Abstract
Background
Avermectin and milbemycin are important 16-membered macrolides that have been widely used as pesticides in agriculture. However, the wide use of these pesticides inevitably causes serious drug resistance, it is therefore imperative to develop new avermectin and milbemycin analogs. The biosynthetic gene clusters of avermectin and milbemycin have been identified and the biosynthetic pathways have been elucidated. Combinatorial biosynthesis by domain swap provides an efficient strategy to generate chemical diversity according to the module polyketide synthase (PKS) assembly line.
Results
The substitution of aveDH2-KR2 located in avermectin biosynthetic gene cluster in the industrial avermectin-producing strain Streptomyces avermitilis NA-108 with the DNA regions milDH2-ER2-KR2 located in milbemycin biosynthetic gene cluster in Streptomyces bingchenggensis led to S. avermitilis AVE-T27, which produced ivermectin B1a with high yield of 3450 ± 65 μg/ml. The subsequent replacement of aveLAT-ACP encoding the loading module of avermectin PKS with milLAT-ACP encoding the loading module of milbemycin PKS led to strain S. avermitilis AVE-H39, which produced two new avermectin derivatives 25-ethyl and 25-methyl ivermectin (1 and 2) with yields of 951 ± 46 and 2093 ± 61 μg/ml, respectively. Compared to commercial insecticide ivermectin, the mixture of 25-methyl and 25-ethyl ivermectin (2:1 = 3:7) exhibited 4.6-fold increase in insecticidal activity against Caenorhabditis elegans. Moreover, the insecticidal activity of the mixture of 25-methyl and 25-ethyl ivermectin was 2.5-fold and 5.7-fold higher than that of milbemycin A3/A4 against C. elegans and the second-instar larva of Mythimna separate, respectively.
Conclusions
Two new avermectin derivatives 25-methyl and 25-ethyl ivermectin were generated by the domain swap of avermectin PKS. The enhanced insecticidal activity of 25-methyl and 25-ethyl ivermectin implied the potential use as insecticide in agriculture. Furthermore, the high yield and genetic stability of the engineered strains S. avermitilis AVE-T27 and AVE-H39 suggested the enormous potential in industrial production of the commercial insecticide ivermectin and 25-methyl/25-ethyl ivermectins, respectively.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0337-y) contains supplementary material, which is available to authorized users.
Keywords: 25-Ethyl ivermectin, 25-Methyl ivermectin, Domain swap, Insecticidal activity
Background
Avermectins and milbemycins (Fig. 1), the 16-membered macrolide antibiotics with potent anthelmintic and insecticidal activity, have been widely used for broad-spectrum parasite control in agricultural, medical, and veterinary fields [1–3]. They are structurally related compounds with structural differences at C25, C22–C23, and C13, leading to their own unique ‘spectral fingerprint’ with various strengths and dosage-limiting species. The subsequent chemically modification of avermectins and milbemycins resulted in series of analogs, some of which are commercially developed as anthelmintics and insecticides, such as ivermectin, selamectin, abamectin, emamectin, doramectin, milbemycin oxime, lepimectin, and latidectin [2]. Among these analogs, ivermectin (22,23-dihydroavermectin B1), showing the same effective antiparasitic activity and lesser toxic side effect than avermectins B1, has been worldwide used as an anthelmintic for livestock and companion animals and as an agricultural insecticide. Moreover, ivermectin has also been applied in human medicine, particularly treatment of onchocerciasis and lymphatic filariasis [4]. In the case of milbemycins, moxidectin is currently undergoing a phase III clinical trial to compare its efficacy with ivermectin in subjects with Onchocerca volvulus infection [1]; milbemycin oxime has been used against intestinal nematodes in dogs and cats, against adult heartworm in dogs, and against ectoparasites in companion animals [5]. Recently, it has been reported that ivermectin, selamectin and moxidectin demonstrated antibacterial activity against Mycobacterium tuberculosis, especially the multidrug-resistant and extensively drug-resistant clinical strains [1]. The approval for clinical and veterinary uses as well as the documented pharmacokinetic and safety profiles of these compounds make them potential therapeutic options for treating M. tuberculosis. The outstanding activities of avermectins and milbemycins together with the potential uses in the filed of human medicine and agriculture stimulate the semisynthetic derivatives of avermectins and milbemycins. On the other hand, the wide use of avermectins in agriculture has inevitably caused serious drug resistance [2], it is therefore imperative to develop novel avermectins.
Fig. 1.
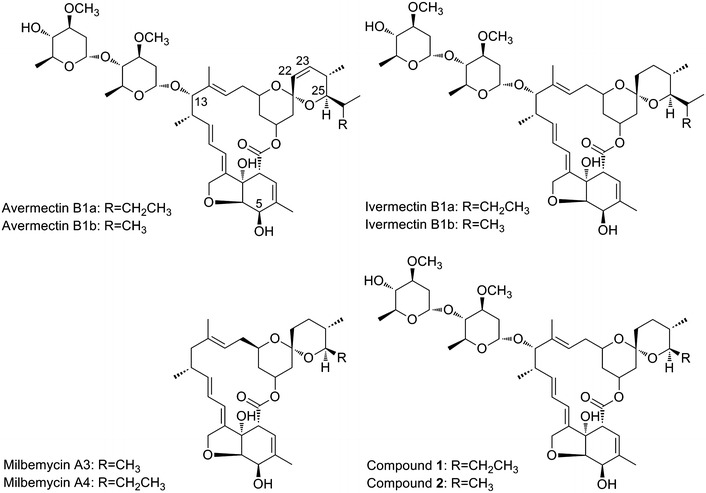
The structures of avermectins, milbemycins, and 25-methyl/25-ethyl ivermectin (1 and 2)
The previously established industrial process for preparing the analogs of avermectins and milbemycins involves extracting avermectins or milbemycins from the fermentation broth and the subsequently chemical modification. However, this process suffers several drawbacks, such as the expensive cost, the heavy metal pollution, and low efficiency [3, 6–8]. Fortunately, the combinatorial biosynthesis and genetic manipulation provide alternative and efficient strategies to generate the known semi-biosynthetic polyketides and new hybrid compounds according to the module polyketide synthase (PKS) assembly line [3, 9, 10]. For example, on the basis of understanding the biosynthetic mechanism of avermectin, 22,23-dihydroavermectins including ivermectin were successfully produced by direct fermentation of engineered S. avermitilis, in which the DNA region encoding the dehydratase (DH) and ketoreductase (KR) domains of module 2 from the avermectin PKS was replaced by the DNA fragment encoding the DH, enoylreductase (ER), and KR domains of module 13 from the rapamycin PKS, module 4 from the pikromycin PKS, or module 3 from the oligomycin PKS [6, 8, 11]. Traditionally, doramectin is produced by a bkd mutant S. avermitilis, which blocks the formation of the starter units (isobutyryl-CoA and 2-methylbutyryl-CoA) of avermectin biosynthesis, with the addition of cyclohexanecarboxylic acid (CHC) to the fermentation broth. While, it can be successfully produced without CHC supplementation if the CHC-CoA biosynthetic gene cassette was introduced into the bkd mutant S. avermitilis [12]. Furthermore, it has been reported that the replacement of the loading module of avermectin PKS by a cyclohexanecarboxylic unique loading module from phoslactomycin PKS combining with the introduction of the CHC-CoA biosynthetic gene cassette into the wild-type strain S. avermitilis led to efficiently produce doramectin without knocking out the bkd gene and CHC supplementation [13]. Recently, the biosynthetic gene cluster of milbemycin was characterized by the whole-genome sequencing of S. bingchenggensis [14]. The subsequent inactivation of the gene encoding a C5-ketoreductase led to the accumulation of 5-oxomilbemycins A3/A4, which can be used as the substrate for the semi-synthesis of milbemycin oxime through one step chemical reaction [7]. The milbemycin biosynthetic gene cluster mil in S. bingchenggensis consists of four large ORFs (milA1–milA4) encoding giant multifunctional polypeptides of PKS, some regulatory genes and genes encoding tailoring enzymes (Additional file 1: Figure S1), which are highly homologous to avermectin biosynthetic gene cluster ave [15]. Both milbemycin and avermectin PKSs consist of 12 modules, each of which contains distinctive active site domains catalyzing a specific round of polyketide chain elongation. However, the differences between the module and its counterpart, such as the module 2, the module 7 and the loading module, lead to the structural diversity of avermectin and milbemycin (Additional file 1: Figure S1). Base on the fact that avermectin and milbemycin are similar molecules, it appeared feasible to generate hybrid compounds sharing the structural features of avermectin and milbemycin by combinatorial biosynthesis.
In order to widen the insecticidal spectra of avermectins and milbemycins, it is desirable to generate new derivatives. Herein, ivermectin B1a was produced by the replacement of aveDH2-KR2 in the avermectin producer S. avermilitis MA-108 with milDH2-ER2-KR2 from S. bingchenggensis. The subsequent substitution of aveLAT-ACP encoding the loading module of avermectin PKS with milLAT-ACP from S. bingchenggensis led to two hybrid compounds 25-methyl and 25-ethyl ivermectin (Fig. 1) simultaneously sharing the structural features of avermectins and milbemycins and showing significantly enhanced insecticidal activity.
Results
Construction of aveDH2-KR2 replacement mutant to yield ivermectin
On the basis of understanding the biosynthetic pathways of avermectin and milbemycin, we attempted to construct an ivermectin-producing strain by replacing the aveDH2-KR2 of avermectin biosynthetic gene cluster in S. avermitilis NA-108 with milDH2-ER2-KR2 of milbemycin biosynthetic gene cluster from S. bingchenggensis (Fig. 2). PCR verification using the primers E1 and E2 (Additional file 2: Table S1) demonstrated that the expected 2.6-kb DNA fragment encoding MilDH2-ER2-KR2 was obtained from the genomic DNA of S. bingchenggensis and the double-crossover mutant S. avermitilis AVE-T27, whereas no PCR product was detected from the genomic DNA of S. avermitilis NA-108 (Fig. 3a, b). The PCR product was then sequenced, and the results further confirmed that aveDH2-KR2 in avermectin biosynthetic gene cluster was successfully replaced with milDH2-ER2-KR2 by double crossover. Compared to the parental strain S. avermitilis NA-108, strain AVE-T27 demonstrated a different metabolic profile. As shown in Fig. 4, in addition to the disappearance of avermectin “a” components and the remarkable decrease of avermectin “b” components, a new compound, which showed identical retention time and molecular mass (m/z = 897, [M+Na]+) to those of the authentic sample ivermectin B1a, was detected in culture extracts of S. avermitilis AVE-T27 by HPLC analysis. Therefore, the compound was considered to be ivermectin B1a, which was consistent with the designed biosynthetic strategy (Fig. 2).
Fig. 2.
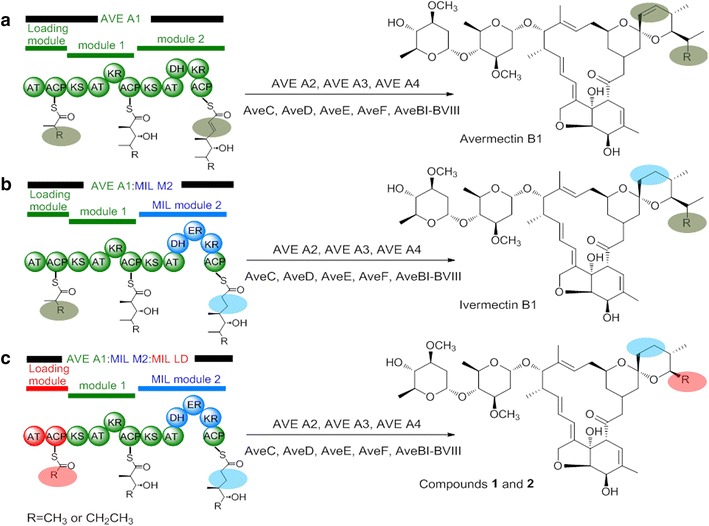
Predicted products of engineered avermectin PKS. a AVE A1 of avermectin PKS. b AveDH2-KR2 in module 2 of avermectin PKS was replaced with MilDH2-ER2-KR2 of milbemycin PKS. c AveDH2-KR2 in module 2 and loading module AveLAT-ACP of avermectin PKS were replaced with MilDH2-ER2-KR2 of and loading module MilLAT-ACP of milbemycin PKS, respectively
Fig. 3.
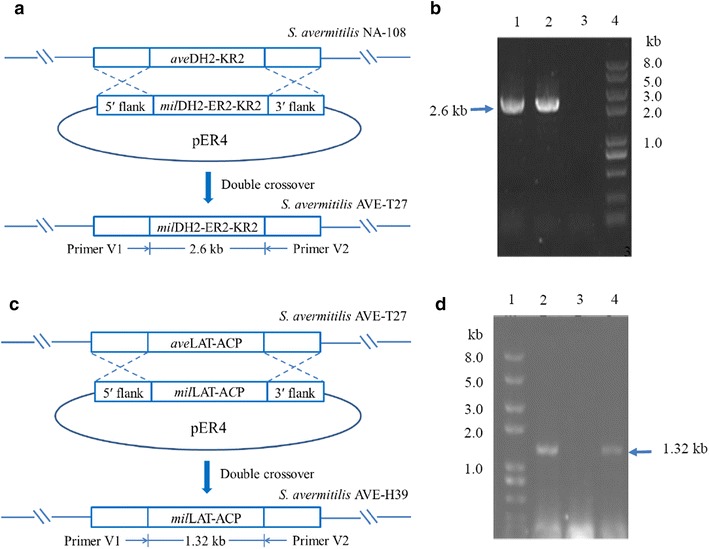
Gene replacement in S. avermitilis NA-108. a Schematic description of the gene replacement of aveDH2-KR2 in S. avermitilis NA-108 with milDH2-ER2-KR2 from S. bingchenggensis via double crossover. b PCR analysis with genomic DNA from S. avermitilis NA-108, S. avermitlis AVE-T27 and S. bingchenggensis, using primers E1 and E2. Lane 1 S. bingchenggensis; Lane 2 S. avermitilis AVE-T27; Lane 3 S. avermitilis NA-108; Lane 4 DNA ladder. c Schematic description of the gene replacement of aveLAT-ACP in S. avermitilis AVE-T27 with milLAT-ACP from S. bingchenggensis via double crossover. d PCR analysis with genomic DNA from S. avermitilis AVE-T27, S. avermitlis AVE-H39 and S. bingchenggensis, using primers V1 and V2. Lane 1 DNA ladder; Lane 2 S. bingchenggensis; Lane 3 S. avermitilis AVE-T27; Lane 4 S. avermitilis AVE-H39
Fig. 4.
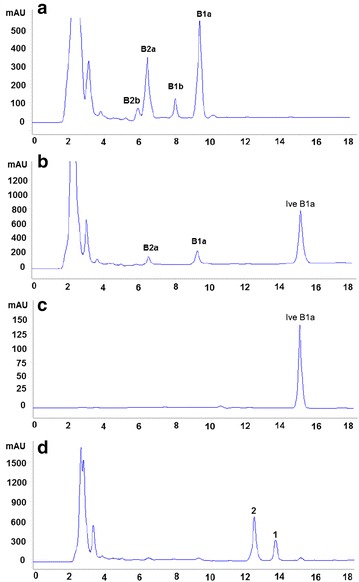
HPLC analysis of the mycelial extracts from the parental and mutant strains. a S. avermitilis NA-108; b S. avermitlis AVE-T27; c standard sample ivermectin B1a; d S. avermitilis AVE-H39. B1a avermectin B1a, B2a avermectin B2a, B1b avermectin B1b, B2b avermectin B2b, Ive B1a ivermectin B1a, 1 compound 1, 2 compound 2
Second domain swap to yield new hybrid antibiotics
In order to obtain more avermectin analogs and further investigate the structure–activity relationship of milbemycins and avermectins, the loading module of avermectin PKS encoding by aveLAT-ACP in the ivermectin-producing strain S. avermitilis AVE-T27 was replaced by that of milbemycin PKS encoding by milLAT-ACP in milbemycin-producing strain S. bingchenggensis (Fig. 2). After the successive treatment of the transformants growing at a non-permissive temperature (39 °C) and under non-selective antibiotic pressure, 100 apramycin-sensitive strains were randomly selected and examined by fermentation experiments and HPLC analysis. Compared to strain AVE-T27, 91 apramycin-sensitive strains demonstrated distinct metabolic profiles, in which ivermectin B1a, avermectins B1a and B2a disappeared but two new compounds 1 and 2 with a molecular ion at m/z = 869.53 [M+Na]+ and 855.30 [M+Na]+, respectively, were detected (Additional file 3: Figure S2). The double crossover event occurred in strain AVE-H39 with the highest yield of the new compounds was then verified by PCR analysis using a pair of primers V1/V2 (Additional file 2: Table S1), which was designed to amplify the internal fragment of milLAT-ACP. As shown in Fig. 3d, the expected 1.32-kb DNA fragment was obtained from the genomic DNA of S. bingchenggensis and the mutant strain AVE-H39, whereas no PCR product was detected from the genomic DNA of AVE-T27. The result implied that the aveLAT-ACP in strain AVE-T27 was successfully replaced with milLAT-ACP by double crossover, which was further confirmed by sequencing the PCR product.
The structure elucidation of hybrid compounds 1 and 2
Compound 1 (Fig. 1) was isolated as a white amorphous powder. Its molecular formula was determined to be C46H70O14 on the basis of HRESIMS at m/z 869.4652 [M+Na]+ (calcd 869.4663 for C46H70O14Na) and 13C NMR data (Additional file 4: Table S2). The 1H NMR spectrum of compound 1 (Additional file 5: Figure S3) displayed four doublet aliphatic methyls at δ 0.82 (d, J = 6.3 Hz), 1.15 (d, J = 6.9 Hz), 1.25 (d, J = 6.1 Hz), 1.27 (d, J = 6.4 Hz); two olefinic methyl signals at δ 1.50 (s), 1.87 (s); two methoxy signals at δ 3.42 (s), 3.43 (s); one trans-double bond at 5.72 (overlap), 5.73 (overlap). Its 13C NMR (Additional file 6: Figure S4) revealed 46 carbon resonances, including an ester carbonyl carbon at δ 174.07, a ketal carbon at 97.63, five sp2 methines, three sp2 quaternary carbons, four secondary methyls, two vinylic methyls, one primary methyl, nine methylenes, seventeen aliphatic methines (fourteen oxygenated), one oxygenated quaternary carbon, and two methoxy groups. Comparison of the 1H and 13C NMR spectral data of 1 with those of avermectin B1b revealed that compound 1 was structurally related to avermectin B1b except for the differences at C22–C23 and C25, where the isopropyl group at C25 and the unsaturated bond between C22 and C23 in avermectin B1b were replaced by an ethyl group and a saturated bond in compound 1, respectively [16]. The correlations of H-25, H-26 and H-27 in the 1H-1H COSY spectrum and the HMBC correlations from H-26 to C-25 and C-27 confirmed the presence of an ethyl group at C-25 in compound 1 (Additional file 7: Figure S5). The HMBC correlation between H-22, C-21, C-23 and C-24 indicated the saturated bond between C-22 and C-23. As a consequence, the gross structure of 1 was established as 25-ethyl ivermectin (Fig. 1). Compound 2 was also obtained as a white amorphous powder. Its molecular formula was determined to be C45H68O14 on the basis of HRESIMS at m/z 855.4508 [M+Na]+ (calcd 855.4507 for C45H68O14Na). Comparison of the 1H and 13C NMR data (Additional file 4: Table S2) with those of 1 indicated that compound 2 and 1 differed only at C25, in that the ethyl group at C-25 in compound 1 was replaced by a methyl group in 2, which was supported by 1H-1H COSY correlations of H-25 and H-26 and HMBC correlations from H26 to C24 and C25 (Additional file 7: Figure S5). As a result, the structure of 2 was established as 25-methyl ivermectin (Fig. 1). The 1D- and 2D-NMR spectra of compounds 1 and 2 can be found in the supplemental material (Additional files 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15: Figures S3–S13).
The bioactivity of hybrid compounds 1 and 2
The insecticidal activity of compounds 1 and 2 against C. elegans and M. separate was evaluated and compared to that of commercial insecticides milbemycin and ivermectin. The results demonstrated that the lethal effects of the tested compounds were enhanced with increased concentrations, indicating that the insecticidal activity was dose-dependent. However, the mixture of 1 and 2 possessed higher insecticidal activities against C. elegans and M. separate than milbemycin and ivermectin. As listed in Table 1, the LC50 of the mixture of 1 and 2 is 2.2 ± 0.7 μg/ml, which is approximately 2.5-fold and 4.6-fold lower than that of milbemycin (5.42 ± 2.1 μg/ml) and ivermectin (10.1 ± 1.3 μg/ml), respectively. Moreover, it exhibited impressive insecticidal activity against M. separata with a LC50 of 19.4 ± 1.3 μg/ml, which is 5.7-fold lower than that of milbemycin (110.7 ± 7.4 μg/ml).
Table 1.
Insecticidal activity of the mixture of 1 and 2 compared with those of milbemycins A3/A4 and ivermectin
| Samples | LC50 (μg/ml)c | |
|---|---|---|
| C. elegans | M. separata | |
| The mixture of 1 and 2 a | 2.2 ± 0.7 | 19.4 ± 1.3 |
| Milbemycin A3/A4b | 5.4 ± 2.1 | 110.7 ± 7.4 |
| Ivermectin | 10.1 ± 1.3 | |
aThe mixture contains 70 % compound 1 and 30 % compound 2
bMilbemycins A3/A4 contains 70 % A4 and 30 % A3
cValues are the means ± SDs of three independent experiments
Genetic stability of AVE-T27 and AVE-H39
The genetic stabilities of strains AVE-T27 and AVE-H39 to produce ivermectin B1a and compounds 1 and 2 were evaluated by five successive subcultivation tests. The shaking flask experiments and HPLC analysis showed that the yield of ivermectin B1a and 25-methyl/25-ethyl ivermectin produced by AVE-T27 and AVE-H39 among five generations ranged from 3450 ± 65 to 3510 ± 37 μg/ml and 3045 ± 78 to 3087 ± 46 μg/ml, respectively. These results suggested that strains AVE-T27 and AVE-H39 were genetically stable and could be applied in industry.
Discussion
Since the discovery of avermectin in 1975, many efforts have been devoted to its structural modification to yield avermectin derivatives. To date, six avermectins (abamectin, ivermectin, doramectin, emamectin, eprinomectin and selamectin) have been commercialized and widely used as anthelmintic and insecticidal drugs in animal health and agriculture [17]. The structural differences of these drugs are located on C25, C22–C23 and C13 positions. For example, ivermectin (22,23-dihydroavermectin B1), which is synthesized by chemical reduction of the double bond between C22 and C23 of avermectins B1, exhibits the same effective antiparasitic activity but lesser toxic side effect than avermectins B1 [18]. Doramectin (C25-cyclohexyl avermectin B1), an avermectin analog with a cyclohexyl group at C25, demonstrates better pharmacokinetic properties and efficacy than avermectin [19]. Compared to avermectin, eprinomectin, emamectin and selamectin possess different groups at C13 and show unique pharmacokinetic characteristics. The various applications of avermectins in agricultural and veterinary fields have attracted much attention, and more avermectin analogs with high insecticidal and acaricidal activity have been reported [20, 21]. Milbemycins are a group of 16-membered macrolides chemically related to avermectins with the major structural differences at C25, C22–C23 and C13. Due to the high and broad-spectrum activity against insects and parasites, low toxicity to mammals, and environment friendliness, milbemycins have received considerable interest in agricultural chemistry, and four kinds of milbemycins including milbemectin (a mixture of milbemycins A3/A4), milbemycin oxime, lepimectin, and latidectin have been marketed and used as a broad-spectrum acaricide, anthelmintic and insecticide [7, 22]. Although the natural avermectins and milbemycins demonstrate structural differences, some commercial avermectins and milbemycins share the same structural features. For example, selamectin and ivermectin contain a saturated bond at C22–C23, which is similar to milbemycins; while latidectin and lepimectin contain 13-substituted groups, the modified pattern of which is different from milbemectin and milbemycin oxime, but similar to avermectins. Inspired by the structure–activity relationship of avermectins and milbemycins, series of compounds with the partial structural features of milbemycins and avermectins were synthesized and showed potential insecticidal activity [23].
The polyketide backbond of avermectin and milbemycin is biosynthesized by the multifunctional and multimodular proteins referred as type I polyketide synthetases (type I PKSs). The type I PKSs consist of several modules, each of which always carries three core domains acyltransferase (AT), acyl carrier protein (ACP) and ketosynthase (KS) as well as other accessory domains KR, DH and ER [24]. The organization of the modular PKSs allows a wide range of complex polyketide products to be assembled from simple precursors. Given the increased understanding of biosynthetic machineries of polyketides, combinatorial biosynthesis by rearranging the domains in PKSs has been developed as an alternative strategy to generate novel unnatural polyketides [25, 26]. Although milbemycin biosynthetic gene cluster mil and avermectin biosynthetic gene cluster ave demonstrate different gene organizations, both of them contain four large ORFs (milA1–milA4/aveA1–aveA4), a regulatory gene (milR/aveR) and some genes encoding tailoring enzymes (milC/aveC, milE/aveE, milF/aveF, milD/aveD) (Additional file 1: Figure S1). The four large ORFs encode the PKS, which is responsible for the biosynthesis of polyketide backbond and consists of a loading module and 12 elongating modules. These homologous ORFs in mil and ave share high identity in protein sequences ranging from 48.41 to 64.78 %, leading to a similar polyketide backbone observed in milbemycin and avermectin. However, the differences at the loading module, module 2 and module 7 are believed to be associated with the structural changes at C25, C22–C23 and C13 positions. Inspired by the structural diversity and impressive bioactivity of milbemycins and avermectins, two new avermectin derivatives were designed by reconstructing the biosynthetic pathway of avermectins (Fig. 2). Firstly, the aveDH2-KR2 of avermectin biosynthetic gene cluster in S. avermitilis NA-108 was replaced by milDH2-ER2-KR2 of milbemycins biosynthetic gene cluster from S. bingchenggensis to yield the ivermectin-producing strain AVE-T27. Compared to the parental strain NA-108, strain AVE-T27 no longer produced “b” components of avermectins (Fig. 4b), and the yield of avermectin B1a and B2a (259 ± 23 and 173 ± 18 μg/ml, respectively)decreased approximately 7 fold and 2.3 fold, respectively. The remarkable decrease in the yield of avermectins was also observed in other ivermectin-producing strains [6, 8, 11]. It is obvious that strain NA-108 only produces four components of avermectins, of which the “a” components B1a and B2a are produced in quantity (Fig. 4a), suggesting that the loading module of the avermectin PKS in strain NA-108 may dominantly utilize 2-methylbutyryl as the starter unit for polyketide assembly. Therefore, the substitution of aveDH2-KR2 by milDH2-ER2-KR2 resulted in the substantial accumulation of ivermectin B1a, whereas the yield of ivermectin B1b may be too low to be detected (Fig. 4b). Before the thorough understanding the function of AveC in avermectin biosynthesis [27], the unsaturated bond of C22–C23 was considered to be formed through the optional dehydration by partially active dehydratase (DH) in module 2 with the assistance of AveC [28]. Therefore, several attempts have been made to construct ivermectin-producing strains by replacing the partially active DH with completely active DH and ER from other PKSs through the genetic manipulation of avermectin biosynthetic gene cluster. However, the yield of ivermectin produced by the genetic engineered strains is too low for large-scale production probably due to the poor folding of the hybrid PKSs and unfavorable protein–protein interactions [6, 8, 11]. Thus, ivermectin is still synthesized by a regiospecific hydrogenation at C22–C23 of avermectins using rhodium chloride as the catalyst, even though this process suffers from expensive cost and heavy metal pollution [6]. It is worth noting that strain AVE-T27 demonstrated excellent performance in producing ivermectin B1a with a yield of 3450 ± 65 μg/ml, which is only slightly lower than the total amount of avermectins (3577 ± 26 μg/ml) produced by the parental strain NA-108. It is commonly reported that PKS bioengineering experiments often produce new compounds at compromised yields [6, 8, 10, 11, 24, 29]. For examples, the total amount of avermectins and ivermectins produced in the ivermectin-producing strains Ive12-4 and OI-31, which were constructed by replacing the AveDH2-KR2 in S. avermitilis Olm73-12 with PikDH4-ER4-KR4 from pikromycin PKS and OlmDH3-ER3-KR3 from oligomycin PKS, respectively, was estimated to be only 1–3 % of the amount of avermectins produced by the parental strain or decreased approximately eightfold [6, 8]. Compared to PikDH4-ER4-KR4 (46.48 %) and OlmDH3-ER3-KR3 (43.06 %), MilDH2-ER2-KR2 demonstrates a higher identity (51.50 %) with AveDH2-KR2. Furthermore, both of MilDH2-ER2-KR2 and AveDH2-KR2 catalyze the reduction of C22–C23 in the similar polyketide backbone. Therefore, it is no surprise that the substitution of AveDH2-KR2 with MilDH2-ER2-KR2 could not lead to the drastic decrease in the total amount of avermectins and ivermectin. Indeed, the use of alternative domains and modules from PKSs of structurally related polyketides is considered as an efficient approach to generate hybrid PKSs with high yield [30]. The high yield of ivermectin B1a produced by strain AVE-T27 implied that the DH2-ER2-KR2 from milbemycin PKS may be more suitable for swapping AveDH2-KR2 than the domains from other PKS reported previously [6, 8, 11]. We speculated that the domain swapping between AveDH2-KR2 and MilDH2-ER2-KR2 does not seriously affect the substrate specificity of the KS domain and dynamic KS:ACP interaction, which are believed to play crucial roles in polyketide chain transfer and elongation [10, 24]. However, strain AVE-T27 still produced a small amount of avermectins B1a and B2a, suggesting the incompletable function of MilDH2-ER2-KR2 in hybrid PKS. Recently, Sun et al. have revealed a dual function of AveC, which works as a spirocyclase and an independent dehydratase, involved in the spiroketal formation and the optional dehydration of C22–C23 during the avermectin biosynthesis [27]. Due to the complete inactivity of AveDH2, AveC was believed to be the only factor responsible for C22-C23 dehydration to yield the “1” components of avermectins. This dehydration reaction is post-PKS but precedes spiroketal formation, and the spirocyclase activity of AveC can tolerate variation of the C22-C23 bond. Therefore, we assumed that parts of avermectin B1a produced by AVE-T27 are formed by the incompletable activity of MilDH2-ER2-KR2 during skeleton assembly process, while the others are probably associated with the optional dehydratase activity of AveC in the post-PKS modification.
Loading modules of type I PKSs catalyze selection and recruitment of starter units into polyketides. Accordingly, starter units are incorporated into polyketides at only one position, and swapping of the loading module can lead to changes in the side chain originating from the starter unit [24, 31]. For example, the replacement of the loading module of the erythromycin PKS or spinosyn PKS by that of avermectin PKS resulted in the production of novel erythromycin or spinosyn analogs [32, 33]. Recently, it has been reported that the replacement of the loading module of avermectin PKS with that of phoslactomycin PKS, which prefers cyclohexylcarbonyl-CoA as the start unit, led to enhanced ratio of doramectin to avermectins produced by genetic engineered S. avermitilis [13]. Therefore, the loading module of avermectin PKS could be a suitable site for module swapping to generate unnatural avermectins. After the replacement of the loading module of avermectin PKS in strain AVE-T27 by that of milbemycin PKS, two new avermectin derivatives with a methyl or ethyl group at C25 position were obtained (Fig. 1). The loading module of avermectin PKS possesses broad substrate specificity, for example, it can use 2-methylbutyryl-CoA, isobutyryl-CoA and cyclohexylcarbonyl-CoA as substrates to biosynthesize avermectin “a” components, avermectin “b” components and doramectin, respectively [2, 34]. However, the loading module of milbemycin in S. bingchenggensis seems to only use acetyl-CoA and propionyl-CoA as substrates to biosynthesize 25-methyl and 25-ethyl milbemycins, respectively [15]. Therefore, it was speculated that the loading module of avermectin PKS possesses a broader substrate specificity than that of milbemycin PKS. Compared to the yield of ivermectin produced by AVE-T27, the total amount of 25-methyl and 25-ethyl ivermectin (3045 ± 78 μg/ml) produced by AVE-H39 is satisfory. However, no avermectins with C25-isopropyl, C25-isobutyl groups were detected (Fig. 4d), which confirmed the relatively narrow substrate specificity of the loading module of milbemycin PKS. Although the loading module of avermectin PKS exhibits an identity of 46.54 % with that of milbemycin PKS on protein level, it seems that the domain swapping between them did not seroiusly affect the biosynthetic ability of the resultant hybrid PKS. The high yield of 25-methyl and 25-ethyl ivermectin again confirmed that the use of alternative modules from PKSs of structurally related polyketides is an efficient approach to generate hybrid polyketides.
The previous structure–activity relationship studies of avermectins suggested that C5, C13 and C22–C23 are essential for the bioactivity, and the subsequent modification at these positions successfully developed several commercial drugs such as ivermectin, emamectin, eprinomectin and selamectin [17]. Although some C25-substitituted avermectins were designed [34–36], only doramectin with a cyclohexyl group at C25 was commercialized. Compared to ivermectin, compounds 1 and 2 exhibited significantly enhanced insecticidal activity, implying that the C25 is a potential target for structural modification, which provides new clues for the structural modification of avermectins.
Conclusions
In conclusion, 25-methyl and 25-ethyl ivermectin were obtained by domain swapping. The impressive insecticidal activity of these two new compounds suggested their promising use as novel insecticides in agriculture. Furthermore, the high yield and genetic stability of the engineered strains S. avermitilis AVE-T27 and AVE-H39 highlight their potential in industrial production of ivermectin B1a and 25-methyl/25-ethyl ivermectin, respectively.
Materials and methods
Strains, vectors, reagents, and cultivation
All bacterial strains and plasmids used in this study are listed in Table 2. The initial strain S. avermitilis NA-108, originating from the mutation of S. avermitilis NEAU1069, can only produces “B” components of avermectins with high yield but does not produce “A” components of avermectins and oligomycin. S. avermitilis NEAU1069 and S. bingchenggensis have been deposited at the China General Microbiology Culture Collection Center, Institute of Microbiology, Chinese Academy of Science with accession numbers of CGMCC2943 and CGMCC1734, respectively. The 16S rDNA sequence of S. avermitilis NEAU1069 and S. bingchenggensis was deposited in GenBank with accession numbers of DQ768097 and DQ449953, respectively. Primers (Additional file 2: Table S1) were synthesized in Sangon Biotech (Shanghai, China). Restriction endonucleases, DNA ligase, and DNA polymerases were purchased from TaKaRa Biotechnology (Dalian, China). DNA sequencing was performed by GenScript (Nanjing, China). MS (mannitol soya flour) medium was used for S. avermilitis NA-108 sporulation and conjugation between Escherichia coli and Streptomyces. YEME (yeast extract-malt extract) medium containing 25 % sucrose was used to grow mycelia for the isolation of total DNA [37]. Luria–Bertani (LB) medium was used for E. coli propagation [38]. All E. coli procedures were performed according to standard protocols [38]. Isolation of genomic DNA from S. avermitilis NA-108 and agarose gel electrophoresis were performed according to the standard methods [37, 38].
Table 2.
Strains and plasmids used in this study
| Strains/plasmids | Relevant characteristicsa | References |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Host strain for cloning | Invitrogen |
| ET12567/pUZ8002 | Donor strain for conjugation | Kieser et al. [37] |
| S. bingchenggensis | The milbemycin-producing strain | Wang et al. [14] |
| S. avermitilis | ||
| NA-108 | Mutant strain of S. avermitilis NEAU1069 producing “B” components of avermectins as main secondary metabolites | This study |
| AVE-T27 | Mutant strain of NA-108 with the replacement of aveDH2-KR2 by milDH2-ER2-KR2 | This study |
| AVE-H39 | Mutant strain of AVE-T27 with the replacement of aveLAT-ACP by milLAT-ACP | This study |
| Plasmids | ||
| pUC19 | Cloning vector for E. coli, Amp R | TaKaRa |
| pKC1139 | E. coli-Streptomyces shuttle vector, Am R | Kieser et al. [37] |
| pER-1 | pUC19 derivative containing aveDH2-KR2 upstream fragment, Amp R | This study |
| pER-2 | pUC19 derivative containing aveDH2-KR2 upstream and downstream fragments, Amp R | This study |
| pER-3 | pUC19 derivative containing milDH2-ER2-KR2 together with aveDH2-KR2 upstream and downstream fragments, Amp R | This study |
| pER-4 | pKC1139 derivative containing milDH2-ER2-KR2 together with aveDH2-KR2 upstream and downstream fragments, Am R | This study |
| pLM-1 | pMD19-T derivative containing milLAT-ACP together with aveLAT-ACP upstream and downstream fragments, Amp R | This study |
| pLM-2 | pKC1139 derivative containing milLAT-ACP together with aveLAT-ACP upstream and downstream fragments, Amp R | This study |
a Amp R ampicillin resistance, Am R apramycin resistance
Construction of aveDH2-KR2 replacement mutant strain
In order to replace the DNA region encoding DH2-KR2 of avermectin PKS (aveDH2-KR2) with the DNA fragment encoding DH2-ER2-KR2 of milbemycin PKS (milDH2-ER2-KR2), recombinant plasmid pER-4 was constructed. Using genomic DNAs of S. avermitilis NA-108 as template, a 1106-bp fragment upstream of the aveDH2-KR2 was amplified with primers a1 and a2. The amplified fragment was digested with HindIII/XbaI and ligated into corresponding sites of pUC19 to generate pER-1. Then, a 1055-bp fragment downstream of the aveDH2-KR2 amplified with primers c1 and c2 was digested with XbaI/EcoRI and then ligated to the corresponding sites of pER-1 to yield pER-2. The introduction of XbaI site did not change the natural amino acids sequence at the linker regions. The milDH2-ER2-KR2 (3120-bp) fragment was amplified with primers b1 and b2 using genomic DNAs of S. bingchenggensis as template, and the resultant PCR products were cloned into the XbaI sites of pER-2 to give pER-3. The 5.27-kb insert was recovered from pER-3 by digesting with HindIII/EcoRI and inserted into the same sites of pKC1139 to generate pER-4. After the verification by PCR amplification and restriction digestion analysis, pER-4 was transformed into the non-methylating E. coli ET12567/pUZ8002. The conjugations were performed using the spores of S. avermitilis NA-108 according to the literature [16] and exconjugants were selected using MS agar containing apramycin. These cultures were grown at 28 °C for 2 days, then at 39 °C for 7–10 days. The colonies that were apramycin-resistant at 39 °C were identified as the integrating mutants, in which a single-crossover homologous recombination event took place. Insertion mutants were confirmed by PCR analysis and inoculated on nonselective MS plates at 28 °C for a second round of recombination. Double crossover mutants were screened by replica from the colonies grown on the MS medium. Mutants that lost resistance to apramycin were selected for further screening and genotypic confirmation by PCR using E1 and E2 as primers. The obtained double-crossover mutant was designated as AVE-T27.
Construction of aveLAT-ACP replacement mutant strain
To construct plasmid pLM-2 for replacement of the gene encoding the loading module of avermectin PKS (aveLAT-ACP) with that of milbemycin PKS (milLAT-ACP), the overlap extension PCR was employed [39]. Firstly, three pairs of primers A1/A2, B1/B2 and C1/C2 were designed to amplified 5′flank of aveLAT-ACP (1008-bp), milLAT-ACP (1407-bp) and 3′flank of aveLAT-ACP (1022-bp), respectively. Then a two-step PCR was employed: in the first round PCR, three fragments were mixed in a ratio of 1:3:1 (total DNA amounts about 250 ng), with 0.2 mM of each dNTP, 0.5 U PrimeSTAR HS DNA polymerase (Takara, Dalian, China), 1× PrimeSTAR Buffer (Mg2+ plus), and 10 % DMSO, the PCR program consisted of 12 repetitive cycles with a denaturation step at 94 °C for 30 s, an annealing step at 57 °C for 15 s and an elongation step at 72 °C for 3 min; in the second round PCR, 4 μl reactants from above was took as template, primers A1/C2 and other reagents were added, the PCR program consisted of 25 repetitive cycles with a denaturation step at 94 °C for 30 s, an annealing step at 60.6 °C for 15 s and an elongation step at 72 °C for 4 min. A 3400-bp fragment was obtained and ligated to pMD19-T (Takara, Dalian, China) to generate pLM-1. After sequencing and PCR analysis, the 3.4-kb fusion fragment was recovered from pLM-1 by digesting with HindIII/XbaI and inserted into the same sites of pKC1139 to generate pLM-2. Following the procedure described above, pLM-2 was introduced into strain AVE-T27 for a second replacement. After the validation by PCR amplification using V1 and V2 as primers, the obtained double-crossover mutant was designated as AVE-H39.
Fermentation and HPLC analysis of antibiotic production
S. avermitilis NA-108 and all genetically engineered strains were fermented under the same culture condition. The strains were firstly cultured in seed medium (corn starch 2 %, glucose 0.5 %, yeast extract 1 %, cottonseed cake 1 %, and CoCl2·6H2O 0.005 %, pH 7.2) at 28 °C for 42 h on a rotary shaker at 250 rpm. Then 2.0 ml of the culture was transferred into 250-ml Erlenmeyer flasks containing 25 ml of the fermentation medium consisting of corn starch 80 g/l, glucose 5 g/l, peptone10 g/l, cottonseed cake 10 g/l, NaCl 1 g/l, K2HPO4·3H2O 1 g/l, MgSO4·7H2O 1 g/l, CaCO3 1 g/l and CoCl2·6H2O 5 mg/l, pH 7.0. Fermentation was carried out at 28 °C for 8 days on a rotary shaker at 250 rpm. After finishing the fermentation, the broths were mixed with an equal volume of methanol. The resultant mixture was then centrifuged at 12,000×g for 20 min, and the supernatant was filtered through a 0.22 μm membrane filter and analyzed by HPLC. HPLC was performed with a Shimadzu LC-2010CHT system (Shimadzu, Koyoto, Japan) by using a NOVA-PAKR C18 column (3.9 × 150 mm, 5 μm, Waters, Milford, MA) at a flow rate of 1.0 ml/min, CH3OH/H2O (85:15, v/v) and detected at 246 nm. For LC/MS analysis, the positive electrospray ionization mass spectra were obtained from a Waters Q-TOF Micro LC–MS-MS spectrometer (electrospray voltage, 3.5 kV; heated capillary temperature, 450 °C; gas flow, 900 l/h) coupled with a Agilent HPLC 1200 system equipped with a Eclipse × DB-C18 column (4.6 × 150 mm, 5 μm) using a mobile phase of CH3OH/H2O (80:20, v/v). The flow rate was 0.3 ml/min, and the detection was at 246 nm.
Purification of hybrid antibiotics
The hybrid antibiotics produced by the recombinant strain AVE-H39 were purified from the culture broth. After the fermentation, the broth (5 l) was centrifugated, and the resulting cake was washed with H2O. MeOH (10 l) was used to extract the washed cake. The MeOH extract was evaporated under reduced pressure to approximately 0.2 l at 45 °C and the resulting concentrate was extracted three times using an equal volume of EtOAc. The combined EtOAc phase was concentrated under reduced pressure to yield oily substances. The residual oily substance was chromatographed on silica gel and eluted with a petroleum ether-acetone mixture (95:5–50:50, v/v). The fractions eluted with the petroleum ether-acetone mixture (95:5–75:25, v/v) were combined and evaporated to obtain a crude mixture. The crude mixture was then subjected to semipreparative HPLC (Agilent 1100, Zorbax SB-C18, 5 μm, 9.4 × 250 mm; 1.5 ml/min; 246 nm) by eluting with CH3OH-H2O (80:20, v/v) to afford compound 1 and 2.
Structural analysis and bioactivity of hybrid antibiotics
The HRESI-MS spectra were taken on a Q-TOF Micro LC–MS-MS spectrometer (Waters, Milford, MA, USA) and an API QSTAR time-of-flight spectrometer, respectively. 1D- and 2D-NMR spectra were recorded on Bruker AM-400 and DRX-600 spectrometers (Bruker, Rheinstetten, Germany) using 2.5 mm microcells (Synthware) for CDCl3. The 7.26 ppm and 77.23 ppm resonance of CDCl3 was used as internal references for 1H NMR spectra and 13C NMR spectra, respectively. The insecticidal activity of the mixture of compounds 1 and 2 against the second-instar larva of M. separata was measured by the leaf-dipping method [17]. Briefly, the test sample was dissolved in methanol at a concentration of 100 μg/ml and diluted to appropriate concentrations with distilled water containing 1 % Tween 80. Wafer discs (1-cm diamemter, 1-mm-thick) made from fresh wild corn leaves were dipped into the diluted solutions of the test sample for 3 s, and then taken out and kept in a conditioned room. The number of dead larvae was recorded at 48 h after treatment, and the corrected mortality was calculated. The insecticidal activity of the test sample against C. elegans was evaluated according to methods described previously [40]. The appropriate amounts of the diluted solutions of the test sample were added to 1-l portion of an aqueous suspension containing living nematodes C. elegans. The mixtures were kept at 25 °C for 15 h after shaking. The number of nematodes that were immobilized and the total number of nematodes tested were counted under a stereoscopic microscope. Immobilized rates against the total number of tested nematodes were calculated. The insecticidal activity was expressed as LC50 values, which were calculated by the LOGIT method. Milbemycin A3/A4 (A3:A4 = 3:7) and ivermectin were used as positive controls.
Authors’ contributions
JZ, YJY, JA carried out experiments and analyzed the primary data. SXH assisted with data analysis of MS and NMR. JZ wrote the manuscript. XJW and WSX supervised the whole research work and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by grants from the National Outstanding Youth Foundation (No. 31225024), the National Key Technology R&D Program (No. 2012BAD19B06), the Program for New Century Excellent Talents in University (NCET-11-0953), the National Natural Science Foundation of China (Nos. 31372006, 31171913, and 31071750), the Outstanding Youth Foundation of Heilongjiang Province (JC201201), and Chang Jiang Scholar Candidates Program for Provincial Universities in Heilongjiang (CSCP).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
Additional file 1: Figure S1. Organizations of avermectin and milbemycin biosynthetic gene clusters.
Additional file 2: Table S1. Primers used for gene cloning, constructing and confirming the mutants.
Additional file 3: Figure S2. The ESI-MS spectra of compounds (a) 1 and (b) 2.
Additional file 4: Table S2. 1H and 13C NMR data of compounds 1 and 2.
Additional file 5: Figure S3. 1H NMR spectrum of compound 1 in CDCl3 (400 MHz).
Additional file 6: Figure S4. 13C NMR spectrum of compound 1 in CDCl3 (100 MHz).
Additional file 7: Figure S5. The 1H-1H COSY and HMBC correlations of compounds 1 and 2.
Additional file 8: Figure S6. COSY spectrum of compound 1.
Additional file 9: Figure S7. HSQC spectrum of compound 1.
Additional file 10: Figure S8. HMBC spectrum of compound 1.
Additional file 11: Figure S9. 1H NMR spectrum of compound 2 in CDCl3 (400 MHz).
Additional file 12: Figure S10. 13C NMR spectrum of compound 2 in CDCl3 (100 MHz).
Additional file 13: Figure S11. COSY spectrum of compound 2.
Additional file 14: Figure S12. HSQC spectrum of compound 2.
Additional file 15: Figure S13. HMBC spectrum of compound 2.
Contributor Information
Ji Zhang, Email: zhangji@neau.edu.cn.
Yi-Jun Yan, Email: quasar110@126.com.
Jing An, Email: Ajsyz1987@163.com.
Sheng-Xiong Huang, Email: sxhuang@mail.kib.ac.cn.
Xiang-Jing Wang, Email: wangneau2013@163.com.
Wen-Sheng Xiang, Email: xiangwensheng@neau.edu.cn.
References
- 1.Lim LE, Vilchèze C, Ng C, Jacobs WR, Jr, Ramón-García S, Thompson CJ. Anthelmintic avermectins kill Mycobacterium tuberculosis, including multidrug-resistant clinical strains. Antimicrob Agents Chemother. 2013;57:1040–1046. doi: 10.1128/AAC.01696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thuan NH, Pandey RP, Sohng JK. Recent advances in biochemistry and biotechnological synthesis of avermectins and their derivatives. Appl Microbiol Biotechnol. 2014;98:7747–7759. doi: 10.1007/s00253-014-5926-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Zhang T, Wang Q, Cruz-Morales P, Zhang B, Liu M, Barona-Gómez F, Zhang L. Synthetic biology of avermectin for production improvement and structure diversification. Biotechnol J. 2014;9:316–325. doi: 10.1002/biot.201200383. [DOI] [PubMed] [Google Scholar]
- 4.Ōmura S, Crump A. Ivermectin: panacea for resource-poor communities? Trends Parasitol. 2014;30:445–455. doi: 10.1016/j.pt.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Lumaret J-P, Errouissi F, Floate K, Römbke J, Wardhaugh K. A review on the toxicity and non-target effects of macrocyclic lactones in terrestrial and aquatic environments. Curr Pharm Biotechnol. 2012;13:1004–1060. doi: 10.2174/138920112800399257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Chen Z, Lin X, Zhang X, Song Y, Wen Y, Li J. Engineering of avermectin biosynthetic genes to improve production of ivermectin in Streptomyces avermitilis. Bioorg Med Chem Lett. 2008;18:5359–5363. doi: 10.1016/j.bmcl.2008.09.061. [DOI] [PubMed] [Google Scholar]
- 7.Wang H-Y, Zhang J, Zhang Y-J, Zhang B, Liu C-X, He H-R, Wang X-J, Xiang W-S. Combined application of plasma mutagenesis and gene engineering leads to 5-oxomilbemycins A3/A4 as main components from Streptomyces bingchenggensis. Appl Microbiol Biotechnol. 2014;98:9703–9712. doi: 10.1007/s00253-014-5970-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Chen Z, Li M, Wen Y, Song Y, Li J. Construction of ivermectin producer by domain swaps of avermectin polyketide synthase in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2006;72:986–994. doi: 10.1007/s00253-006-0361-2. [DOI] [PubMed] [Google Scholar]
- 9.Frasch H-J, Medema MH, Takano E, Breitling R. Design-based re-engineering of biosynthetic gene clusters: plug-and-play in practice. Curr Opin Biotechnol. 2013;24:1144–1150. doi: 10.1016/j.copbio.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Wong FT, Khosla C. Combinatorial biosynthesis of polyketides—a perspective. Curr Opin Chem Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaisser S, Kellenberger L, Kaja AL, Weston AJ, Lill RE, Wirtz G, Kendrew SG, Low L, Sheridan RM, Wilkinson B, Galloway IS, Stutzman-Engwall K, McArthur HAI, Staunton J, Leadlay PF. Direct production of ivermectin-like drugs after domain exchange in the avermectin polyketide synthase of Streptomyces avermitilis ATCC31272. Org Biomol Chem. 2003;1:2840–2847. doi: 10.1039/b304022d. [DOI] [PubMed] [Google Scholar]
- 12.Cropp TA, Wilson DJ, Reynolds KA. Identification of a cyclohexylcarbonyl CoA biosynthetic gene cluster and application in the production of doramectin. Nat Biotechnol. 2000;18:980–983. doi: 10.1038/79479. [DOI] [PubMed] [Google Scholar]
- 13.Wang J-B, Pan H-X, Tang G-L. Production of doramectin by rational engineering of the avermectin biosynthetic pathway. Bioorg Med Chem Lett. 2011;21:3320–3323. doi: 10.1016/j.bmcl.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Wang X-J, Yan Y-J, Zhang B, An J, Wang J-J, Tian J, Jiang L, Chen Y-H, Huang S-X, Yin M, Zhang J, Gao A-L, Liu C-X, Zhu Z-X, Xiang W-S. Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol. 2010;192:4526–4527. doi: 10.1128/JB.00596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X-J, Zhang B, Yan Y-J, An J, Zhang J, Liu C-X, Xiang W-S. Characterization and analysis of an industrial strain of Streptomyces bingchenggensis by genome sequencing and gene microarray. Genome. 2013;56:677–689. doi: 10.1139/gen-2013-0098. [DOI] [PubMed] [Google Scholar]
- 16.Albers-Schönberg G, Arison BH, Chabala JC, Douglas AW, Eskola P, Fisher MH, Lusi A, Mrozik H, Smith JL, Tolman RL. Avermectins. Structure determination. J Am Chem Soc. 1981;103:4216–4221. doi: 10.1021/ja00404a040. [DOI] [Google Scholar]
- 17.Wang X-J, Zhang J, Wang J-D, Huang S-X, Chen Y-H, Liu C-X, Xiang W-S. Four new doramectin congeners with acaricidal and insecticidal activity from Streptomyces avermitilis NEAU1069. Chem Biodivers. 2011;8:2117–2125. doi: 10.1002/cbdv.201000295. [DOI] [PubMed] [Google Scholar]
- 18.Chabala JC, Mrozik H, Tolman RL, Eskola P, Lusi A, Peterson LH, Woods MF, Fisher MH. Ivermectin, a new broad-spectrum antiparasitic agent. J Med Chem. 1980;23:1134–1136. doi: 10.1021/jm00184a014. [DOI] [PubMed] [Google Scholar]
- 19.Lanusse C, Lifschitz A, Virkel G, Alvarez L, Sánchez S, Sutra JF, Galtier P, Alvinerie M. Comparative plasma disposition kinetic of ivermectin, moxidectin and doramectin in cattle. J Vet Pharmacol Ther. 1997;20:91–99. doi: 10.1046/j.1365-2885.1997.00825.x. [DOI] [PubMed] [Google Scholar]
- 20.dos Santos AR, Falcäo CAB, Muzitano MF, Kaiser CR, Rossi-Bergmann B, Férézou J-P. Ivermectin-derived leishmanicidal compounds. Bioorg Med Chem. 2009;17:496–502. doi: 10.1016/j.bmc.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Pitterna T, Cassayre J, Hüter OF, Jung PMJ, Maienfisch P, Kessabi FM, Quaranta L, Tobler H. New ventures in the chemistry of avermectins. Bioorg Med Chem. 2009;17:4085–4095. doi: 10.1016/j.bmc.2008.12.069. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, An J, Wang J-J, Yan Y-J, He H-R, Wang X-J, Xiang W-S. Genetic engineering of Streptomyces bingchenggensis to produce milbemycins A3/A4 as main components and eliminate the biosynthesis of nanchangmycin. Appl Microbiol Biotechnol. 2013;97:10091–10101. doi: 10.1007/s00253-013-5255-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J-H, Xu X-J, Ji M-H, Cheng J-L, Zhu G-N. Design, synthesis, and biological activities of milbemycin analogues. J Agric Food Chem. 2011;59:4836–4850. doi: 10.1021/jf2001926. [DOI] [PubMed] [Google Scholar]
- 24.Williams GJ. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr Opin Struct Biol. 2013;23:603–612. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Li L, Feng C, Chen Y, Tan H. Novel polyoxins generated by heterologously expressing polyoxin biosynthetic gene cluster in the sanN inactivated mutant of Streptomyces ansochromogenes. Microb Cell Fact. 2012;11:135. doi: 10.1186/1475-2859-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 27.Sun P, Zhao Q, Yu F, Zhang H, Wu Z, Wang Y, Wang Y, Zhang Q, Liu W. Spiroketal formation and modification in avermectin biosynthesis involves a dual activity of AveC. J Am Chem Soc. 2013;135:1540–1548. doi: 10.1021/ja311339u. [DOI] [PubMed] [Google Scholar]
- 28.Yoon YJ, Kim E-S, Hwang Y-S, Choi C-Y. Avermectin: biochemical and molecular basis of its biosynthesis and regulation. Appl Microbiol Biotechnol. 2004;63:626–634. doi: 10.1007/s00253-003-1491-4. [DOI] [PubMed] [Google Scholar]
- 29.Yoon YJ, Beck BJ, Kim BS, Kang H-Y, Reynolds KA, Sherman DH. Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem Biol. 2002;9:203–214. doi: 10.1016/S1074-5521(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 30.Gregory MA, Kaja AL, Kendrew SG, Coates NJ, Warneck T, Nur-Alam M, Lill RE, Sheehan LS, Chudley L, Moss SJ, Sheridan RM, Quimpere M, Zhang M-Q, Martin CJ, Wilkinson B. Structure guided design of improved anti-proliferative rapalogs through biosynthetic medicinal chemistry. Chem Sci. 2013;4:1046–1052. doi: 10.1039/C2SC21833J. [DOI] [Google Scholar]
- 31.Park SR, Han AR, Ban Y-H, Yoo YJ, Kim EJ, Yoon YJ. Genetic engineering of macrolide biosynthesis: past advances, current state, and future prospects. Appl Microbiol Biotechnol. 2010;85:1227–1239. doi: 10.1007/s00253-009-2326-8. [DOI] [PubMed] [Google Scholar]
- 32.Marsden AF, Wilkinson B, Cortés J, Dunster NJ, Staunton J, Leadlay PF. Engineering broader specificity into an antibiotic producing polyketide synthase. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 33.Sheehan LS, Lill RE, Wilkinson B, Sheridan RM, Vousden WA, Kaja AL, Crouse GD, Gifford J, Graupner PR, Karr L, Lewer P, Sparks TC, Leadlay PF, Waldron C, Martin CJ. Engineering of the spinosyn PKS: directing starter unit incorporation. J Nat Prod. 2006;69:1702–1710. doi: 10.1021/np0602517. [DOI] [PubMed] [Google Scholar]
- 34.Dutton CJ, Gibson SP, Goudie AC, Holdom KS, Pacey MS, Ruddock JC, BuʼLock JD, Richards MK. Novel avermectins produced by mutational biosynthesis. J Antibiot. 1991;44:357–365. doi: 10.7164/antibiotics.44.357. [DOI] [PubMed] [Google Scholar]
- 35.Meinke PT, O’Connor SP, Mrozik H, Fisher MH. Synthesis of ring-contracted, 25-nor-6,5-spiroketal-modified avermectin derivatives. Tetrahedron Lett. 1992;33:1203–1206. doi: 10.1016/S0040-4039(00)91896-3. [DOI] [Google Scholar]
- 36.Shih TL, Mrozik H, Holmes MA, Fisher MH. A witting approach to novel C24 and C25-substituted avermectins. Tetrahedron Lett. 1990;31:3529–3532. doi: 10.1016/S0040-4039(00)94434-4. [DOI] [Google Scholar]
- 37.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. Norwich: John Innes Foundation Norwich; 2000. [Google Scholar]
- 38.Sambrook J, Fritsch T, Maniatis EF. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 39.Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotecnhol Tech. 1998;12:653–657. doi: 10.1023/A:1008848517221. [DOI] [Google Scholar]
- 40.Xiang W-S, Wang J-D, Wang M, Wang X-J. New nemadectin congener from Streptomyces microflavus neau3: fermentation, isolation, structure elucidation and biological activities. J Antibiot. 2010;63:171–175. doi: 10.1038/ja.2010.12. [DOI] [PubMed] [Google Scholar]


