Abstract
Transcription and splicing are intrinsically linked, as splicing needs a pre-mRNA substrate to commence. The more nuanced view is that the rate of transcription contributes to splicing regulation. On the other hand there is accumulating evidence that splicing has an active role in controlling transcription elongation by DNA-dependent RNA polymerase II (RNAP II). We briefly review those mechanisms and propose a unifying model where splicing controls transcription elongation to provide an optimal timing for successive rounds of splicing.
Keywords: chromatin and transcription, chromatin marks, co-trancriptional splicing, Eukaryotic transcription, PolII, splicing, transcription and RNA processing, transcription elongation
Speed of Transcription Regulates Alternative Splice Site Selection
Splicing often occurs co-transcriptionally,1 so that at the time of splicing the mRNA molecule is attached to the RNAP II complex which is still engaged in transcription.2 This observation is the foundation of models explaining the effect of transcription elongation on splicing. Most studies on the role of transcription in regulation of splicing focused on alternative splice sites, as their splicing is easily disturbed by modulation of the RNAP II transcription elongation rate.3
Based on those data a kinetic coupling model has been proposed.4 This model predicts that when 2 alternative splice sites (SS) of similar strength compete, the slowdown of RNAP II elongation would favor the upstream SS while the faster elongation would favor the downstream SS. This model has gained strong support from studies using fast/slow RNAP II mutants,5 chemical modulators of RNAP II elongation rate6 and alternative splice site selection defects in elongation factors mutants7 (Fig. 1 upper panel). We have recently inhibited in Arabidopsis RNAP II elongation by 6AU treatment and interference of RNAP II endonucleolytic cleavage by mutations in Transcription Elongation Factor S-II (TFIISmut). As predicted by the kinetic coupling model we observed a preferential selection of upstream splices sites.8 In addition, the analysis of RNAP II occupancy in TFIISmut background revealed a localized increase of RNAP II levels at the alternative splice sites rather than a uniform effect on RNAP II occupancy.8
Figure 1.
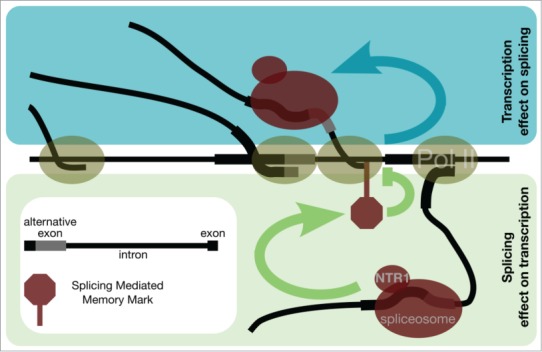
Schematic representation of the transcription–splicing mutual dependency. Splicing controls transcription elongation of consecutive RNAP II molecules to provide an optimal timing for successive rounds of splicing. Blue arrow represents RNAP II transcription elongation rate effect on splice site selection by spliceosome. Green arrow represents spliceosome effect on transcription.
Splicing-Induced Transcriptional Pausing
Many different factors have been discovered that help polymerase to elongate efficiently.9 In spite of that, recent data have shown that RNAP II elongation rate is far from uniform.10 One of the major transcription elongation blocks within the gene body turned out to be splicing junctions.10-12 This is further supported by enhanced RNAP II accumulation on alternatively spliced exons in humans.13,14 Consistent with this model research carried out in yeast has shown that splicing leads to transient polymerase pausing at the splice sites.15 In addition, recently circular ncRNA produced by back-splicing have been proposed to feedback to locally control transcription elongation of RNAP II at the splice site.16,17 All this strongly suggests that splicing, especially alternative splicing control RNAP II elongation at splice sites (lower panel of Fig. 1).
Therefore, on one hand splicing is regulated by the speed of transcription, while on the other hand splicing itself can enforce a transient pausing/slowdown of the elongating polymerase.
Is Kinetic Coupling of Splicing and Transcription a Consequence of Splicing-Mediated Transcriptional Pausing?
As mentioned above the transcription elongation rate is not uniform and active splice sites pose a significant barrier to RNAP II. In agreement with this Arabidopsis plants with indirectly compromised RNAP II endonucleolytic cleavage showed a restricted increase in the RNAP II signal at alternative splice sites.8 We interpret this localized increase in RNAP II occupancy as a transient RNAP II elongation pause site. We therefore propose that RNAP II elongation impediment may perturb the ability of RNAP II to negotiate an elongation roadblock posed by splicing. As a result, regulation of splicing by transcription elongation may to some extent be a consequence of splicing-mediated transcriptional roadblock.
Mutation analysis in yeast has shown that the act of splicing rather than splice sites themselves are the cause of pausing.15,18 Correspondingly we have recently identified Arabidopsis homolog of NTR1, a spliceosome accessory factor, as required for transcriptional pausing.8 This supports the view that the process of splicing and spliceosome, rather than a DNA/RNA sequences, are required for RNAP II pausing.
A rational consequence of this model is some type of a splicing memory. We envisage that after a round of transcription and splicing, splice sites are marked depending on their splicing efficiency. Our anticipation is that this splicing memory mark impedes RNAP II elongation during the next round of transcription to increase the splicing effectiveness. We hypothesize that although this memory mark is related to splicing, it should be stable enough to withstand a temporal shutdown of transcription and splicing (Fig. 1).
Splicing memory in the context of tissue specific alternative splicing has been previously proposed to rely on histone posttranslational modifications.19 In addition in plants alternative splicing of FLC antisense transcript COOLAIR has been shown to affect FLC gene expression.20,21 This effect is correlated with a concomitant change in positive/negative histone marks ratio and transcription elongation change that potentially feed on FLC capping efficiency.21,22 This provides another example of how alternative splicing can affect gene expression presumably thru splicing memory.
The epigenetic nature of histone modifications makes them likely candidates for the proposed splicing memory marks. However, there are many other possible candidates, including proteins deposited during splicing at the nascent-RNA/DNA, DNA methylation, circular RNA, histone variants etc.
The function of splicing-dependent RNAP II pausing may go beyond feeding back to increase the splicing efficiency. Aberrant splicing often leads to NMD (Nonsense-mediated decay) mediated RNA degradation.23 In addition, aberrant splicing may prime shutdown of gene expression. In agreement with this we reported transcriptional downregulation of Delay of Germination 1 (DOG1) expression in response to aberrant splicing in Arabidopsis.8 One possible mechanism may involve a crosstalk with the RNAi pathway, as recently reported in Cryptococcus neoformans.24 However, other pathways are clearly operating as a transcriptional downregulation by aberrant splicing has been observed in Saccharomyces cerevisiae, which lack RNAi machinery.18
In support of these observations the presence of the correctly spliced introns has an activating effect on transcription in many different systems.25,26 Even so, molecular mechanisms behind these observations remain unclear.
To conclude, efficient splicing emerges as one of the main quality checkpoints in the production of mRNA, and splicing and transcription appear to be mutually dependent: splicing is regulated by the transcription elongation rate but the outcome of splicing modulates transcription elongation. Therefore the presence of introns not only represents an evolutionary conserved way to enhance transcription but may also serve as a quality checkpoint in the course of gene expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Ministry of Science and Higher Education, Poland (grant No. IdP2011 000461), National Science Centre, Poland (DEC-2011/01/D/NZ8/03690), Foundation for Polish Science (grant No. TEAM2010-5/9).
References
- 1.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep 2013; 5:9; PMID:23638305; http://dx.doi.org/ 10.12703/P5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 1988; 2:754-65; PMID:3138163; http://dx.doi.org/ 10.1101/gad.2.6.754 [DOI] [PubMed] [Google Scholar]
- 3.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009; 15:1896-908; PMID:19656867; http://dx.doi.org/ 10.1261/rna.1714509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 2003; 12:525-32; PMID:14536091; http://dx.doi.org/ 10.1016/j.molcel.2003.08.001 [DOI] [PubMed] [Google Scholar]
- 5.Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al.. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 2013; 154:775-88; PMID:23932120; http://dx.doi.org/ 10.1016/j.cell.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 2011; 21:390-401; PMID:21163941; http://dx.doi.org/ 10.1101/gr.111070.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe KJ, Kane CM, Ares M Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 2003; 9:993-1006; PMID:12869710; http://dx.doi.org/ 10.1261/rna.5390803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolata J, Guo Y, Ko owerzo A, Smolinski D, Brzyzek G, Jarmo owski A, Swiezewski S. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J 2015; 34:544-58; PMID:25568310; http://dx.doi.org/ 10.15252/embj.201489478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16:167-77; PMID:25693130; http://dx.doi.org/ 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 2013; 339:950-3; PMID:23430654; http://dx.doi.org/ 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, Oren M. 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol 2014; 15:R69; PMID:24887486; http://dx.doi.org/ 10.1186/gb-2014-15-5-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 2014; 24:896-905; PMID:24714810; http://dx.doi.org/ 10.1101/gr.171405.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 2005; 13:22-9; http://dx.doi.org/ 10.1038/nsmb1030 [DOI] [PubMed] [Google Scholar]
- 14.Saint-André V, Batsché E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol 2011; 18:337-44; PMID:21358630; http://dx.doi.org/ 10.1038/nsmb.1995 [DOI] [PubMed] [Google Scholar]
- 15.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in Yeast. Mol Cell 2010; 40:582-93; PMID:21095588; http://dx.doi.org/ 10.1016/j.molcel.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, Zhu S, Yang L, Chen L-L. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792-806; PMID:24035497; http://dx.doi.org/ 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al.. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256-64; PMID:25664725; http://dx.doi.org/ 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 18.Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 2014; 53:779-90; PMID:24560925; http://dx.doi.org/ 10.1016/j.molcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative Pre-mRNA splicing. Cell 2011; 144:16-26; PMID:21215366; http://dx.doi.org/ 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swiezewski S, Crevillen P, Liu F, Ecker JR, Jerzmanowski A, Dean C. Small RNA-mediated chromatin silencing directed to the 3' region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci U S A 2007; 104:3633-8; PMID:17360694; http://dx.doi.org/ 10.1073/pnas.0611459104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 2014; 54:156-65; PMID:24725596; http://dx.doi.org/ 10.1016/j.molcel.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Tao Z, Dean C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev 2015; 29(7):696-701; [cited 2015 Mar 25]; Available from; PMID:25805848; http://dx.doi.org/ 10.1101/gad.258814.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 2003; 100:189-92; PMID:12502788; http://dx.doi.org/ 10.1073/pnas.0136770100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR 3rd, Bartel DP, Madhani HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 2013; 152:957-68; PMID:23415457; http://dx.doi.org/ 10.1016/j.cell.2013.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ares M, Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA 1999; 5:1138-9; PMID:10496214; http://dx.doi.org/ 10.1017/S1355838299991379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 2003; 9:618-30; PMID:12702820; http://dx.doi.org/ 10.1261/rna.5260303 [DOI] [PMC free article] [PubMed] [Google Scholar]


