the biology of the organisms. Adaptions for massive maternal contribution during meiosis seem to have co-evolved with the requirement for the maternal deposition. For example, the late onset of transcription during Xenopus embryogenesis requires a large amount of maternal deposition and complex gene regulation during development. To achieve this, Xenopus has evolved months-long diplotene stages, developed an rRNA gene amplification mechanism,3 and diverse mechanisms of post-transcriptional gene regulation during oogenesis and embryo development.39,40 Increasing evidence suggests that paternal contributions may also play an important role during early development.41 With the advancement of new technologies enabling the use of limited amounts of samples (single-cell technology) and the whole genome identification of nascent transcription (such as GRO-seq and RNA polymerase II ChIP-seq), additional examples of the dynamic balance between maternal deposition, post-transcriptional regulation, and new transcription are likely to be defined at high-resolution in various organisms. Recent studies have also demonstrated the importance of transcriptional regulation in embryonic stem cell maintenance and its differentiation in early development in mammals.42 Transcription is regulated by the dynamic structural changes of chromatin, such as histone modifications, DNA methylation, and chromatin remodeling, that effect access of RNA polymerase and general and specific transcription factors.43 With differences in the timing and contributions of transcription in early development, chromatin regulatory mechanisms must also change, be dynamic, and be specifically adapted to each organism. Understanding the balance and interplay of transcriptional and post-transcriptional regulation during early development will shed new light on one of the most complex transformations in biology: the fusion of oocyte and sperm and subsequent development to generate a new life.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ashley Neff for discussions and suggestions.
Funding
This work was supported by NIH Grant AI0149558.
References
- 1.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999; 126:1011–22; PMID:9927601. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert SF. Developmental Biology. Sinauer Associates. 2010. [Google Scholar]
- 3.Dawid IB, Brown DD, Reeder RH. Composition and structure of chromosomal and amplified ribosomal DNA's of Xenopus laevis. J Mol Biol 1970; 51:341–60; PMID:5485907; http://dx.doi.org/ 10.1016/0022-2836(70)90147-6 [DOI] [PubMed] [Google Scholar]
- 4.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science 2007; 316:407–8; PMID:17446393; http://dx.doi.org/ 10.1126/science.1138236 [DOI] [PubMed] [Google Scholar]
- 5.Vasudevan S, Seli E, Steitz JA. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev 2006; 20:138–46; PMID:16418480; http://dx.doi.org/ 10.1101/gad.1398906 [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 2013; 4:437–61; PMID:23776146; http://dx.doi.org/ 10.1002/wrna.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 1994; 79:617–27; PMID:7954828; http://dx.doi.org/ 10.1016/0092-8674(94)90547-9 [DOI] [PubMed] [Google Scholar]
- 8.Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell 1999; 4:1017–27; PMID:10635326; http://dx.doi.org/ 10.1016/S1097-2765(00)80230-0 [DOI] [PubMed] [Google Scholar]
- 9.Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 2007; 12:143–55; PMID:17199047; http://dx.doi.org/ 10.1016/j.devcel.2006.10.005 [DOI] [PubMed] [Google Scholar]
- 10.Stoeckius M, Grun D, Kirchner M, Ayoub S, Torti F, Piano F, Herzog M, Selbach M, Rajewsky N. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. Embo J 2014; 33:1751–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312:75–9; PMID:16484454; http://dx.doi.org/ 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- 12.Robertson S, Lin R. The oocyte-to-embryo transition. Adv Exp Med Biol 2013; 757:351–72; PMID:22872483; http://dx.doi.org/ 10.1007/978-1-4614-4015-4_12 [DOI] [PubMed] [Google Scholar]
- 13.Li L, Zheng P, Dean J. Maternal control of early mouse development. Development 2010; 137:859–70; PMID:20179092; http://dx.doi.org/ 10.1242/dev.039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development 2009; 136:3033–42; PMID:19700615; http://dx.doi.org/ 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- 15.Wei Z, Angerer RC, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Dev Biol 2006; 300:476–84; PMID:17007833; http://dx.doi.org/ 10.1016/j.ydbio.2006.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, Stolc V. The transcriptome of the sea urchin embryo. Science 2006; 314:960–2; PMID:17095694; http://dx.doi.org/ 10.1126/science.1131898 [DOI] [PubMed] [Google Scholar]
- 17.Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 2003; 130:889–900; PMID:12538516; http://dx.doi.org/ 10.1242/dev.00302 [DOI] [PubMed] [Google Scholar]
- 18. De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol 2007; 5:e117; PMID:17456005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali-Murthy Z, Lott SE, Eisen MB, Kornberg TB. An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLoS Genet 2013; 9:e1003428; PMID:23593026; http://dx.doi.org/ 10.1371/journal.pgen.1003428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. . Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 2011; 21:1328–38; PMID:21555364; http://dx.doi.org/ 10.1101/gr.116012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. . Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 2012; 22:577–91; PMID:22110045; http://dx.doi.org/ 10.1101/gr.133009.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, Smith JC. Identification of the zebrafish maternal and paternal transcriptomes. Development 2013; 140:2703–10; PMID:23720042; http://dx.doi.org/ 10.1242/dev.095091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The Earliest Transcribed Zygotic Genes Are Short, Newly Evolved, and Different across Species. Cell Rep 2014; 6:285–92; PMID:24440719; http://dx.doi.org/ 10.1016/j.celrep.2013.12.030 [DOI] [PubMed] [Google Scholar]
- 24.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell 2011; 20:483–96; PMID:21497761; http://dx.doi.org/ 10.1016/j.devcel.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res 2013; 23:201–16; PMID:22960373; http://dx.doi.org/ 10.1101/gr.141424.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJ. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics 2013; 14:762; PMID:24195446; http://dx.doi.org/ 10.1186/1471-2164-14-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6:117–31; PMID:14723852; http://dx.doi.org/ 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- 28.Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, et al. . Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 2010; 20:804–15; PMID:20219939; http://dx.doi.org/ 10.1101/gr.100594.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, et al. . Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013; 500:593–7; PMID:23892778; http://dx.doi.org/ 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Liu Z, Zeng Q, Cheng L, Sun YE, et al. . Gene expression divergence recapitulates the developmental hourglass model. Nature 2010; 468:811–4; PMID:21150996; http://dx.doi.org/ 10.1038/nature09634 [DOI] [PubMed] [Google Scholar]
- 31.Levin M, Hashimshony T, Wagner F, Yanai I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell 2012; 22:1101–8; PMID:22560298; http://dx.doi.org/ 10.1016/j.devcel.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Garrey J, Davis RE. Transcription in pronuclei and one- to four-cell embryos drives early development in a nematode. Curr Biol 2014; 24:124–33; PMID:24374308; http://dx.doi.org/ 10.1016/j.cub.2013.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol 2011; 357:478–91; PMID:21741375; http://dx.doi.org/ 10.1016/j.ydbio.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laugsch M, Schierenberg E. Differences in maternal supply and early development of closely related nematode species. Int J Dev Biol 2004; 48:655–62; PMID:15470638; http://dx.doi.org/ 10.1387/ijdb.031758ml [DOI] [PubMed] [Google Scholar]
- 35.Powell-Coffman JA, Knight J, Wood WB. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev Biol 1996; 178:472–83; PMID:8812143; http://dx.doi.org/ 10.1006/dbio.1996.0232 [DOI] [PubMed] [Google Scholar]
- 36.Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 1991; 67:303–10; PMID:1680567; http://dx.doi.org/ 10.1016/0092-8674(91)90182-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swinburne IA, Silver PA. Intron delays and transcriptional timing during development. Dev Cell 2008; 14:324–30; PMID:18331713; http://dx.doi.org/ 10.1016/j.devcel.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science 2007; 316:406–7; PMID:17446392; http://dx.doi.org/ 10.1126/science.1140693 [DOI] [PubMed] [Google Scholar]
- 39.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 2008; 1779:217–29; PMID:18316045; http://dx.doi.org/ 10.1016/j.bbagrm.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heasman J. Patterning the early Xenopus embryo. Development 2006; 133:1205–17; PMID:16527985; http://dx.doi.org/ 10.1242/dev.02304 [DOI] [PubMed] [Google Scholar]
- 41.Neff AT, Wang J, Davis RE. "Father knows best?." Embo J 2014; 33:1729–31; PMID:25061225; http://dx.doi.org/ 10.15252/embj.201489490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young RA. Control of the embryonic stem cell state. Cell 2011; 144:940–54; PMID:21414485; http://dx.doi.org/ 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707–19; PMID:17320508; http://dx.doi.org/ 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]


