Abstract
Background
In this study, we aimed to identify microRNA from urine of multiple myeloma patients that could serve as a biomarker for the disease.
Results
Analysis of urine samples was performed using Serum/Plasma Focus PCR MicroRNA Panel (Exiqon) and verified using individual TaqMan miRNA assays for qPCR. We found 20 deregulated microRNA (p < 0.05); for further validation, we chose 8 of them. Nevertheless, only differences in expression levels of miR-22-3p remained close to statistical significance.
Conclusions
Our preliminary results did not confirm urine microRNA as a potential biomarker for multiple myeloma.
Background
Multiple myeloma (MM) is a malignancy of plasma cells (PCs) that manifests also by renal insufficiency [1]. MicroRNAs (miRNAs) are small non-coding single stranded RNAs and important regulators of gene expression involved in MM pathogenesis [2]. Currently, research attention is focused on circulating miRNAs which can be detected in various body fluids, also in urine. Circulating miRNAs are highly stable and have the potential to become easily available minimally invasive biomarkers of the disease [3]. For MM, they would represent a new more convenient approach since painful and invasive bone marrow (BM) biopsy is used for disease diagnostics and monitoring. We hypothesized that miRNAs identified in urine of MM patients could become a completely non-invasive biomarker potentially distinguishing MM patients from healthy donors (HD).
Results and discussion
Screening of miRNA in urine revealed 20 deregulated miRNAs (p < 0.05 for all miRNAs) between MM and HD (Fig. 1). For further validation, we chose 8 most deregulated miRNAs between MM and HD: miR-200c-3p, miR-29b-3p, miR-29c-3p, miR-22-3p, miR-29a-3p, miR-25-3p, miR-106b-5p, miR-18a-3p (Table 1). Expression levels of these miRNAs were further verified on a larger cohort of MM patients, RCC patients and HD. Results from validation revealed that expression levels of chosen miRNAs from urine of MM patients are not statistically different (p = 0.05) from HD and RCC patients (Fig. 2). Results from validation did not confirm statistical significance from the screening. Only differences in expression levels of miR-22-3p between MM patients and HD in urine (p = 0.090) remained close to statistical significance (Table 2).
Fig. 1.
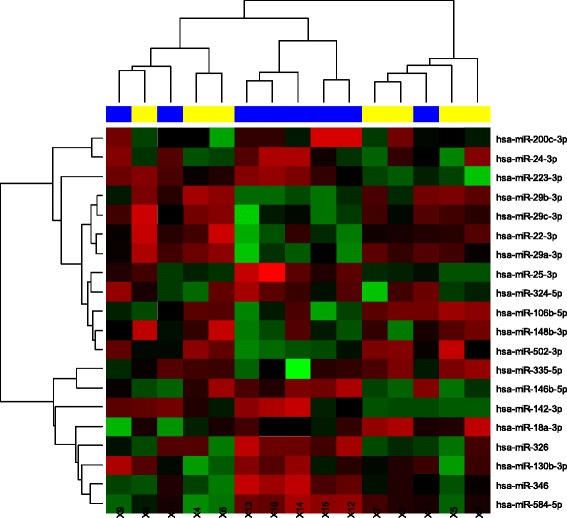
Hierarchical clustering of differentially expressed miRNAs from urine between MM patients (yellow) and HD (blue) using heatmaps. Different expression is expressed by range of red (overexpression) vs green (low expression) colors
Table 1.
P values and sample size estimation for differentially expressed miRNAs in the screening phase
| miRNA | P value | adj.P value | Sample size |
|---|---|---|---|
| hsa-miR-29b-3p | 0.0043 | 0.2074 | 25.14 |
| hsa-miR-29a-3p | 0.0060 | 0.2074 | 81.74 |
| hsa-miR-22-3p | 0.0062 | 0.2074 | 58.68 |
| hsa-miR-29c-3p | 0.0111 | 0.2074 | 36.19 |
| hsa-miR-25-3p | 0.0176 | 0.2074 | 55.67 |
| hsa-miR-106b-5p | 0.0195 | 0.2074 | 32.32 |
| hsa-miR-18a-3p | 0.0303 | 0.2784 | 125.39 |
| hsa-miR-200c-3p | 0.0426 | 0.3121 | 29.50 |
Fig. 2.
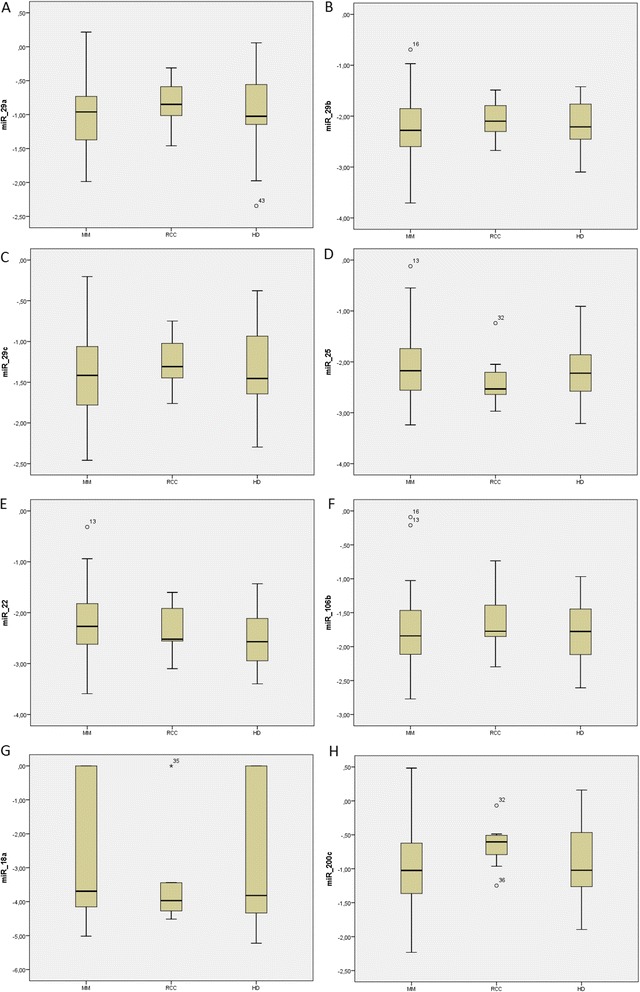
Comparison of expression levels of validated miRNAs defined as logarithmic values of 2-ΔCt normalized to cel-miR-39 expression levels for MM, RCC patients and HD. a – miR-29a-3p; b – miR-29b-3p; c – miR-29c-3p; d miR-25-3p; e – miR-22-3p; f – miR-106b-5p; g – miR-18a-3p; h – miR-200c-3p
Table 2.
Validated microRNAs
| miRNA | HD | MM | RCC | P value | ||
|---|---|---|---|---|---|---|
| mean | mean | mean | ||||
| (25–75) | (25–75) | (25–75) | HD vs. MM | RCC vs. MM | HD vs. RCC vs. MM | |
| Std. deviation | Std. deviation | Std. deviation | ||||
| miR-29a | −1.007 | −0.984 | −0.833 | 0.863 | 0.466 | 0.739 |
| (−1.145) - (−0.556) | (−1.384) - (−0.715) | (−1.172) - (−0.475) | ||||
| 0.559 | 0.506 | 0.393 | ||||
| miR-29b | −2.204 | −2.248 | −2.064 | 0.570 | 0.394 | 0.581 |
| (−2.458) - (−1.751) | (−2.598) - (−1.848) | (−2.352) - (−1.493) | ||||
| 0.446 | 0.580 | 0.438 | ||||
| miR-29c | −1.386 | −1.426 | −1.252 | 0.663 | 0.329 | 0.565 |
| (−1.656) - (−0.933) | (−1.796) - (−1.040) | (−1.570) - (−0.856) | ||||
| 0.482 | 0.518 | 0.361 | ||||
| miR-25 | −2.224 | −2.106 | −2.347 | 0.692 | 0.329 | 0.608 |
| (−2.593) - (−1.848) | (−2.595) - (−1.717) | (−2.742) - (−2.049) | ||||
| 0.598 | 0.634 | 0.567 | ||||
| miR-22 | −2.532 | −2.247 | −2.310 | 0.090 | 0.790 | 0.225 |
| (−2.954) - (−2.084) | (−2.744) - (−1.782) | (−2.566) - (−1.676) | ||||
| 0.538 | 0.689 | 0.536 | ||||
| miR-106b | −1.788 | −1.765 | −1.612 | 0.958 | 0.496 | 0.776 |
| (−2.120) - (−1.419) | (−2.138) - (−1.423) | (−1.901) - (−1.102) | ||||
| 0.460 | 0.555 | 0.524 | ||||
| miR-18a | −2.736 | −2.824 | −3.416 | 0.763 | 0.528 | 0.802 |
| (−4.359) - (0.000) | (−4.171) - (0.000) | (−4.303) - (−3.437) | ||||
| 2.101 | 1.874 | 1.563 | ||||
| miR-200c | −0.909 | −0.990 | −0.645 | 0.543 | 0.088 | 0.237 |
| (−1.273) - (−0.449) | (−1.405) - (−0.588) | (−0.963) - (−0.487) | ||||
| 0.610 | 0.595 | 0.374 | ||||
Expression levels defined as logarithmic values of 2-ΔCt normalized to cel-miR-39 expression levels as mean value and interquantile range with standard deviation. Nonparametric Kruskal-Wallis or Mann–Whitney U test was used to compare the values
Nowadays, there is increasing evidence of miRNAs importance in MM pathogenesis. MiRNA expression profiles could be useful for MM stratification, prognostic estimation, prediction of therapeutic effectiveness or disease relapse [2, 4]. Moreover, miRNAs could potentially provide deeper insight into molecular nature of the disease and help to develop miRNAs-based therapeutic agents subsequently improving patients’ outcome [5]. It has been shown that circulating miRNAs found in various body fluids may serve as a new class of powerful and minimally invasive biomarkers for MM. Although several studies describing circulating miRNAs in peripheral blood of MM patients were published [6–8], no study focused on miRNA expression profiles in urine of MM patients has been published so far.
As one of the typical clinical manifestations of MM is renal insufficiency, included as one of the ‘CRAB’ criteria for organ damage in MM [1], we hypothesized that miRNAs found in urine of MM patients could serve as diagnostic biomarkers of the disease. Twenty differentially expressed miRNAs between MM and HD urine samples were identified using Serum/Plasma Focus PCR MicroRNA Panel; out of them, eight miRNAs were chosen for further analysis. Unfortunately, none of them was assessed as significantly deregulated in a larger cohort of MM, RCC patients and HD using individual TaqMan miRNA assays. In our study, we included not only MM patients and HD, but also RCC patients to exclude miRNA that are related to kidney damage.
As no differences in miRNA expression were found in the validation phase of the study, we considered several reasons for this outcome. First reason is possible analytical difference between the screening and validation phase of our study. We believe that using different detection approaches (Serum/Plasma Focus PCR MicroRNA Panel versus individual TaqMan miRNA assays) was not the reason for discordance between the screening and validation phase since they are both reliable methods verified by many researchers [9, 10]. Second reason is differences in sample collection and processing. We believe that the discordance was not caused by disunity of samples as they were all collected and processed in the same manner. Third possibility is using cel-miR-39 as a spike-in control for normalization. This approach is now considered a suitable approach in relative quantification [11], and it was applied in both phases of the study. On the other hand, a small cohort of patients used in the screening phase may be considered a major limitation as it may not be powerful enough for identification of a biomarker. Although the cohort seems to be small, it is a standard way of identifying possible differences of miRNA expression [8, 10]. While it is possible that we could have missed some significantly differently expressed miRNAs, we found twenty miRNAs to be deregulated (p < 0.05 for all miRNAs). In the design of our study, the screening phase was not a test cohort, but a way of finding significant miRNAs which should be studied further; we believe that the screening phase fulfilled this purpose.
Another possibility to be considered is the existence of previously published work that identified urine miRNAs as markers of various diseases – such work has been done, for example in urologic cancers (reviewed in [12]). Urinary miRNAs are easily accessible and quantifiable and thus have a great potential to become biomarkers in oncology and nephrology [12]. It has been previously published that urinary miRNAs may be used for diagnosis and monitoring of urothelial carcinoma (UC). Deregulated levels of miR-126, miR-96, miR-200 family and miR-183 family were repeatedly observed in UC patients. Also, it was observed that increased expression levels of urinary miR-15a could serve as a biomarker for benign/malignant RCC differentiation [12].
Moreover, some of studied urinary miRNAs were previously described in MM PCs [4, 13, 14]. Members of the miR-29 family (miR-29a-3p, miR-29b-3p and miR-29c-3p) were found in MM PCs with decreased expression levels; also, presence of circulating miR-29a was detected in serum of MM patients [13, 15]. Apart from hematological malignancies, increased expression levels of these miRNAs were found in RCC and decreased expression levels in diabetic nephropathy (DN) [16].
Overexpression of cluster miR-106b-25 seems to be contributing to transformation of monoclonal gammopathy of undetermined significance (MGUS) patients to MM as its expression levels are increased in both, MGUS and MM PCs compared to HD [2]. Increased expression levels of miR-106b occur in RCC cells as well. It was suggested that expression levels of this miRNA could become a predictive biomarker for metastasis formation after surgical removal of kidneys [14]. On the other hand, miR-22 expression levels were found to be decreased in MM cell lines and were associated with 17p deletion [4].
MiR-18a is member of the miR-17-92 cluster which is present in PCs of MM patients with higher expression levels. MiRNAs originating from this cluster of genes promote leukemogenesis. The miR-17-92 cluster is activated by Myc and subsequently downregulates proapoptotic protein Bim thus promoting MM cell proliferation and inhibition of cell apoptosis. Moreover, the miR-17-92 cluster is linked to MM progression and poor prognosis [17]. On contrary, decreased expression levels of miR-18a was detected in bladder cancer [14, 18]. MiR-200c has not been found in MM PCs so far. However, its decreased expression levels were shown in RCC and DN [19].
Conclusions
In conclusion, we have identified several miRNAs in urine of MM patients that were previously described to be involved in MM pathogenesis or kidney-associated diseases but are not disease-specific. Thus, we could not confirm our hypothesis that there is a set of circulating urinary miRNA that could serve as a non-invasive marker of MM.
Methods
In total, 85 urine samples were included in the study (Table 3). Samples were collected as 8 mL of urine stabilized by 0.149 g of EDTA. MiRNAs from 1 mL of urine were isolated using Urine MicroRNA Purification Kit (Norgen Biotek, Canada) according to manufacturer’s recommendations and quantified using Nanodrop-ND1000 spectrophotometer. 40 ng of isolated miRNA was reverse transcribed by Universal cDNA Synthesis Kit (Exiqon, Denmark). Analysis of potentially biologically significant miRNAs in urine was performed using Serum/Plasma Focus PCR MicroRNA Panel (Exiqon, Denmark) determining expression levels of 179 miRNAs of 7 urine samples of newly diagnosed MM patients and 8 HD. Normalized expression data from the screening phase of the study were statistically analyzed by freeware R/Bioconductor and its additional packages. LIMMA approach was used to identify differentially expressed miRNAs with Benjamini-Hochberg adjustment of P values. To clarify similarity of samples, hierarchical clustering was applied. Individual TaqMan miRNAs assays for 8 differentially expressed miRNAs (hsa-miR-200c-3p, hsa-miR-29b-3p, hsa-miR-29c-3p, hsa-miR-22-3p, hsa-miR-29a-3p, hsa-miR-25-3p, hsa-miR-106b-5p, hsa-miR-18a-3p, Life Technologies, USA) were used for qPCR on 7500 Real-Time PCR System. qPCR and reverse transcription using TaqMan MicroRNA Reverse Transcription Kit (Life Technologies, USA) was performed following manufacturer’s recommendations. Results were obtained by relative quantification using spike-in controls cel-miR-39 in 49 newly diagnosed MM, 20 HD and 7 patients with renal cell carcinoma (RCC) in order to distinguish miRNAs associated with MM. Analytical performance of the study was assessed by intraplate and interplate controls. Standard descriptive statistics were applied in the analysis; median supplemented by interquartile range for continuous variables. Statistical significance of differences in continuous variables among groups of patients was analyzed using nonparametric Kruskal-Wallis or Mann–Whitney U test. Statistical analysis of data from validation phase of the study was performed using IBM SPSS Statistics, v. 20. The study was approved by the Ethics committee of the University Hospital Brno. All patients were included into the study only after they signed the informed consent form.
Table 3.
Patients’ characteristics
| HD | RCC | MM | |
|---|---|---|---|
| No. of patients/donors | 22 | 7 | 56 |
| Gender: males-females | 9-13 | 6-1 | 28-28 |
| Age median (min-max) [years] | 57 (50–90) | 61 (27–83) | 69 (49–89) |
| ISS stage: I-II-III | ND | ND | 16-12-19 |
| D-S stage: I-II-III | ND | ND | 3-7-40 |
| D-S substage: A-B | ND | ND | 39-11 |
| Ig isotype: IgG-IgA-FLC | ND | ND | 34-11-10 |
| Light chains: kappa-lambda | ND | ND | 40-15 |
| Amount of M-Ig/FLC g/l median (min-max) | ND | ND | 25,45 (0,17-65,4) |
| PCs infiltration in BM | ND | ND | 13,1 (0,14-87,4) |
| No treatment | ND | ND | 56 (100 %) |
ND – not done, ISS stage – International Staging System stage, D-S stage – Durie-Salmon stage, M-Ig – monoclonal immunoglobulin, FLC – Free Light Chains
Acknowledgements
This work was supported by grant of the Ministry of Health, Czech Republic IGA NT14575. We would like to thank Marcela Stouracova, Bozena Hanakova and Barbora Sablikova for technical assistance.
Abbreviations
- BM
bone marrow
- DN
diabetic nephropathy
- HD
healthy donors
- miRNAs
microRNAs
- MGUS
monoclonal gammopathy of undetermined significance
- MM
multiple myeloma
- PCs
plasma cells
- RCC
renal cell carcinoma
- UC
urothelial carcinoma
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LS carried out the experiments, analyzed the data and wrote the manuscript. LB performed miRNA screening and helped to draft the manuscript. SN and VK performed all miRNA isolations. LR helped with statistical analyses. MK supervised collection of the multiple myeloma samples and clinical data. MS supervised collection of the renal cell carcinoma samples and clinical data. SS participated in design of the study and helped with manuscript preparation. RH provided critical revision of the results. All authors read and approved the final manuscript.
Contributor Information
Lenka Sedlaříková, Email: lena.sedlarikova@gmail.com.
Lenka Bešše, Email: lenka.kubiczek@gmail.com.
Soňa Novosadová, Email: 357892@mail.muni.cz.
Veronika Kubaczková, Email: veronika.kubaczkova@seznam.cz.
Lenka Radová, Email: avodar@gmail.com.
Michal Staník, Email: stanik@mou.cz.
Marta Krejčí, Email: mkrejci@fnbrno.cz.
Roman Hájek, Email: rom.hajek@seznam.cz.
Sabina Ševčíková, Email: sevcik@med.muni.cz.
References
- 1.Rajkumar SV. Multiple Myeloma. Curr Probl Cancer. 2009;33:7–64. doi: 10.1016/j.currproblcancer.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lionetti M, Agnelli L, Mosca L, Fabris S, Andronache A, Todoerti K, et al. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes Chromosomes Cancer. 2009;48:521–531. doi: 10.1002/gcc.20660. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28:732–738. doi: 10.1038/leu.2013.262. [DOI] [PubMed] [Google Scholar]
- 6.Jones CI, Zabolotskaya MV, King AJ, Stewart HJ, Horne GA, Chevassut TJ, et al. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. 2012;107:1987–1996. doi: 10.1038/bjc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa S, Ohyashiki JH, Ohyashiki M, Umezu T, Suzuki K, Inagaki A, et al. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012;2 doi: 10.1038/bcj.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubiczkova L, Kryukov F, Slaby O, Dementyeva E, Jarkovsky J, Nekvindova J, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99:511–518. doi: 10.3324/haematol.2013.093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 10.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, et al. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheffer AR, Holdenrieder S, Kristiansen G, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs in serum: novel biomarkers for patients with bladder cancer? World J Urol. 2014;32:353–358. doi: 10.1007/s00345-012-1010-2. [DOI] [PubMed] [Google Scholar]
- 12.Mlcochova H, Hezova R, Stanik M, Slaby O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Oncol. 2014;32:41. doi: 10.1016/j.urolonc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Fišerová B, Kubiczková L, Sedlaříková L, Hájek R, Ševčíková S. The miR-29 family in hematological malignancies. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2014 Jul 4. doi: 10.5507/bp.2014.037. [Epub ahead of print]. [DOI] [PubMed]
- 14.Slabý O, Svoboda M. [MicroRNA in oncology]. Praha: Galén; 2012. ISBN 9788072625871 807262587X.
- 15.Sevcikova S, Kubiczkova L, Sedlarikova L, Slaby O, Hajek R. Serum miR-29a as a marker of multiple myeloma. Leuk Lymphoma. 2012;54:189–191. doi: 10.3109/10428194.2012.704030. [DOI] [PubMed] [Google Scholar]
- 16.Schena FP, Serino G, Sallustio F. MicroRNAs in kidney diseases. New promising biomarkers for diagnosis and monitoring. Nephrol Dial Transplant. 2014;29:755–763. doi: 10.1093/ndt/gft223. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Tao J, Wu D, Li P, Xu B, Lu Q, Zhang W. microRNA-18a, a member of the oncogenic miR-17-92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol Med Rep. 2012;5:167–172. doi: 10.3892/mmr.2011.591. [DOI] [PubMed] [Google Scholar]
- 19.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, et al. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol. 2008;216:418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]


