Summary
Bioluminescence imaging (BLI) has emerged as a powerful tool in the study of animal models of viral disease. BLI enables real-time in vivo study of viral infection, host immune response and the efficacy of intervention strategies. Substrate dependent light emitting luciferase enzyme when incorporated into a virus as a reporter gene enables detection of bioluminescence from infected cells using sensitive charge-coupled device (CCD) camera systems. Advantages of BLI include low background, real-time tracking of infection in the same animal and reduction in the requirement for larger animal numbers. Transgenic luciferase-tagged mice enable the use of pre-existing nontagged viruses in BLI studies. Continued development in luciferase reporter genes, substrates, transgenic animals and imaging systems will greatly enhance future BLI strategies in viral research.
Keywords: bioluminescence imaging, BLI, CCD camera, cytomegalovirus, herpes simplex, influenza, luciferase, pathogenesis, viral dissemination, virus imaging
This review explores current strategies utilized in the study of virus infection via bioluminescence imaging (BLI) in tissue culture and in various animal models of human disease. BLI has become a highly effective technology for the study of viral pathogenesis, host immune response and antiviral strategies (Table 1). Noninvasive and biocompatible BLI enables real-time in vivo analysis of viral replication and dissemination in an animal model. BLI takes advantage of the light emitting luciferase enzymes as novel reporter genes. Although there are many different luciferases [1], only a few are used for in vivo imaging: Photinus pyralis (firefly) luciferase (FLuc); Pyrophorus plagiophthalamus click beetle green luciferase (CBGLuc); and click beetle red luciferase (CBRLuc); Renilla reniformis (sea pansy) luciferase (RLuc); Gaussia princeps (marine copepod) luciferase (GLuc); and Oplophorus gracilirostris (deep sea shrimp) derived luciferase (NanoLuc) (Table 2). Typically an animal inoculated with a luciferase reporter gene tagged recombinant virus, or expression plasmid, is subsequently injected with a luciferase substrate, sedated and placed in a light proof specimen chamber for measurement of light emission. Gray-scale photographic, or a laser scan topography image, of the animal is superimposed onto the bioluminescent pseudocolor images using specialized software. Although the pseudocolors depend on the vendors' choice, generally areas showing the highest level of light (photons) emitted are depicted with red, and lowest emission regions are represented in blue. The bioluminescence signal is expressed in units of photons per second per cm2/steradain (reviewed in [2]). Figure 1A shows the overall strategy for BLI, which can be applied to tissue culture plates as well as animal models. Unlike fluorophores, such as green fluorescent protein (GFP), luciferase does not require excitation light for photon emission but instead luciferase metabolizes a specific substrate. Depending on the luciferase specific cofactors such as ATP, Mg2+ and oxygen are also utilizes to produce light (photons), the example shown in Figure 1 is for FLuc (Figure 1A). Since cells do not normally emit light, BLI has a good signal to background ratio [3,4]. BLI has several advantages over alternative more conventional strategies for detection of reporter gene expression: the technique is extremely sensitive (as low as 10–17 moles of luciferase/L [5]); it is faster and less expensive than other imaging strategies; it does not have a problem of background signal common to other approaches; and the assay can be performed multiple times on the same animal. The latter removes problems of variation between animals, as well as the time consuming process of gathering data through sequential sacrificing of animals at specific time points. High sensitivity, ease of use, high throughput and minimal post image analysis makes BLI preferable to other forms of in vivo imaging. In living animal tissue, the rate limiting factor for the detection of luciferase is an optimal supply of the substrate to maximize the signal strength. When conditions are at an optimal level the intensity of the bioluminescence should be directly proportional to the amount of luciferase. If the substrate is at a suboptimal level then substrate dosage is directly proportional to bioluminescence detected [2]. Animal imaging studies should always be performed with substrate at as near optimal level as possible. Initial studies with a new animal model or recombinant Luc-tagged virus should initially go through a series of empirical studies to determine optimal substrate levels for BLI.
Table 1.
Summary of major viral bioluminescence applications.
| In vivo BLI application | Method | Ref. |
|---|---|---|
| Antiviral discovery high throughput | Correlating BLI signal to real-time effects of antiviral strategies on viral load | [6] |
| Gene therapy | Coexpression with gene of interest to verify expression and location | [7] |
| Viral replication | Recombinant luciferase-expressing viruses in vivo or in vitro | [8–10] |
| Innate immune pathways | Either through the use of luciferase-tagged viral infection in transgenic mice containing knock-out immune components, or imaging luciferase-tagged immune components in transgenic mice in response to wild-type or mutant viral infection | [11,12] |
| Immune cell response to viral infection | Adoptive transfer of luciferase-expressing cells from transgenic animals into viral infected animals for cell type tracking | [13] |
| Oncolytic cancer therapy | Demonstrating tumor targeting and/or effects of therapy on tumor | [7] |
| Viral pathogenies/disease model | Identifying specific role of viral select proteins | [12] |
BLI: Bioluminescence imaging.
Table 2.
Summary of commonly used luciferase.
| Luciferase | Sequence length (bp) | Protein size (kDa) | Substrate | Cofactors | Emission wavelength (at 37 ° C); nm | Signal type | Time until signal (postsubstrate addition); min | Ref. |
|---|---|---|---|---|---|---|---|---|
| FLuc | 1650 | ∼61 | D-Luciferin | O2, ATP, Mg2+ | 612 | Glow | 5 | [14,15] |
| Gluc | 557 | ∼20 | coelenterazine | O2 | 485 | Flash | 1–2 | [1,16] |
| Rluc | 936 | ∼36 | coelenterazine | O2 | 480 | Flash | 1–2 | [1,16] |
| NanoLuc | 513 | ∼19 | Furimazine | O2 | 460 | Glow | 5 | [17] |
| CBRLuc | 1630 | ∼64 | D-Luciferin | O2, ATP, Mg2+ | 615 | Glow | 5 | [18] |
| CBGLuc | 1630 | ∼64 | D-Luciferin | O2, ATP, Mg2+ | 544 | GLow | 5 | [18] |
Figure 1. Bioluminescence imaging in tissue culture and an animal model.
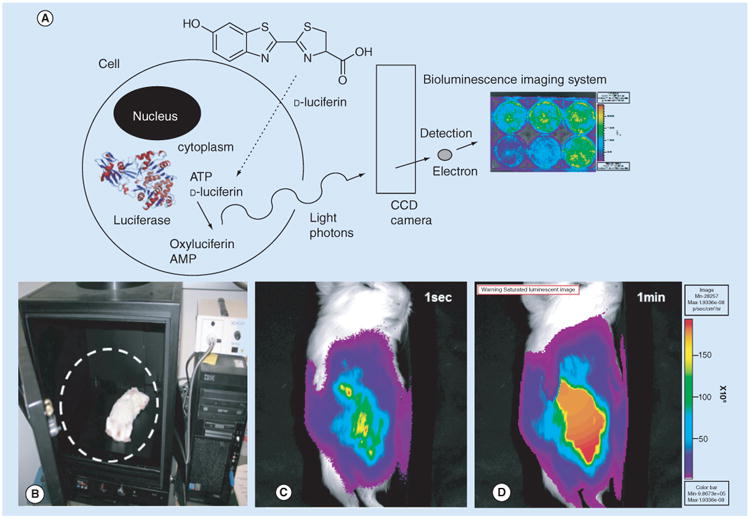
(A) Diagram of a single cell expressing FLuc, which enzymatically acts on d-Luciferin (substrate) to produce oxyluciferin + AMP and a photon of light. The photons are detected by the CCD camera, which produces the image, example shows tissue culture plate with FLuc expression virus. (B) IVIS-50 imaging system (Xenogen/Perkin Elmer., CA, USA) and BLI of FLuc expression from recombinant guinea pig CMV (GPCMV) in infected guinea pig pup. (C) The overlay image of the bioluminescence detected from the Fluc catalyzing D-Luciferin after 1 s and (D) 1 min onto image of the subject guinea pig pup.
CCD: Charge-coupled device.
In vivo detection of luciferase bioluminescence can potentially be less useful in larger animal model as light is attenuated by approximately tenfold for every centimeter of tissue through which it passes [2].
Although bioluminescent detection in vivo from recombinant plasmids expressing luciferase has been well described, the use of recombinant viruses tagged with luciferase is less well studied but is a growing field. The luciferase gene can be expressed under any promoter. The human cytomegalovirus (HCMV) immediately early promoter is most commonly used as it has a high level of expression that is universally strong across a broad range of host species. One herpesvirus that has proven amenable to luciferase tagging and BLI in the mouse model is herpes simplex type 1 (HSV-1) [6,8,11,19]. Studies by Burgos et al. [6] were able to show, via BLI and real-time PCR viral detection assays, that there is a direct correlation between viral DNA load and bioluminescence for HSV-1 infection in a mouse model, which demonstrates that this approach can be both qualitative and quantitative for viral studies. Studies with HSV will be discussed in more detail in the ‘DNA viruses’ section.
FLuc is a 61 kDa protein and emits a yellow-green λmax = 562 nm ‘glow-like’ signal at room temperature, however, at 37°C the wavelength shifts to 612 nm. FLuc can produce a signal for extended periods of time (t1/2 = ∼3 h), assuming continuous supply of substrate. The substrate that FLuc utilizes is D-luciferin (t1/2 ≈ 30 min), which requires ATP and Mg2+ as cofactors in order to produce a signal [14,15,20]. Notably, D-luciferin has good distribution in the animal model as it is capable of crossing the brain [8] and placental barriers [21]. The strong signal produced in the yellow-green spectrum along with the good distribution of its substrate has made FLuc the luciferase of choice for in vivo studies. Importantly, the ATP dependency of FLuc ensures only actively metabolizing cells produce a signal, which reduces false positive results. By mutating the FLuc gene, it has been possible to create an enzyme that produces red-shifted light [22], which can potentially improve sensitivity in larger animals as the longer wavelength of light is able to penetrate through more tissue.
The 19 kDa GLuc emits light at λmax = 485 nm and the 36 kDa RLuc produces light at λmax = 480 nm [1,16]. Both produce an ATP independent ‘flash-type’ bioluminescence reaction by which the signal peaks 10 s following substrate addition and declines to background levels rapidly over 10 min in tissue culture. In animals, GLuc and RLuc produce signal 1–2 min postintravenous injection of substrate (coelenterazine) [16]. Consequently, obtaining quantitative results from these luciferases is greatly affected by small variations in the time between substrate administration and imaging. The GLuc and RLuc substrate coelenterazine is not favorable for in vivo animal studies as serum can cause auto-oxidation, which increases background noise giving poorer signal. Additionally, coelenterazine cannot cross the blood–brain barrier [23] and needs to be directly injected into the brain for BLI studies. An additional drawback of GLuc and RLuc is the relatively short wavelength of light emitted, which is particularly problematic for in vivo experiments as shorter wavelengths have a reduced ability to penetrate tissues. Red-shift variants of GLuc and RLuc have been developed [24,25] with improved in vivo imaging. The benefits of using GLuc and RLuc for in vivo BLI is their relatively small size compared with FLuc. GLuc is naturally secreted and can be detected in the blood and urine. Therefore, it can be used to monitor ex vivo biological processes [26,27]. However, viruses are intracellular pathogens and the secreted GLuc is less desirable for bioluminescence but is useful for luminescence studies of viral replication. A nonsecreted version of GLuc is required for adequate imaging of intracellular pathogens [28]. FLuc and RLuc are both cell associated and their requirement for different substrates potentially allows both to be utilized within a single subject (tissue culture or animal model) [29]. Dual imaging has been implemented to investigate bacteria pathogens and cancer studies [30,31] but this strategy has not been fully explored for virus studies.
Another luciferase, NanoLuc, is a relatively small (19.1 kDa) protein that produces a glow-type signal, which is over 150× greater than FLuc or RLuc. As with RLuc and GLuc, NanoLuc can use coelenterazine as a substrate, however, bioluminescence is optimized when the coelenterazine analogue furimazine is used [32], which is administered intravenously [17]. NanoLuc is one-third the size of FLuc and does not require ATP, which makes it suitable for use as an ex vivo biomarker. However, NanoLuc has an emission peak wavelength of 460 nm, which hampers the effectiveness of signal in deep tissue studies [17].
Overall, Fluc is the more effective luciferase for in vivo BLI studies due to its long wavelength and the ability of D-luciferin to diffuse effectively throughout the animal. However, other experimental constraints, such as the size of the virus genome, may necessitate the use the smaller luciferase.
Bioluminescence imaging equipment
The light emitted by the luciferase–substrate reaction cannot be observed by the naked eye or by conventional microscopy. Highly sensitive imaging systems are required for bioluminescence as the signal reaching the surface of the animal is very weak. In mice, a bioluminescence source of 1 mm3 cells will only produce signal in the order of nW and pW. Therefore, the animal is placed in a light proof specimen chamber where a highly sensitive charge-coupled device (CCD) camera is used to detect photons emitted (Figure 1B). The CCD camera converts photons that strike silicon wafers into electrons and records the intensity, longer exposure allows for stronger signal detection (Figure 1C & D). The CCD is supercooled to -90°C in order to reduce thermal noise from the charge carrier, and increasing the CCD sensitivity to detect very low level light. Image acquisition is controlled by software that enables analysis and modification of the final image. Some imaging systems are capable of utilizing laser scanning to render a 3D surface topography, which is used to map internal source of the bioluminescent signal [33]. The development of bioluminescence topography systems is being carried out in order to give 3D reconstructions of the signal source [34,35].
Combining BLI with other imaging modalities such as x-ray computed tomography (CT; IVIS-200 system and living image software), ultrasonography, PET, single photon emission CT and MRI can produce a detailed and accurate identification of tissues that are producing bioluminescence [36]. A list of commercially available in vivo imaging systems is presented in Box 1. The Xenogen IVIS systems (Perkin Elmer, CA, USA) are the most sensitive for BLI dedicated studies.
Box 1. Commercial manufactures of in vivo imaging systems.
Quantifying bioluminescence images
To quantify the photons emitted from an anatomical site/region of interest the number of photons emitted per unit time from a fixed area (photon flux) are measured using image analysis systems. By using the photon flux, the differences in image acquisition time and field of view are normalized between exposures, enabling easier comparison of the data between experiments. During repeat imaging of the animal, it is important to position the animal accurately in reproducible positions for image acquisition. By this method BLI can be used as a direct correlation with viral load, provided that substrate is at optimal level. However, animal model BLI data is hampered by absorption of light by mammalian tissues, therefore similar levels of signal originate from different tissue depths appear different at the surface thereby quantitation relies on the tissue depth being known and uniform. Consequently, more accurate results may require conventional strategy such as real-time PCR of extracted DNA for viral load [44].
Bioluminescence tissue optics
The major difficulty of in vivo BLI is signal attenuation (∼tenfold per cm of tissue) [2]. In theory, the larger the animal model the more tissue between the light source and the detector, which results in reduced signal [2]. The most commonly used animal is the mouse, but other animals are being effectively explored (such as rats [45], guinea pigs (Figure 1) [46], and ferrets [47]). The specific best-fit model is dependent upon the specific viral pathogens being studied. Pigmentation of the animal can absorb light and modify the strength of the signal. Animal fur can cause light scattering, which also affects the signal produced, with white fur quenching signal significantly less than dark fur [48]. Localized shaving of the animal can improve image sensitivity, although it may lead to change in skin pigmentation, which in turn alters the signal [5,49]. Skin and muscle are better at transmitting light than highly vascular tissues/organs, therefore difference in bioluminescence between organs may not necessarily be due to differences in viral load, but instead result of variation in tissue optics [50].
Substrate administration
In order to optimize the BLI signal there should be saturating level of substrate within the animal at the time of imaging. The route of substrate administration is critical to insure adequate distribution [51,52]. The most commonly used routes of substrate administration are intraperitoneal (IP) and intravenous (IV). IP-delivered substrate leads to a lag time (∼10 min) for maximal signal production but results in uniform distribution in the animal, including the brain, and has an extended duration of maximum bioluminescence. The IP route is commonly used for Fluc substrate (D-luciferin at 150 mg/kg [8]). IV injection has a much faster biodistribution resulting in a much stronger signal produced within seconds of substrate administration. However, the clearance of the substrate from the animal is considerably faster, and this route is typically used for coelenterazine for Gluc/RLuc (at 4 μg/g [8]). Working stock solutions can be stored for a limited time at -80°C in light proof container but substrate should be stored as a powder long term. Substrates are administered at a weight-dependent dose [53]. The increase in demand for BLI has increased the number of suppliers of substrate, which has reduced the initial prohibitive high cost.
As animals need to be immobilized when imaging, isoflurane is most commonly used as an anesthetic. The subject is usually anesthetized with 4% isoflurane mixed with oxygen and maintained with a constant supply of 2% isoflurane, which can be piped into the imaging chamber. Animals can be sedated for the duration of the imaging procedure, up to 40 min per session or longer as required. For optimal bioluminescence, the D-Luciferin substrate should be injected before sedation [54]. If isoflurane cannot be used then other drugs such as ketamine can be considered. Ketamine is administered through intramuscular injection, and produces anesthesia in a dose-dependent manner, with a minimum sedation time of 15–20 min [55]. However, dosage should be optimized by individual investigators and varies between animal models. Notably anesthetics can have different effects on the blood system, which can alter the BLI results [53]. Anesthetics can also potentially have an effect on the long-term health of the animal. Isoflurane causes the least reduction in cardiac output, and typically allows the animal to recover more rapidly, compared with injected anesthetics [55].
Luciferase-tagged viruses for BLI studies
Recombinant viruses are commonly engineered to express a luciferase reporter gene under control of a viral promoter to maximize gene expression [6,56]. Although it is possible to incorporate a luciferase gene into large DNA virus, for example, HSV-1 [8], insertion of the reporter gene into a small RNA virus, for example., Influenza A virus, is not always practical because of limited ability to accommodate extra genes without resulting in viral attenuation. Therefore, alternative strategies are required for BLI [28]. Below is a review of successful BLI studies of luciferase-tagged DNA and RNA viruses.
DNA viruses
Human herpesvirus 1 (HHV-1)
One of the most extensively studied viruses via BLI is HSV-1 an alphaherpesvirus, which has a large dsDNA genome. HSV-1 typically initiates lytic infection at a mucosal epithelial sites before entering peripheral nerve terminals and then migrates to sensory nerve ganglia where it establishes latency [11]. The virus can cause serious life-threatening disease in the immunecompromised individual. Recombinant HSV-1 encoding FLuc or RLuc were initially generated for BLI studies [57]. BLI experiments in mice determined that the FLuc was more effective than RLuc [8,23]. Treatment of virus infected mice with the antiviral valacyclovir produced a dose-dependent decrease in FLuc signal. This demonstrated that in vivo bioluminescence could be utilized to quantify/monitor HSV-1 infection and efficacy of antiviral therapy in an animal model [8]. However, to obtain fully quantitative data the BLI was combined with ex vivo imaging of organs and real-time PCR of extracted tissue DNA to determine absolute viral load [6].
Combination studies of recombinant luciferase-tagged viruses with various transgenic knockout mice models has enabled investigation into the influence of the host innate immune system on viral infection/dissemination. Several transgenic mice with knockout interferon (IFN) components (such as, type 1 and type 2 IFN, Stat1, IFNαβγR IRF-3 and IRF-7) were studied with FLuc-expressing HSV-1 via corneal infection.
Bioluminescence imaging of recombinant Fluc-tagged HSV-1 demonstrated that HSV-1, in mice lacking type 1 IFN receptors, infected the lungs, liver, spleen and regional lymph nodes, unlike wild-type mice where the virus was confined to site of infection before being cleared. By contrast, mice with type 2 IFN receptor knockout showed no systemic spread of HSV-1. However, mice with both type1 and type 2 IFN receptor knockouts showed rapid systemic viral spread with the mouse dying after only a few days postinfection [9], which demonstrated an interdependence on both receptors. Stat1 and IFN together help to regulate infection by mediating IFN-dependent gene expression. Interestingly, BLI of HSV-1 dissemination in mice with an N-terminal deletion in Stat1 demonstrated transient infection of the liver and spleen. However, in IFNαβγR knockout mice, BLI of HSV-1 showed viremia and sudden infection of the liver and spleen [10]. In order to investigate the difference in results, transgenic mice carrying a Stat-1 deletion in the DNA-binding domain demonstrated, via BLI, lethal viremia and sudden infection of the liver and spleen of HSV-1. These results demonstrated the advantage of BLI as a rapid screen for changes in viral tropism dissemination in vivo [10].
Interferon regulatory factors IRF-3 and IRF-7 are thought to be central to the innate antiviral response. HSV-1 pathogenesis studies using transgenic IRF-3, and/or IRF-7 knockout mice demonstrated via BLI that viral replication, spread in the CNS and throughout the body occured in the double-knockout mice as opposed to individual knockouts. Additionally, several days postinfection the IRF-7 knockout mice demonstrated significantly greater bioluminescence in lymph nodes compared with IRF-3 knockout mice whilst the animal survival was greatly shortened compared with wild-type and IRF-3-deficient mice. These results demonstrated that the IRF-7 is the dominant innate immune factor and that IRF-3 and IRF-7 act together to prevent viremia. Further investigation showed high levels of IL-6 transcripts and G-CSF indicating systemic inflammatory response, which explains the systemic spread of HSV-1 observed in the BLI, and therefore the increased mortality [58].
In additional experiments using immunocompromised mice, lacking IFN-α/β and -γ receptors (AG129 mice), it was possible to observe the requirement of HSV-1 viral host shutoff (vhs) function for innate immune evasion. BLI imaging of AG129 mice with recombinant FLuc-tagged HSV-1 demonstrated rapid viremia and death. In contrast, mutant (vhs-) Fluc-tagged HSV-1 showed severe attenuation and impaired viral dissemination [59]. These results demonstrate that BLI can be a critical tool in investigating the innate immune response to viral infection [9–10,58–59] and the study of specific viral genes associated with pathogenicity/immune evasion.
Human herpesvirus 3 (HHV-3)
The alphaherpesvirus HHV-3, or Varicella-Zoster virus (VZV) is the causative agent of chicken-pox and shingles. Unlike HSV, VZV can only be studied in a limited capacity in an animal model. However, BLI has been used to great effect in screening the viral genome for essential genes that are involved in replication and tropism in a high-throughput tissue culture system. Systematic global knockout of every open reading frame in the genome of an infectious recombinant FLuc-tagged VZV (vaccine strain) was used to assay virus growth via bioluminescence in MeWo cells and human fetal skin cell culture. These studies demonstrated the successful use of BLI as an efficient assay to compare viral growth of the mutant viruses in vitro, and thereby identify essential viral gene for infection [60,61]. For in vivo BLI VZV studies, a humanized severe combined immunodeficient (hu-SCID) mouse model was developed using grafts of implanted human tissue. The xenograft was infected with FLuc-tagged VZV and imaged at specific time points postinfection. The results from BLI demonstrated a daily increase in bioluminescence from infected xenografts related to viral spread [62]. This approach holds significant promise for the study of VZV epithelial and neuronal tropism [63].
Reactivation of latent herpes viruses
Alphaherpesvirus latency and reactivation have not been investigated by BLI. However, BLI has successfully been utilized to study viral latency and reactivation of other herpesviruses. For example, a FLuc-tagged murine gamma-herpesvirus-68 (MHV-68) was investigated as a model human gammaherpesviruses. BLI of mice infected with the FLuc-tagged MHV-68 identified initial sites of lytic replication. Additionally, after initial clearance of an acute infection, BLI demonstrated real-time reactivation of latent virus, induced by treating the animal with proteasome inhibitor (Velcade), or an immunosuppressant (cyclosporine A) [64]. Recent BLI studies demonstrated reactivation of MHV-68 as a result of an acute infection of parasitic worms, and a mechanism of herpesvirus reactivation related to specific cytokines [65].
Reactivation of CMV, a betaherpesvirus, was studied in mice with a recombinant mouse CMV (MCMV) expressing GLuc and red fluorescent protein mCherry. The GLuc and mCherry genes were synthesized from a single open reading frame (ORF) with the proteins separated by picornavirus p2A site. The HCMV Immediate early (IE) promoter allowed simultaneously high levels of expression of both reporter proteins [66]. In an animal model, poor spatial resolution impairs imaging of single infected cells. Therefore, individual cells were visualized by fluorescence imaging of histological tissue sections using mCherry to identify individual virus-positive cells. Additionally, secreted GLuc from infected cells made it possible to quantify virus reactivation in explants by measuring luminescence in culture supernatants [66].
Poxviruses
Among the Poxviridae only Variola virus is a human-specific pathogen but other poxviruses can be transmitted from animals to humans. Although Variola virus, the causative agent of smallpox, was eradicated in 1977, the study of various poxviruses remains important. One of the most commonly studied poxviruses is vaccinia virus because of interest in development as a vaccine platform. In order to investigate the effects of interferon on viral replication and dissemination, FLuc was inserted into the viral genome [67]. In transgenic interferon type I receptor knockout mice compared with wild-type mice, viral replication and dissemination to the liver and spleen of vaccinia virus was significantly greater. Vaccinia virus infected the lungs and brain in both IFN I receptor knockout and wild-type mice after intranasal inoculation [67]. Subsequently, BLI studies in mice depleted of alveolar macrophages, via liposomal clodronate, demonstrated the importance of the macrophages in limiting vaccinia virus infection in the lung [68].
Monkeypox virus (MPXV) causes lesions similar to that of smallpox. However, where smallpox can only infect humans, monkeypox is capable of infecting a wide range of mammals [69]. Black-tailed prairie dogs are most susceptible to monkeypox and are therefore a good potential model [69,70]. Recombinant Fluc-tagged monkeypox virus was generated and BLI of adult black-tailed prairie dogs infected with the virus enabled identification of viral replication around the area of inoculation [70]. However, the liver, lung, brain, spleen, intestines and kidney were also positive for virus although no bioluminescence was detected in these tissues in vivo [70,71]. These studies demonstrated the potential limitations of BLI in larger animal models.
RNA viruses
Influenza A virus
Viruses with smaller genomes such as RNA viruses are limited in their ability to accommodate relatively large reporter genes because of restricted coding capacity. Influenza A virus, an orthomyxovirus, causes respiratory tract infection commonly referred to as flu. The influenza A genome is relatively small and segmented. In order to investigate the effectiveness of cross-protective monoclonal antibodies against influenza A in vivo, a GLuc-tagged virus was developed [28]. However, despite using one of the smallest luciferases the reporter gene insertion attenuated the virus [28]. The GLuc gene was linked to other viral genes using foot and mouth disease virus (FMDV) 2A site to restore virus growth to near wild-type levels [28]. Additionally, in order to convert the usually secreted luciferase GLuc to a cell-associated luciferase, a C-terminal endoplasmic reticulum retention sequence (KDEL) was added to the GLuc [28]. The BLI images of infected mice showed that the lungs of the mice had reduced bioluminescence to varying degrees depending on the therapeutic antibody used.
In an alternative approach, a recombinant Influenza A virus was generated to express NanoLuc. The recombinant virus replicated with near wild-type kinetics and showed very little attenuation in culture or in vivo. Furthermore, this virus was capable of causing lethal infection in mice [72]. Additionally, recombinant NanoLuc-tagged virus could be used to track the airborne dissemination of the virus between animal (ferret) and demonstrated by lung infection [47]. This approach holds significant promise in the future studies of intervention strategies in real time in an animal model.
Dengue virus
Dengue virus (DENV) is a single-stranded RNA flavivirus that is mosquito-borne and is prevalent in southwest Asia, the pacific and the Americas. DENV causes hemorrhagic fever and significant morbidity and mortality. In order to study DENV infection in the brain, a recombinant RLuc-tagged DENV was generated and used in an AG129 mouse model that lacks IFN-α/β and -γ receptors. Since the RLuc substrate, coelenterazine, cannot cross the blood–brain barrier an intracranial inoculation was required to enable BLI brain infection studies [73]. The results showed a strong correlation between the signal produced and the viral titer, thereby validating this technique as a measure of monoclonal antibody antiviral strategies for DENV [73]. A FLuc-recombinant DENV was also generated but, unfortunately this virus was attenuated in vivo [73,74]. However, the FLuc-tagged DENV did identify the lymph nodes, spleen and gut-associated tissues as sites of viral replication [74]. Potentially, a recombinant DENV encoding NanoLuc might be a more effective in a future approach based on the success of NanoLuc-tagged Influenza A studies [72].
High-throughput antiviral screening & iloviruses
The recent Ebola virus (EBOV) disease outbreak in west Africa 2014 has dwarfed previous epidemics and threatens to spread throughout neighboring countries [75]. A case fatality rate of 50–70% highlights the necessity for effective intervention strategies and development of rapid screening protocols to evaluate new antivirals. A high-throughput antiviral assay using luciferase-tagged Ebola virus has been developed. The 19 kb single-stranded negative sense RNA genome, was modified by a reverse genetics system to incorporate a GLuc gene into both recombinant infectious Ebola and another closely related filovirus, Marburg viruses (MARV) [76]. Antiviral studies performed in African green monkey kidney (Vero-E6) cells with GLuc-tagged virus evaluated potential antiviral strategies by measuring bioluminescence signal from a cell monolayer [76,77]. These experiments highlight the effectiveness of rapid quantification of viral replication through the use of luciferase-tagged virus. Potentially, the ability to study Ebola virus in a guinea pig [78] or hamster [79] animal models in conjunction with luciferase-tagged viruses could produce highly informative data on viral disease and efficacy of intervention strategies.
Gene-therapy delivery systems, oncolytic viruses & BLI
Gene therapy
Specific viruses can be modified for use as a therapeutic gene delivery vector. BLI and a luciferase-tagged viral vector can be useful to verify gene expression in targeted cells/tissues. Defective recombinant lentiviruses [80] and adenovirus [81–83] have been used to deliver tagged reporter genes into mice either by or into specific cells for transplantation into tissues [21]. These experiments used BLI to investigate specific viral routes for potential delivery vectors for corrective agents [19]. Large DNA viruses have the potential to act as a gene-delivery vector, such as Bovine Herpesvirus 4 (BoHV-4) which demonstrated specific tissue targeting in vivo. BoHV-4 shows moderate or no pathogenicity and no oncogenicity and can accommodate large amounts of genetic material without appreciable effect on it replication. A recombinant virus expressing FLuc demonstrated that the virus was located only in the liver of mice [84].
Oncolytic viruses
Oncolytic viruses show potential as novel therapy strategies against specific cancers [85]. There are several viruses currently being studied via BLI for their ability to target cancer cells. BLI has been utilized to investigate the effectiveness of mutant HSV-1 in targeting and lytically replicating within tumors [86]. In this study, dual imaging of the virus and the tumor was performed by establishing an RLuc-expressing cancer cell line. The Rluc-expressing cancer cells were injected into each flank of BALB/c mice. When the tumors had grown to 5 mm in diameter recombinant Fluc-tagged HSV-1 was injected directly into one of the tumors. The results showed that in the uninfected tumor the RLuc signal grew, whereas the infected tumor showed a decrease in RLuc signal. The Fluc signal peaked after 24 h postinfection and reduced over time, presumably in response to the reduced number of cancer cells in which to replicate. In conjunction with PET, which is a technique used to identify and stage tumors, it was possible to demonstrate replicating virus in the tumor along with cancer cell death [86]. Vaccinia virus also has potential as an oncolytic vector. Since vaccinia virus has no known specific receptor for entry it can infect almost any tumor cell type. FLuc-tagged vaccinia virus has been used to track the virus to specific tumor sites in SCID mice [7]. Using GFP-tagged tumor and FLuc plus red fluorescent protein-tagged virus it was possible to demonstrate virus replication and tumor destruction in vivo following IV administration [87].
An attenuated Measles virus (MV), a paramyxovirus, is currently in clinical trials as an oncolytic vector [85]. A recombinant MV was generated which was unable to utilize the signaling lymphocyte activation molecule which the virus uses to enter lymphocyte cells [88]. By these means, the mutant virus was restricted to entering cells via other receptors, predominantly using Polio virus receptor-related 4 protein (PVRL4) which is expressed in breast, lung and ovarian cancer cells. In order to verify breast cancer targeting, nude mice, xenografted with breast cancer cells, were infected with recombinant MV expressing FLuc which was unable to recognize the cellular receptor signaling lymphocyte activation molecule. The virus was administered directly into the tumor and detection of bioluminescence from FLuc combined with MRI and demonstrated that the viral replication was confined to the tumor [88].
Bioluminescent transgenic mouse models & viruses
As an alternative strategy to inserting a luciferase gene into a recombinant virus genome the luciferase reporter gene can be introduced into transgenic animals for BLI studies. A luciferase-tagged animal model enables the study of preexisting wild-type or mutant viruses in vivo in conjunction with BLI technology. Predominantly, all transgenic mice are FLuc tagged [89]. However, leaky FLuc expression in transgenic animals can potentially produce a background in BLI studies.
Mice encoding FLuc, under IFN-β promoter control, were generated and used in influenza A virus studies. Infected mice did not show significant IFN-β gene activation during the first 1–2 days postinfection. The luciferase activity increased in lungs when the animal showed signs of disease. A mutant influenza A virus lacking IFN-antagonistic factor NS1 (a nonstructural viral protein) showed luciferase activity peaking 24 h postinfection. These results suggest that the viral NS1 protein inhibits IFN-β gene expression in the lung epithelial cells. Viral loads were quantified using qRT-PCR in homogenized lung 24 h postinfection. This approach successfully characterized a specific viral immune suppression protein without attenuating the virus by insertion of a luciferase cassette into the viral genome [90].
Investigation into the humoral immune response in the central nervous system to MCMV brain infections used transgenic BALB/c mice expressing FLuc under β-actin promoter control. MCMV-primed CD19+ B-cells from the transgenic mice were transferred into MHC-matched B-cell deficient recipient mice, via tail vein injection, one day before intracerebral inoculation with MCMV. By this means it was possible to track the primed CD19+ response from the spleen to the brain as MCMV infection developed. BLI results demonstrated that a neuroimmune response is elicited in the mouse brain when challenged with MCMV [13].
Luciferase-tagged transgenic mice can additionally be controlled by the site-specific recombinase Cre-loxP system and the use of Cre-expressing virus. Transgenic mice that encode the luciferase gene inserted into the generalized expression locus ROSA26 are prevented from expressing luciferase by a loxP-flanked STOP fragment inserted between the ROSA26 promoter and luciferase gene. Recombinant virus expressing the Cre enzyme removes the stop cassette resulting in luciferase expression and therefore enables bioluminescence. This method has been utilized in humanized mice to characterize recombinant Cre-expressing HCV [81]. HCV only infects humans and nonhuman primates. However, four cellular proteins have been identified as being all that is required to enable HCV infection. Therefore, mice were further humanized by using recombinant adenoviruses encoding the four proteins to act as a gene-delivery vector and transfect the genes into transgenic mice expressing lox-P flanked FLuc in the ROSA26 locus. When the mice were infected recombinant Cre-expressing HCV, bioluminescence could be detected in the liver of the mice [81]. This strategy requires the recombinant virus to encode a Cre enzyme, which has the advantage of being smaller than FLuc (38 kDa vs 61 kDa) and therefore capable of being incorporated into viruses with smaller genomes.
Future of bioluminescence imaging & animal models
BLI equipment
Further improvements in the accuracy of qualitative and quantitative data gathering and analysis are still possible for BLI. Advances in the CCD cameras used in BLI such as intensified CCD (ICCD) and electron-multiplying CCD (EMCCD) will reduce imaging times to millisecond durations and potentially allow imaging of real-time tracking of moving objects such as blood. Initial capital cost of the equipment remains high and restricts the common use of BLI systems. However, core facilities can overcome prohibitive costs to individual labs.
Combining BLI with other imaging modalities such as x-ray C T, ultrasonography, PET, single photon emission CT and MRI can produce a detailed and accurate identification of the source of the bioluminescence signal and thereby the infected tissue.
Improvements in luciferase & substrate
Enzyme
The development of red-shifted variants of the luciferases will improve signal penetration from deep tissue providing more sensitive measurements. There have been several luciferases that produce light at red/infrared (e.g., railroad worm luciferase) but these have activity and ability issues [22,91]. Mutations in the FLuc, RLuc and GLuc have created variants that produce red-shifted light, which have greater photon output compared with the native luciferase making detecting the emitted light easier in larger animals [22,92]. New luciferases have been identified that are more cell compatible such as a variant of GLuc that emits more sustained light in mammalian cells [93,94]. Notably, by covalently labeling FLuc with near infrared dyes it has been demonstrated that signal of 705 and 783 nm can be produced via bioluminescence resonance energy transfer [95].
The development of a small Luciferase that produces high yield signal in the far red spectrum would allow for a more accurate study of the virus. The smaller luciferase would, in theory, have minimal impact on the virus because of size constraints affecting replication. Additionally the longer wavelength would allow for deeper tissue penetration and therefore more accurate detection in the animal model.
Substrate
Development of a more efficient enzyme substrate will greatly aid in assay sensitivity [32, 96–97]. Synthetic luciferin substrates are also being developed that are more stable and produce brighter signal [98]. For example, synthetic luciferin cyclic alkylaminoluciferin (CycLuc1) has been demonstrated to significantly improved in vivo BLI using FLuc. Data showed a greater than tenfold higher bioluminescence was detected using CycLuc1 compared to equivalent dose of D-Luciferin, therefore allowing for less substrate to be used in order to produce detectable signal, therefore cutting costs [20]. The real advances in BLI will most likely come from advancements in the substrate to produce light at near infrared [99]. Notably, it has been possible to attach small molecule fluorophores (AlexaFluor 650 and 680) to luciferins, and thereby use the bioluminescence from the luciferase–luciferin reaction to stimulate the attached fluorophore, through a process called bioluminescence resonance energy transfer, to produce bioluminescence at near infrared wavelength [100]. However, these constructs are bulky and native luciferases were unable to effectively utilize the substrate resulting in reduced photon output and are not amenable for long term in vivo study.
Luciferases are available that do not require substrate to be added to the cell. The lux operons from bacteria such as photorhabdus luminescens, encode all proteins required for bioluminescence, including the luciferase, substrate and substrate-regenerating enzymes, producing light at 490 nm. The bacteria lux operon has been demonstrated to function in human cells [101] and allows for possible use not only in bacterial/animal studies but also recombinant virus pathogenesis model.
Transgenic animals
Adoptive transfer of luciferase-expressing cell types can be used to study the adaptive [102] and innate immune system in response to existing mutant viruses. Notably, defective recombinant Fluc-tagged adenovirus was used to infect zona-free blastocysts, which were implanted into pseudo-pregnant recipients. The results showed trophectoderm specific infection with only bioluminescent signal detected in the placenta [21], demonstrating that specific organs can be made to express luciferase genes, therefore it is possible to have selective luciferase-tagged organs and not the entire animal.
Imaging protein–protein interactions/viral tropism/animal model
Imaging protein–protein interactions in vivo is possible by utilizing the split luciferase assay whereby inactive N-term luciferase (NLuc) and C-term luciferase (CLuc) fragments tagged to two different proteins of interest. Combination of the fragments forms a functional luciferase, moreover dissociation of the tagged proteins results in separation of the fragments and loss of signal, allowing quantitative analysis of real-time protein–protein interactions. This can be utilized to study the mechanism of specific viruses tropism or any viral–host cell protein binding [103].
Split luciferase can also be used to investigate the activity of proteases. By separating the NLuc and CLuc with the specific cut site for the protease only allows for bioluminescence if the separating segment is cleaved. In this manner the activity of specific viral proteases, such as in Hepatitis C, can be analyzed in vivo [104].
Caspase 3 dependent pro-luciferase are available that are only active if caspase 3 cleaves the cleavage site (DEVD), which separates the N-luc from the CLuc. Consequently, luciferase can only be activated during apoptosis. In a study on mouse hepatitis virus 3 (MHV-3) and apoptosis inhibitors pathogen-free mice were hydrodynamically injected into the tail vein with plasmid containing the proluciferase 14 days prior to infection by lethal dose of MHV-3. The BLI showed apoptosis of the liver after 48 h, which peaked at 72 h (all mice died within 96 h). This model demonstrates a sensitive strategy of measuring apoptosis during viral infection [105]. Apoptosis associated virus replication can alternatively be assayed by the use of a proluciferin substrate that carries the caspase 3/7 cleavage site, which is similarly only active in apoptotic cells [106].
Conclusion & future perspective
Bioluminescent imaging is an important development in the study of virology and disease in animal models. Improvements to luciferase, substrate and imaging equipment will result in highly sensitive tools for investigating viral replication, host immune response and therapeutic gene in larger animal models. Over the next 5–10 years the field of BLI will expand with the development of more stable, higher yielding luciferases and substrates capable of producing a palette of signal wavelengths enabling multi-source imaging. More powerful imaging equipment capable of multimodal and 3D imaging will combine to minimize spatial resolution resulting in more accurate data collection and the use in large animal models. In short the future is looking bright for BLI in viral research.
Executive Summary.
Bioluminescent reporter genes
Bioluminescent imaging has become an effective strategy for tracking viral infection in real time in an animal model through the use of a dedicated imaging system. Although various luciferase reporter genes are available for bioluminescence imaging (BLI) studies, the firefly luciferase (FLuc) is most commonly used as it has the best signal production for in vivo imaging. Viruses with limited space in their genome cannot accommodate such a large reporter gene, therefore, smaller luciferase must be used or an alternative strategy of using luciferase-tagged transgenic animals employed.
DNA viruses & bioluminescence
Various DNA viruses have been successfully studied using BLI, HSV-1 being perhaps the most studied virus using BLI strategy. Imaging of recombinant viruses in a variety of transgenic knockout animal models has provided novel insights into the host immune response to viral infection. BLI has enabled the coherent tracking of the viral life cycle including the reactivation of various herpesviruses.
RNA viruses & bioluminescence
Small RNA viruses, especially those with segmented genomes (e.g., influenza A virus) cannot accommodate large luciferases (e.g., FLuc) without attenuating the virus. Smaller luciferases are potentially effective but alternative approaches using Cre virus and loxP luciferase transgenic animals are also effective strategies. BLI has been successfully employed in the study of various RNA viruses in a number of animal models.
High-throughput antiviral screening using bioluminescence
Bioluminescent imaging can be used as a real-time strategy to measure viral load and is therefore an effective tool for rapid high throughput antiviral screening in tissue culture and efficacy studies of antivirals in animals.
Oncolytic virus & gene therapy vectors & bioluminescence
The developing field of virotherapy and oncolytic viruses is aided by the ability to track viruses to specific tissues/tumors. Dual tagging of both the tumor and the virus means that the regression of the tumor and the viral load can be measured simultaneously. For viruses used as gene-delivery vectors, BLI can be used to confirm gene delivery systemically or to a specific tissue.
Virus-infected transgenic animals & bioluminescence
Many investigators have generated untagged libraries of mutants that they would like to study via BLI. An alternative strategy to luciferase tagging the virus is to insert the luciferase tag into transgenic animals, which enables observation in vivo of wild-type or mutant viral infection using preexisting viruses. Experiments have been designed such that luciferase is expressed in direct response of viral infection. The luciferase can also be used to observe the host innate and adaptive immune response. However, using luciferase-tagged transgenic animals can have potential background issues due to leaky expression.
Future developments in virology & bioluminescence imaging
Future developments in luciferases and substrates such as red-shifted variants along with better imaging systems are overcoming much of the current imaging constraints of BLI and enabling more sensitive in vivo study of virus replication in larger animal models.
Acknowledgments
The authors apologize to all their colleagues whose important work could not be directly cited.
Footnotes
Financial & competing interests disclosure: The authors obtained research funding by grants from NIH/NIAID R21AI090156, RO1AI100933. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10(4):41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 2.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 3.Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat Protoc. 2009;4:582–591. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurdinger T. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 6.Burgos JS, Guzman-Sanchez F, Sastre I, Fillat C, Valdivieso F. Non-invasive bioluminescence imaging for monitoring herpes simplex virus type 1 hematogenous infection. Microbes Infect. 2006;8(5):1330–1338. doi: 10.1016/j.micinf.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Thorne SH. Design and testing of novel oncolytic vaccinia strains. Methods Mol Biol. 2009;542:635–647. doi: 10.1007/978-1-59745-561-9_32. [DOI] [PubMed] [Google Scholar]
- 8.Luker GD, Bardill JP, Prior JL, Pica CM, Piwnica-Worms D, Leib DA. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J Virol. 2002;76(23):12149–12161. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luker GD, Prior JL, Song J, Pica CM, Leib DA. Bioluminescence imaging reveals systemic dissemination of herpes simplex virus type 1 in the absence of interferon receptors. J Virol. 2003;77(20):11082–11093. doi: 10.1128/JVI.77.20.11082-11093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasieka TJ, Collins L, O'connor MA, et al. Bioluminescent imaging reveals divergent viral pathogenesis in two strains of Stat1-deficient mice, and in alphassgamma interferon receptor-deficient mice. PLoS ONE. 2011;6(9):e24018. doi: 10.1371/journal.pone.0024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luker KE, Schultz T, Romine J, Leib DA, Luker GD. Transgenic reporter mouse for bioluminescence imaging of herpes simplex virus 1 infection in living mice. Virology. 2006;347(2):286–295. doi: 10.1016/j.virol.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81(13):7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mutnal MB, Hu S, Lokensgard JR. Persistent humoral immune responses in the CNS limit recovery of reactivated murine cytomegalovirus. PLoS ONE. 2012;7(3):e33143. doi: 10.1371/journal.pone.0033143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wet JR, Wood KV, Helinski DR, Deluca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci USA. 1985;82(23):7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lembert N, Idahl LA. Regulatory effects of ATP and luciferin on firefly luciferase activity. Biochem J. 1995;305(Pt 3):929–933. doi: 10.1042/bj3050929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99(1):377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacer AC, Nyati S, Moudgil P, et al. NanoLuc reporter for dual luciferase imaging in living animals. Mol Imaging. 2013;12(7):1–13. [PMC free article] [PubMed] [Google Scholar]
- 18.Villalobos V, Naik S, Bruinsma M, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol. 2010;17(9):1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimitsu M, Sato T, Tao K, et al. Bioluminescent imaging of a marking transgene and correction of Fabry mice by neonatal injection of recombinant lentiviral vectors. Proc Natl Acad Sci USA. 2004;101(48):16909–16914. doi: 10.1073/pnas.0407572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans MS, Chaurette JP, Adams ST, Jr, et al. A synthetic luciferin improves bioluminescence imaging in live mice. Nat Methods. 2014;11(4):393–395. doi: 10.1038/nmeth.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X, Ren P, Dhal S, et al. Noninvasive monitoring of placenta-specific transgene expression by bioluminescence imaging. PLoS ONE. 2011;6(1):e16348. doi: 10.1371/journal.pone.0016348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branchini BR, Ablamsky DM, Murtiashaw MH, Uzasci L, Fraga H, Southworth TL. Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2007;361(2):253–262. doi: 10.1016/j.ab.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Doyle TC, Wong RJ, et al. Characterization of coelenterazine analogs for measurements of Renilla luciferase activity in live cells and living animals. Mol Imaging. 2004;3(1):43–54. doi: 10.1162/15353500200403181. [DOI] [PubMed] [Google Scholar]
- 24.Kim SB, Suzuki H, Sato M, Tao H. Superluminescent variants of marine luciferases for bioassays. Anal Chem. 2011;83(22):8732–8740. doi: 10.1021/ac2021882. [DOI] [PubMed] [Google Scholar]
- 25.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4(8):641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 26.Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat Protoc. 2009;4(4):582–591. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wurdinger T, Badr C, Pike L, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P. In vivo bioluminescent imaging of influenza A virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol. 2013;87(15):8272–8281. doi: 10.1128/JVI.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maguire CA, Bovenberg MS, Crommentuijn MH, et al. Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol Ther Nucleic Acids. 2013;2:e99. doi: 10.1038/mtna.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wendt MK, Molter J, Flask CA, Schiemann WP. In vivo dual substrate bioluminescent imaging. J Vis Exp. 2011;(56):e3245. doi: 10.3791/3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cronin M, Akin AR, Collins SA, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS ONE. 2012;7(1):e30940. doi: 10.1371/journal.pone.0030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall MP, Unch J, Binkowski BF, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo C, Coquoz O, Troy TL, Xu H, Rice BW. Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging. J Biomed Opt. 2007;12(2):024007. doi: 10.1117/1.2717898. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Machado HB, Bao Q, Stout D, Herschman H, Chatziioannou AF. In vivo mouse bioluminescence tomography with radionuclide-based imaging validation. Mol Imaging Biol. 2011;13(1):53–58. doi: 10.1007/s11307-010-0332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J, Qin C, Jia K, Zhu S, Yang X, Tian J. Bioluminescence tomography imaging in vivo: recent advances. J Sel Topics Quant Electr. 2012;18(4) [Google Scholar]
- 36.Bray M, Lawler J, Paragas J, Jahrling PB, Mollura DJ. Molecular imaging of influenza and other emerging respiratory viral infections. J Infect Dis. 2011;203(10):1348–1359. doi: 10.1093/infdis/jir038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.In vivo imaging technology. www.perkinelmer.com
- 38.Berthold. www.berthold.com/en
- 39.Photometrics. www.photometrics.com
- 40.Li-Cor Biosciences. http://licor.com
- 41.Spectral Instruments. www.specimg.com
- 42.TriFoil Imaging. www.trifoilimaging.com
- 43.Bruker. www.bruker-preclinical.com
- 44.Luker KE, Luker GD. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral Res. 2008;78(3):179–187. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeamari S, Rumping G, Floot B, Lyons S, Stewart FA. In vivo bioluminescence imaging of locally disseminated colon carcinoma in rats. Br J Cancer. 2004;90(6):1259–1264. doi: 10.1038/sj.bjc.6601637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mcgregor A, Mcvoy MA, Schleiss MR. The guinea pig model of congenital cytomegalovirus infection. In: Reddehase MJ, editor. Cytomegaloviruses. Caister Academic Press; Norfolk, UK: 2013. pp. 88–118. [Google Scholar]
- 47.Karlsson EA, Meliopoulos VA, Tran V, Mehle A, Schultz-Cherry S. Real-time in vivo imaging of influenza virus transmission dynamics in ferrets; Presented at: American Society for Virology 33rd Annual Meeting; Fort Collins, CO, USA. Jun, 2014. pp. 21–25. [Google Scholar]
- 48.Edinger M, Cao YA, Hornig YS, et al. Advancing animal models of neoplasia through in vivo bioluminescence imaging. Eur J Cancer. 2002;38(16):2128–2136. doi: 10.1016/s0959-8049(02)00410-0. [DOI] [PubMed] [Google Scholar]
- 49.Curtis A, Calabro K, Galarneau JR, Bigio IJ, Krucker T. Temporal variations of skin pigmentation in C57BL/6 mice affect optical bioluminescence quantitation. Mol Imaging Biol. 2011;13(6):1114–1123. doi: 10.1007/s11307-010-0440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'neill K, Lyons SK, Gallagher WM, Curran KM, Byrne AT. Bioluminescent imaging: a critical tool in pre-clinical oncology research. J Pathol. 2010;220(3):317–327. doi: 10.1002/path.2656. [DOI] [PubMed] [Google Scholar]
- 51.Berger F, Paulmurugan R, Bhaumik S, Gambhir SS. Uptake kinetics andbiodistribution of 14C-D-luciferin – a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur J Nucl Med Mol Imaging. 2008;35(12):2275–2285. doi: 10.1007/s00259-008-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue Y, Kiryu S, Izawa K, Watanabe M, Tojo A, Ohtomo K. Comparison of subcutaneous and intraperitoneal injection of D-luciferin for in vivo bioluminescence imaging. Eur J Nucl Med Mol Imaging. 2009;36(5):771–779. doi: 10.1007/s00259-008-1022-8. [DOI] [PubMed] [Google Scholar]
- 53.Keyaerts M, Remory I, Caveliers V, et al. Inhibition of firefly luciferase by general anesthetics: effect on in vitro and in vivo bioluminescence imaging. PLoS ONE. 2012;7(1):e30061. doi: 10.1371/journal.pone.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aswendt M, Adamczak J, Couillard-DespRes S, Hoehn M. Boosting bioluminescence neuroimaging: an optimized protocol for brain studies. PLoS ONE. 2013;8(2):e55662. doi: 10.1371/journal.pone.0055662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hildebrandt IJ, Su H, Weber WA. Anesthesia and other considerations for in vivo imaging of small animals. ILAR J. 2008;49(1):17–26. doi: 10.1093/ilar.49.1.17. [DOI] [PubMed] [Google Scholar]
- 56.Cook SH, Griffin DE. Luciferase imaging of a neurotropic viral infection in intact animals. J Virol. 2003;77(9):5333–5338. doi: 10.1128/JVI.77.9.5333-5338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Summers BC, Leib DA. Herpes simplex virus type 1 origins of DNA replication play no role in the regulation of flanking promoters. J Virol. 2002;76(14):7020–7029. doi: 10.1128/JVI.76.14.7020-7029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy AA, Rosato PC, Parker ZM, Khalenkov A, Leib DA. Synergistic control of herpes simplex virus pathogenesis by IRF-3, and IRF-7 revealed through non-invasive bioluminescence imaging. Virology. 2013;444(1–2):71–79. doi: 10.1016/j.virol.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasieka TJ, Menachery VD, Rosato PC, Leib DA. Corneal replication is an interferon response-independent bottleneck for virulence of herpes simplex virus 1 in the absence of virion host shutoff. J Virol. 2012;86(14):7692–7695. doi: 10.1128/JVI.00761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Selariu A, Warden C, et al. Genome-wide mutagenesis reveals that ORF7 is a novel VZV skin-tropic factor. PLoS Pathog. 2010;6:e1000971. doi: 10.1371/journal.ppat.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Huang Y, Zhu H. A highly efficient protocol of generating and analyzing VZV ORF deletion mutants based on a newly developed luciferase VZV BAC system. J Virol Methods. 2008;148(1–2):197–204. doi: 10.1016/j.jviromet.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z, Rowe J, Wang W, et al. Genetic analysis of varicella-zoster virus ORF0 to ORF4 by use of a novel luciferase bacterial artificial chromosome system. J Virol. 2007;81(17):9024–9033. doi: 10.1128/JVI.02666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zerboni L, Arvin A. Investigation of varicella-zoster virus neurotropism and neurovirulence using SCID mouse-human DRG xenografts. J NeuroVirol. 2011;17(6):570–577. doi: 10.1007/s13365-011-0066-x. [DOI] [PubMed] [Google Scholar]
- 64.Hwang S, Wu TT, Tong LM, et al. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol. 2008;82(24):12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reese TA, Wakeman BS, Choi HS, et al. Coinfection Helminth infection reactivates latent gamma-herpes virus via cytokine competition at a viral promoter. Science. 2014;345(6196):573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marquardt A, Halle S, Seckert CK, et al. Single cell detection of latent cytomegalovirus reactivation in host tissue. J Gen Virol. 2011;92(Pt 6):1279–1291. doi: 10.1099/vir.0.029827-0. [DOI] [PubMed] [Google Scholar]
- 67.Luker KE, Hutchens M, Schultz T, Pekosz A, Luker GD. Bioluminescence imaging of vaccinia virus: effects of interferon on viral replication and spread. Virology. 2005;341(2):284–300. doi: 10.1016/j.virol.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 68.Rivera R, Hutchens M, Luker KE, Sonstein J, Curtis JL, Luker GD. Murine alveolar macrophages limit replication of vaccinia virus. Virology. 2007;363(1):48–58. doi: 10.1016/j.virol.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hutson CL, Lee KN, Abel J, et al. Monkeypox zoonotic associations: insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am J Trop Med Hyg. 2007;76(4):757–768. [PubMed] [Google Scholar]
- 70.Falendysz EA, Londono-Navas AM, Meteyer CU, et al. Evaluation of monkeypox virus infection of black-tailed prairie dogs (Cynomys ludovicianus) using in vivo bioluminescent imaging. J Wildl Dis. 2014;50(3):524–536. doi: 10.7589/2013-07-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osorio JE, Iams KP, Meteyer CU, Rocke TE. Comparison of monkeypox viruses pathogenesis in mice by in vivo imaging. PLoS ONE. 2009;4(8):e6592. doi: 10.1371/journal.pone.0006592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tran V, Moser LA, Poole DS, Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J Virol. 2013;87(24):13321–13329. doi: 10.1128/JVI.02381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li XF, Deng YQ, Zhao H, et al. Noninvasive bioluminescence imaging of dengue virus infection in the brain of A129 mice. Appl MicroBiol Biotechnol. 2013;97(10):4589–4596. doi: 10.1007/s00253-013-4799-8. [DOI] [PubMed] [Google Scholar]
- 74.Schoggins JW, Dorner M, Feulner M, et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci USA. 2012;109(36):14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.WHO. Ebola virus disease update – West Africa. 2014 www.who.int.
- 76.Uebelhoer LS, Albarino CG, Mcmullan LK, et al. High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res. 2014;106:86–94. doi: 10.1016/j.antiviral.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 77.Hoenen T, Groseth A, Callison J, Takada A, Feldmann H. A novel Ebola virus expressing luciferase allows for rapid and quantitative testing of antivirals. Antiviral Res. 2013;99(3):207–213. doi: 10.1016/j.antiviral.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mcgregor A, Choi KY. Cytomegalovirus antivirals and development of improved animal models. Expert Opin Drug Metab Toxicol. 2011;7(10):1245–1265. doi: 10.1517/17425255.2011.613824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakayama E, Saijo M. Animal models for Ebola and Marburg virus infections. Front MicroBiol. 2013;4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deroose CM, Reumers V, Gijsbers R, et al. Noninvasive monitoring of long-term lentiviral vector-mediated gene expression in rodent brain with bioluminescence imaging. Mol Ther. 2006;14(3):423–431. doi: 10.1016/j.ymthe.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474(7350):208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson M, Huyn S, Burton J, Sato M, Wu L. Differential biodistribution of adenoviral vector in vivo as monitored by bioluminescence imaging and quantitative polymerase chain reaction. Hum Gene Ther. 2006;17(12):1262–1269. doi: 10.1089/hum.2006.17.1262. [DOI] [PubMed] [Google Scholar]
- 83.Niu G, Xiong Z, Cheng Z, et al. In vivo bioluminescence tumor imaging of RGD peptide-modified adenoviral vector encoding firefly luciferase reporter gene. Mol Imaging Biol. 2007;9(3):126–134. doi: 10.1007/s11307-007-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franceschi V, Stellari FF, Mangia C, et al. In vivo image analysis of BoHV-4-based vector in mice. PLoS ONE. 2014;9(4):e95779. doi: 10.1371/journal.pone.0095779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Msaouel P, Opyrchal M, Domingo Musibay E, Galanis E. Oncolytic measles virus strains as novel anticancer agents. Expert Opin Biol Ther. 2013;13(4):483–502. doi: 10.1517/14712598.2013.749851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuruppu D, Brownell AL, Shah K, Mahmood U, Tanabe KK. Molecular imaging with bioluminescence and PET reveals viral oncolysis kinetics and tumor viability. Cancer Res. 2014;74(15):4111–4121. doi: 10.1158/0008-5472.CAN-13-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lun X, Ruan Y, Jayanthan A, et al. Double-deleted vaccinia virus in virotherapy for refractory and metastatic pediatric solid tumors. Mol Oncol. 2013;7(5):944–954. doi: 10.1016/j.molonc.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sugiyama T, Yoneda M, Kuraishi T, et al. Measles virus selectively blind to signaling lymphocyte activation molecule as a novel oncolytic virus for breast cancer treatment. Gene Ther. 2013;20(3):338–347. doi: 10.1038/gt.2012.44. [DOI] [PubMed] [Google Scholar]
- 89.LPTA Models. www.taconic.com/find-your-model
- 90.Lienenklaus S, Cornitescu M, Zietara N, et al. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol. 2009;183(5):3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 91.Branchini BR, Southworth TL, Khattak NF, Michelini E, Roda A. Red- and green-emitting firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2005;345(1):140–148. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 92.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7(1):5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Degeling MH, Bovenberg MS, Lewandrowski GK, et al. Directed molecular evolution reveals Gaussia luciferase variants with enhanced light output stability. Anal Chem. 2013;85(5):3006–3012. doi: 10.1021/ac4003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bovenberg MS, Degeling MH, Hejazi S, et al. Multiplex blood reporters for simultaneous monitoring of cellular processes. Anal Chem. 2013;85(21):10205–10210. doi: 10.1021/ac401798v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Branchini BR, Ablamsky DM, Rosenberg JC. Chemically modified firefly luciferase is an efficient source of near-infrared light. Bioconjug Chem. 2010;21(11):2023–2030. doi: 10.1021/bc100256d. [DOI] [PubMed] [Google Scholar]
- 96.Morse D, Tannous BA. A water-soluble coelenterazine for sensitive in vivo imaging of coelenterate luciferases. Mol Ther. 2012;20(4):692–693. doi: 10.1038/mt.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reddy GR, Thompson WC, Miller SC. Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J Am Chem Soc. 2010;132(39):13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kheirolomoom A, Kruse DE, Qin S, et al. Enhanced in vivo bioluminescence imaging using liposomal luciferin delivery system. J Control Release. 2010;141(2):128–136. doi: 10.1016/j.jconrel.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Conley NR, Dragulescu-Andrasi A, Rao J, Moerner WE. A selenium analogue of firefly D-luciferin with red-shifted bioluminescence emission. Angew Chem Int Ed Engl. 2012;51(14):3350–3353. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kojima R, Takakura H, Ozawa T, Tada Y, Nagano T, Urano Y. Rational design and development of near-infrared-emitting firefly luciferins available in vivo. Angew Chem Int Ed Engl. 2013;52(4):1175–1179. doi: 10.1002/anie.201205151. [DOI] [PubMed] [Google Scholar]
- 101.Xu T, Ripp S, Sayler GS, Close DM. Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS ONE. 2014;9(5):e96347. doi: 10.1371/journal.pone.0096347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Billerbeck E, Horwitz JA, Labitt RN, et al. Characterization of human antiviral adaptive immune responses during hepatotropic virus infection in HLA-transgenic human immune system mice. J Immunol. 2013;191(4):1753–1764. doi: 10.4049/jimmunol.1201518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Massoud TF, Paulmurugan R, Gambhir SS. Molecular imaging of homodimeric protein-protein interactions in living subjects. FASEB J. 2004;18(10):1105–1107. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Fu Q, Dong Y, et al. Bioluminescence imaging of Hepatitis C virus NS3/4A serine protease activity in cells and living animals. Antiviral Res. 2010;87(1):50–56. doi: 10.1016/j.antiviral.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 105.Fu Q, Duan X, Yan S, et al. Bioluminescence imaging of caspase-3 activity in mouse liver. Apoptosis. 2013;18(8):998–1007. doi: 10.1007/s10495-013-0849-z. [DOI] [PubMed] [Google Scholar]
- 106.Scabini M, Stellari F, Cappella P, Rizzitano S, Texido G, Pesenti E. In vivo imaging of early stage apoptosis by measuring real-time caspase-3/7 activation. Apoptosis. 2011;16(2):198–207. doi: 10.1007/s10495-010-0553-1. [DOI] [PubMed] [Google Scholar]


