Abstract
RNA polymerase (RNAP) performs various tasks during transcription by changing its conformational states, which are gradually becoming clarified. A recent study focusing on the conformational transition of RNAP between the ratcheted and tight forms illuminated the structural principles underlying its functional operations.
Keywords: RNA polymerase, structure-function relationship, transcription, transcription factor, transcription regulation
Abbreviations
- BC
backtracked complex
- BH
bridge helix
- CPX
Cys-pair crosslinking
- EC
elongation complex
- NTP
nucleoside triphosphate
- RNAP
DNA-dependent RNA polymerase
- TL
trigger loop.
Multi-subunit DNA-dependent RNA polymerase (RNAP) is a huge protein complex responsible for gene transcription. It accomplishes multiple tasks required for transcription initiation, elongation, and termination, often with assistance from various regulatory factors. The versatile nature of RNAP is supported by its conformational plasticity. Recent studies are starting to reveal the relationships between particular RNAP conformational states and functions, paving the way toward understanding the general principles of the functions and regulation of RNAP.
The bacterial RNAP core is composed of at least 5 subunits, while the eukaryotic RNAPs I, II, and III comprise 12 or more subunits, and their total masses are over 400 kDa. From bacteria to eukaryotes, RNAP adopts a similar “crab-claw” shape, which can be divided into 4 massive blocks called “modules”.1,2 The central part of RNAP is composed of the “shelf” and “core” modules, which form the primary nucleic-acids-binding channel and the secondary channel, a likely path for the substrate nucleoside triphosphates (NTPs) (Fig. 1A, B). The “clamp” and “jaw-lobe” modules protrude from the shelf and core modules, respectively, to complete the primary channel. The active site is formed in the middle of the primary channel, and includes Mg2+ ions and flexible structural elements, such as the “trigger loop (TL)” and the “bridge helix (BH).”
Figure 1.
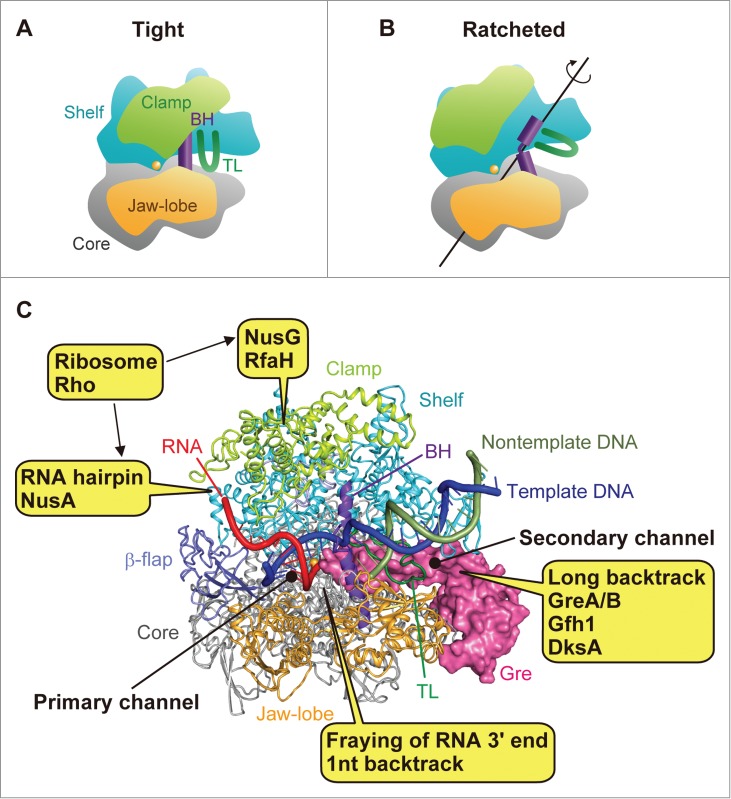
RNAP conformations and points of regulation. Schematic representations of (A) the tight form (with the closed clamp) and (B) the ratcheted form (with the open clamp). The structural elements of RNAP (4) are colored as follows: core module, gray; shelf module, cyan; clamp module, yellow-green; jaw-lobe module (β domains), light orange; BH, purple; TL, green. The active site is represented as an orange sphere. (C) Interaction sites for transcription factors and RNA elements are depicted on the structure of Thermus thermophilus RNAP bound with a Gre protein in the open-clamp ratcheted form (10). The RNAP is shown as a ribbon model, and the Gre protein (a hybrid of GreA and Gfh1 (16)) is shown as a magenta-colored surface model. The β-flap domain is colored light blue. The nucleic acids were modeled based on those in the backtracked complex structure (10), and the DNA template strand, non-template strand, and RNA are colored blue, green, and red, respectively.
The modules can move relative to each other around a particular rotational axis formed between them. Therefore, RNAP has a rich conformational space produced by multiple combinations of module orientations, in addition to the conformations of the flexible elements. For example, the clamp module can swing relative to the shelf module, allowing the opening and closing of the primary channel.3,4 The clamp module is closed upon DNA binding by the RNAP holoenzyme for initiation complex formation,3 while it is opened during transcriptional pausing dependent on a hairpin structure formed upstream of the nascent transcribed RNA.5 While the flexible TL assumes the “straight” or trigger-helices conformation for NTP incorporation into RNA in the transcription elongation complex (EC),6,7 it assumes a “bent” conformation in a paused or backtracked RNAP.8-10
The shelf module can also be rotated or “ratcheted” relative to the core module (Fig. 1B). We previously identified the ratcheted form of RNAP in the complex of Thermus thermophilus RNAP, bound with its inhibitor protein Gfh1 and a nucleic-acid scaffold with RNA bearing a hairpin structure.4 In contrast to the “tight” form seen in the EC,6,11 the active site of the ratcheted form exhibits extensive remodeling, including kinking of the BH, bending of the TL, and expansion of the DNA/RNA binding site. The reorganization of the active site led us to hypothesize that the ratcheted form is involved in various transcriptional functions other than nucleotide addition, beyond the transcription inhibition by Gfh1.4,12,13 Our recent study based on Cys-pair crosslinking (CPX) analyses combined with X-ray crystallography has revealed that the ratcheted form indeed participates in many essential functions of RNAP.10 Thus, the tight-ratcheted transition added a new axis to the conformational space of RNAP, and provided deep insights into the structurally-embedded principles underlying its operations. Here we discuss this ratcheted form and its roles in RNAP regulation, with future perspectives.
The ratcheted form in RNAP backtracking and transcript cleavage
Transcription is unidirectional (5′ to 3′), but the EC does not always move forward along the DNA. The EC sometimes pauses on the DNA, and even moves backward (or backtracks) along it, due to a transcriptional error (incorporation of a mismatched nucleotide into RNA), a lesion in the DNA, sequence contexts, etc.14 We demonstrated that T. thermophilus RNAP adopts the tight form in a one-nucleotide backtracked complex (BC), while it assumes the ratcheted form in a longer, 8-nucleotide BC.10 This is presumably because the enlarged secondary channel in the ratcheted form is more suitable to accommodate a long backtracked stretch of the RNA 3′ end, as compared to the narrower channel in the tight form. Thus, the extent of backtracking is critical for the tight-ratcheted transition.
In the BC, the RNA elongation function is inactive, and instead RNAP exhibits hydrolytic activity to cleave the extruded part of the RNA 3′ end at the same active site. The RNA cleavage contributes to transcription proofreading, in the case of a transcriptional error. The transcription factor GreA dramatically enhances the RNA cleavage activity by a 1–2 nucleotide backtracked RNAP.15 T. thermophilus GreA was revealed to induce the ratcheted form in T. thermophilus RNAP, in a one-nucleotide backtracked state.10 This is because the bulky coiled-coil domain of the Gre protein is compatible with the enlarged secondary channel in the ratcheted form, but not with the narrower channel in the tight form. These data, together with activity measurements of an S-S crosslinked CPX variant (fixed in the ratcheted form), suggested that the GreA-dependent RNA cleavage occurs in the ratcheted form of RNAP, while the GreA-independent, intrinsic RNA cleavage occurs in the tight form of RNAP. Consistently, in the structure of RNAP bound with a Gre protein (a chimera of GreA and Gfh1),16 the RNAP assumed the ratcheted form (Fig. 1C)10. This structure also exhibited an open clamp conformation, which is probably because the DNA/RNA scaffold included an RNA hairpin, as described below. The structure of the ratcheted form with a closed clamp has not been solved. The ratcheting would loosen the grip on the DNA/RNA, and facilitate the clamp opening. However, the clamp module can swing independently of the shelf module.4 Therefore, in the long backtracked or Gre-bound complex (without a hairpin), the clamp may not necessarily be opened, as proposed previously.4,17
In addition to GreA, E. coli possesses another Gre factor homolog, GreB, which can act on a longer backtracked state of RNAP.15 As mentioned above, the long backtracked RNAP tends to assume the ratcheted form, which can accommodate GreB (Fig. 1C). DksA is another coiled-coil protein in E. coli, and is considered to interact with the secondary channel, although it lacks the cleavage stimulating function. It cooperates with the bacterial alarmone guanosine tetraphosphate (ppGpp) to suppress transcription initiation from many genes, as well as to increase fidelity during transcription elongation.18,19 Although the mechanism underlying the DksA activity is elusive, the ratcheted form may support it. It was previously hypothesized that ppGpp could shift E. coli RNAP to the ratcheted form.20
The ratchetable tight form in RNAP backtracking and pausing
Interestingly, the one-nucleotide BC readily transitioned to the ratcheted form with the aid of GreA, whereas ECs exhibited resistance to the conformational shift.10 This suggested that GreA selectively acts on the BC to help the stalled RNAP cleave the RNA and thus resume transcription, without interfering with the transcribing ECs. This is quite reasonable for efficient and accurate transcription. The difference between the EC and the BC probably correlates with their TL conformations. In contrast to the straight conformation of the TL in the EC, it assumes a “bent” conformation in the BC, due to the extrusion of the RNA 3′ end. The bent TL partly opens the path from the secondary channel through the active site, and thus probably facilitates the docking of the GreA coiled-coil domain.10 Similarly to the BC, a paused complex due to a mismatch-frayed RNA 3′ end is also readily transitioned to the ratcheted form. Thus, the tight form can adopt either the “ratchetable” or “unratchetable” state, according to the status of the RNA 3′ end. We speculate that the ratchetable state is an unfixed state in equilibrium between the tight and partly ratcheted forms, and thereby it could serve as the precedent state for switching to the ratcheted form underlying various RNAP functions. In this regard, the ratchetable state may correspond to the so-called “elemental pause” state.21
The ratcheted form in transcriptional pausing and termination
The hairpin structure formed in the nascent RNA is another important element that governs the fate of transcription. In the hairpin-dependent transcriptional pause, such as the well characterized his pause, an RNA hairpin formed upstream of the DNA/RNA hybrid stabilizes the paused state of RNAP.21 In the hairpin-dependent, intrinsic transcription termination, the U-rich tract at the RNA 3′ end causes transcriptional pausing, and a hairpin structure formed just upstream of the U-rich tract facilitates a conformational change of RNAP for dissociation from DNA/RNA.22 We found that the RNA hairpin induces the ratcheted form of RNAP, which may underlie the transcriptional pause and termination.10 Recent studies have revealed that the presence of the RNA hairpin in the his-paused complex is coupled with the clamp opening and the TL bending.5,23 These observations have clarified how the signal from the hairpin formation at the periphery of RNAP is transmitted to the active site. The bulky RNA hairpin in the RNA exit channel causes not only the clamp opening, but also the shelf/core ratcheting, which is inevitably accompanied by the TL bending and transcriptional pausing or complete inactivation (trapping) during termination 22 (Fig. 1C). Consistently, in the structures of RNAP bound with Gre proteins, RNAP assumes the ratcheted form with an opened clamp, probably due to the accommodation of the RNA hairpin, although it was not visible.4,10
NusA is an essential transcription factor involved in the hairpin-dependent transcriptional pausing, termination, and anti-termination. It binds near the RNA exit site including the β-flap domain, facilitates RNA duplex formation in the RNA exit channel, and slows RNAP translocation.5,24,25 Therefore, NusA may maintain the open-clamp ratcheted form induced by the hairpin. NusA is also involved in transcription-coupled DNA repair, where it helps UvrD helicase to backtrack RNAP and expose the DNA lesion for access by repair enzymes.26 Here, the ratcheted form of RNAP could support the RNAP backtracking and DNA repair.
Besides the hairpin-dependent, intrinsic termination, Rho-dependent termination is another major mechanism of bacterial transcription termination. Rho is a hexameric ATPase/helicase that binds to the nascent RNA at a specific Rho-utilization site, where it accesses the transcribing EC and causes termination, probably by inducing a conformational change in the RNAP. Although the exact structural basis of the conformational change is unknown, the Rho-dependent termination was suggested to proceed in a similar manner to the intrinsic termination.27 Therefore, we hypothesize that the ratcheted form with the opened clamp is the common intermediate for both the Rho-dependent and independent terminations. Similarly to the Rho-dependent termination by a bacterial RNAP, the Rat1 and Sen1-dependent terminations by eukaryotic RNAPs I and II are also proposed to be accompanied by an allosteric change in the EC.28 Again, the open-clamp ratcheted form could be the intermediate for the eukaryotic transcription termination mechanism.
Insights into transcriptional regulation
Our study revealed that many transcription functions, such as nucleotide addition, RNA cleavage, RNAP backtracking, pausing, termination, and inhibition, are accomplished by one of the 2 alternative forms of RNAP: the tight and ratcheted forms. As the active site of RNAP is formed between the shelf and core modules, switching between the tight and ratcheted forms could be the major regulation point of the functions and activities of RNAP. Taking into account other changes, including the clamp swinging and the conformational changes in the TL and the BH, the operational code of RNAP in terms of its conformational space has emerged (Table 1). This code may also represent the basis for the regulation of RNAP functions by various elements and transcription factors.
Table 1.
Structure-function relationships of bacterial RNAP (“conformational code”)
| Shelf/Core | Clamp | Trigger loop | Bridge helix | Functions/states | TFs | References |
|---|---|---|---|---|---|---|
| Tight | Closed | Straight | Straight | Nucleotide addition | NusG/RfaH | (6) |
| |
|
|
|
|
Ribosome |
|
| Tight | Closed | Mobile | Straight | Pre-translocation state | NusG/RfaH | (11) |
| |
|
|
|
Post-translocation state |
Ribosome |
|
| Tight | Closed | Bent | Straight | Paused (frayed)Paused (hairpin-dependent)Short backtracked | Entry point for the actions by Gre, Rho and RNA elements (hairpin and long backtracking) | (10) |
| |
|
|
|
Intrinsic RNA cleavage |
|
|
| Tight |
Open |
Straight/bent |
Straight/kinked |
? |
? |
|
| Ratcheted | Closed? | Bent | Kinked | Gre-dependent RNA cleavage | GreA/B | (4, 10) |
| Long backtracked | DksA + ppGpp? | |||||
| Inhibited | UvrD + NusA? | |||||
| |
|
|
|
|
Gfh1 |
|
| Ratcheted | Open | Bent | Kinked | Free RNAP | (3, 5, 10, 26, 27) | |
| Hairpin-dependent pause | Hairpin + NusA | |||||
| Hairpin-dependent termination | Rho? | |||||
| |
|
|
|
Rho-dependent termination? |
|
|
| Tight or ratcheted? | Closed or open? | Bent | Kinked | Translocation intermediate | NusG/RfaH Ribosome |
As described above, the nascent RNA (the 3′ end and hairpin) plays a key role in the tight-ratcheted transition and/or clamp swinging, and determines the fate of transcription (Fig. 1C, Table 1). GreA/B, Gfh1, and possibly DksA, which bind to the secondary channel, stabilize the ratcheted form for their functions. The functions of NusA and Rho could be based on the ratcheted form with an opened clamp. These RNA elements and transcription factors are all related to the secondary channel or the RNA exit channel of RNAP. In contrast, the universal transcription elongation factor NusG (Spt5 in Archaea/Eukarya) and its paralog RfaH bind to a part of the clamp module (clamp coiled-coil) to maintain the closed-clamp tight form5, and thus counteract the ratcheting to ensure processive transcription elongation. NusG is also known as the EC interface for the interactions with the ribosome and Rho.29 RfaH reportedly serves as a physical bridge between RNAP and ribosome, facilitating expression of horizontally transferred genes.30 The translating ribosome may cooperate with NusG/RfaH to maintain the closed-clamp tight form during the translation-coupled transcription, whereas Rho should counteract NusG to destabilize the closed-clamp tight form for termination. Further structural and functional studies will clarify these points.
RNAP ratcheting and translocation
In every nucleotide addition cycle of transcription elongation, RNAP moves or translocates by a one-nucleotide step along DNA/RNA. After nucleotide addition to the nascent RNA 3′ end, the pre-translocation state (the RNA 3′ end residing in the i + 1 position relative to the active site) is shifted to the post-translocation state (the RNA 3′ end residing in the i position). In previous studies, we hypothesized that the ratcheted form of RNAP could also correlate with an intermediary state for RNAP translocation.4,12,13 The ratcheted form represents a kinked BH, which sterically prevents the incorporation of the i + 1 DNA template nucleotide into the active site. The kinking of the BH has been suggested to correlate with RNAP translocation, as it could bias the translocation state to the post-translocation state.31-33 Therefore, it seems reasonable that the ratcheted form could mediate the shift from the pre-translocation state to the post-translocation state, in the Brownian-ratchet equilibrium.34 The ratcheted form also possesses the enlarged DNA/RNA binding site, which would have a looser grip on the DNA/RNA and thus could facilitate RNAP translocation. However, it is not clear whether this hypothesis is correct.
The question could be answered, for example, by restricting the RNAP conformation to the tight form by crosslinking or other techniques. If the RNAP translocation actually accompanies the ratcheting motion, then the restriction of the RNAP conformation would impair the RNA elongation rate, without affecting the rate of the chemical step. It was recently reported that RNAP elongates RNA in crystals,35 in which the spatial constraints should limit the RNAP conformational changes, including the module movements. An estimation of the RNA elongation rate in the crystal and a comparison of the rate with that in solution might provide new insights into the translocation mechanism.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grant Number 15H04344 (to SS) and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to SY), and grants from the National Institutes of Health and the Howard Hughes Medical Institute (to EN).
References
- 1. Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution, Cell 1999; 98, 811-824; PMID:10499798; http://dx.doi.org/ 10.1016/S0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 2. Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution, Science 2001; 292, 1863-76; PMID:11313498; http://dx.doi.org/ 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 3. Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH. Opening and closing of the bacterial RNA polymerase clamp, Science 2012; 337, 591-5; PMID:22859489; http://dx.doi.org/ 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, and Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein, Nature 2010; 468, 978-982; PMID:21124318; http://dx.doi.org/ 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 5. Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement, Nat. Struct. Mol. Biol. 2014; 21, 794-802; PMID:25108353; http://dx.doi.org/ 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase, Nature 2007; 448, 163-8; PMID:17581591; http://dx.doi.org/ 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 7. Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis, Cell 2006; 127, 941-954; PMID:17129781; http://dx.doi.org/ 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA, 2009; 34, 710-21; PMID:19560423. [DOI] [PubMed] [Google Scholar]
- 9. Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 Å resolution, Science 2009; 324, 1203-6; PMID:19478184; http://dx.doi.org/ 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural States of RNA polymerase underlie multiple transcriptional functions, Mol. Cell 2015; 57, 408-21; PMID:25601758; http://dx.doi.org/ 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 11. Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase, Nature 2007; 448, 157-62; PMID:17581590; http://dx.doi.org/ 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 12. Tagami S, Sekine SI, Yokoyama S. A novel conformation of RNA polymerase sheds light on the mechanism of transcription, Transcription 2011; 2, 162-7; PMID:21922057; http://dx.doi.org/ 10.4161/trns.2.4.16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sekine S, Tagami S, Yokoyama S. Structural basis of transcription by bacterial and eukaryotic RNA polymerases, Curr. Opin. Struct. Biol 2012; 22, 110-18; PMID:22155178; http://dx.doi.org/ 10.1016/j.sbi.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 14. Nudler E. RNA polymerase backtracking in gene regulation and genome instability, Cell 2012; 149, 1438-45; PMID:22726433; http://dx.doi.org/ 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action, Mol. Microbiol 2005; 55, 1315-24; PMID:15720542; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 16. Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, Berenguer J, Kong XP, Borukhov S. pH-dependent conformational switch activates the inhibitor of transcription elongation, EMBO J 2006; 25, 2131-41; PMID:16628221; http://dx.doi.org/ 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria, Cell 2013; 152, 431-41; PMID:23374340; http://dx.doi.org/ 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP, Cell 2004; 118, 311-22; PMID:15294157. [DOI] [PubMed] [Google Scholar]
- 19. Roghanian M, Zenkin N, Yuzenkova Y. Bacterial global regulators DksA/ppGpp increase fidelity of transcription, Nucleic Acids Res 2015; 43, 1529-36; PMID:25605801; http://dx.doi.org/ 10.1093/nar/gkv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex, Mol. Cell 2013; 50, 430-6; PMID:23623685; http://dx.doi.org/ 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landick R. The regulatory roles and mechanism of transcriptional pausing, Biochem. Soc. Trans 2006; 34, 1062-66; PMID:17073751. [DOI] [PubMed] [Google Scholar]
- 22. Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination, Mol. Cell 2007; 28, 991-1001; PMID:18158897. [DOI] [PubMed] [Google Scholar]
- 23. Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase, Mol. Cell 2013; 50, 882-893; PMID:23769674; http://dx.doi.org/ 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, Hankamer B, Lewis RJ, Lewis PJ. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA, EMBO Rep 2009; 10, 997-1002; PMID:19680289; http://dx.doi.org/ 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms, Cell 2001; 107, 437-49; PMID:11719185. [DOI] [PubMed] [Google Scholar]
- 26. Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards, Nature 2014; 505, 372-77; PMID:24402227; http://dx.doi.org/ 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination, Nature 2010; 463, 245-9; PMID:20075920; http://dx.doi.org/ 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop, Nat. Rev. Mol. Cell Biol 2015; 16, 190-202; PMID:25650800; http://dx.doi.org/ 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- 29. NandyMazumdar M, Artsimovitch I. Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins - Shifting shapes and paradigms, BioEssays 2015; 37, 324-34; PMID:25640595; http://dx.doi.org/ 10.1002/bies.201400177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, Artsimovitch I, Rosch P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor, Cell 2012; 150, 291-303; PMID:22817892; http://dx.doi.org/ 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution, Science 2001; 292, 1876-82; PMID:11313499. [DOI] [PubMed] [Google Scholar]
- 32. Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution, Nature 2002; 417, 712-9; PMID:12000971; http://dx.doi.org/ 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 33. Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases, J. Biol 2008; 7, 40; PMID:19055851; http://dx.doi.org/ 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control, Cell 2005; 120, 183-93; PMID:15680325; http://dx.doi.org/ 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 35. Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernandez-Tornero C, Kulbachinskiy A, Murakami KS. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme, J. Biol. Chem 2014; 289, 24549-59; PMID:24973216; http://dx.doi.org/ 10.1074/jbc.M114.584037. [DOI] [PMC free article] [PubMed] [Google Scholar]


