Abstract
Elevations in the intracellular Ca2+ concentration are a phenomena commonly observed during stem cell differentiation but cease after the process is complete. The Transient Receptor Potential Melastatin 4 (TRPM4) is an ion channel that controls Ca2+ signals in excitable and non-excitable cells. However, its role in stem cells remains unknown. The aim of this study was to characterize TRPM4 in rat dental follicle stem cells (DFSCs) and to determine its impact on Ca2+ signaling and the differentiation process. We identified TRPM4 gene expression in DFSCs, but not TRPM5, a closely related channel with similar function. Perfusion of cells with increasing buffered Ca2+ resulted in a concentration-dependent activation of currents typical for TRPM4, which were also voltage-dependent and had Na+ conductivity. Molecular suppression with shRNA decreased channel activity and cell proliferation during osteogenesis, but not adipogenesis. As a result, enhanced mineralization and alkaline phosphatase enzyme activity were observed during osteoblast formation, although DFSCs failed to differentiate into adipocytes. Furthermore, the normal agonist-induced first and secondary phases of Ca2+ signals were transformed into a gradual and sustained increase which confirmed the channels’ ability to control Ca2+ signaling. Using whole genome microarray analysis, we identified several genes impacted by TRPM4 during DFSC differentiation. These findings suggest an inhibitory role for TRPM4 on osteogenesis while it appears to be required for adipogenesis. The data also provide a potential link between the Ca2+ signaling pattern and gene expression during stem cell differentiation.
Keywords: TRPM4, dental follicle stem cells, calcium signaling, differentiation
Introduction
Stem cell therapy offers a promising approach to providing an advanced and reliable therapeutic strategy for tissue regeneration and disease treatment. However, fundamental processes controlling the fate of stem cells are not well understood. Dental follicle stem cells (DFSCs) are derived from the neural crest and give origin to the periodontal ligament (1, 2). These cells are multipotent and can differentiate into various cell types including osteoblasts, adipocytes and neurons (3). In addition, they can be easily obtained from extracted third molars that are usually discarded as medical waste. Therefore, DFSCs represent an alternative source of stem cells for tissue regeneration. The Transient Receptor Potential (TRP) proteins are a family of ion channels initially identified in the Drosophila melanogaster visual system (4). Despite intensive research, information regarding their function in stem cells remains largely unknown. The melastatin subfamily of TRP channels is comprised of eight members (TRPM1-8), with TRPM4 and TRPM5 being the only non-calcium conducting channels (5, 6). Both are permeable mainly to Na+, resulting in depolarization upon channel activation. The ability of TRPM4 to depolarize cells transforms the normal intracellular Ca2+ oscillations into sustained Ca2+ increases in T-lymphocytes (7). This is due to a decrease in the driving force for Ca2+ entry via store-operated Ca2+ channels (SOCs), the main pathway for Ca2+ entry in non-excitable cells, such as dental follicle stem cells (DFSCs) of mesenchymal origin (3). Of the TRPMs, only the TRPM7 has been reported in stem cells. It is essential for bone marrow-derived mesenchymal stem cell proliferation and survival and is required for early embryonic development (8, 9).
Oscillations in the intracellular Ca2+ concentration ([Ca2+]i) are commonly observed during stem cell differentiation and there is evidence that they may control the differentiation process. Physical manipulation of Ca2+ signals with non-invasive electrical stimulation enhances Ca2+ entry and osteodifferentiation of human mesenchymal stem cells (hMSCs; (10)). That study suggests that increased Ca2+ entry is a result of activation of G-protein coupled receptors and the opening of Ca2+ channels. In addition, activation of gene transcription by NFAT in immune cells appears to be controlled by the shape and frequency of the Ca2+ signals (7, 11). Interestingly, both Ca2+ signals and NFAT-activated gene transcription disappear at the completion of adipogenesis in hMSCs (12). Similar observations have been made during the terminal stages of osteoblast differentiation (10), implying that Ca2+ signals may be important for directing and terminating the process. Furthermore, oscillations in the [Ca2+]i control the transition from the G1 phase to the S phase of the cell cycle to preserve embryonic stem cell (ESC) pluripotency (13). Therefore, the question of how Ca2+ signals control stem cell differentiation is fundamentally important.
The TRPM4 channel is a widely expressed protein present in both electrically excitable and non-excitable cells. Patch-clamp recordings revealed that it is a Ca2+-Activated Non-selective cation (CAN) channel, inhibited by nucleotides and polyamines (5, 14). Although not permeable to Ca2+, TRPM4 has a significant impact on Ca2+ signals because it provides a mechanism that allows cells to depolarize in a Ca2+-dependent manner. In non-excitable cells such as undifferentiated stem cells, TRPM4-mediated depolarization decreases the driving force for Ca2+ entry through SOCs, whereas in excitable cells (e.g. neuron, endocrine or cardiac muscle), TRPM4 has the opposite effect by providing the depolarization necessary for the opening of voltage dependent Ca2+ channels (VDCCs). Previous studies identified SOCs in hMSCs and mESCs (15, 16). In fact, molecular suppression of TRPM4 increases both Ca2+ entry via SOCs and IL-2 production in non-excitable T-lymphocytes(7). Studies in excitable cells revealed a significant reduction in insulin secretion during glucose stimulation in pancreatic β-cells after TRPM4 knockdown (17); this reduction results from a decrease in the magnitude of the Ca2+ signals (18). A similar observation was made in glucagon secreting α-cells (19). In addition to the effects in immune and islet cells, the control of Ca2+ signals by TRPM4 is critical for myogenic constriction of cerebral arteries, migration of dendritic cells and cardiac function (20–22). Given the importance of Ca2+ signals for stem cell differentiation, it is possible that ion channels such as TRPM4 could be involved in their regulatory mechanism.
In this study, we investigated the role of TRPM4 in differentiation of rat DFSC, a mesenchymal stem cell from the first molar tooth. We examined TRPM4 gene expression by RT-PCR and tested whether currents with the characteristics of those known for this channel could be detected using the patch-clamp technique. To gain insight into TRPM4 function, we generated stable knockdown cells via shRNA. These cells were then used in cell proliferation, Ca2+ imaging analysis and differentiation experiments. Finally, we performed whole genome microarray analysis to examine potential genes regulated by TRPM4 during DFSC differentiation.
Materials and Methods
Reagents
All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA.), except for fura-2 acetoxymethyl ester (Fura-2AM), which was from Molecular Probes (Eugene, OR, USA).
Cell culture
Dental follicle stem cells of the first mandibular molar were harvested from Sprague-Dawley rat pups 5–7 days post-natal and cultured according to published methods (23). Cells were grown in α-MEM medium supplemented with 20% FBS and aerated with 5% CO2 and 95% air at 37°C. Experiments were performed with cells from passages 2 through 8. MC3T3-E1preosteoblast cells were grown in α-MEM medium supplemented with 10% FBS and cultured as described for DFSCs. Human adipocyte stem cells (hACSs) were maintained in Dulbecco’s Modified Eagles Medium (DMEM)/Ham’s F-12 medium (Sigma-Aldrich, St. Louis, MO, USA) with 10% FBS and aerated with 5% CO2 at 37°C. All experiments were performed with cells from passages 6 through 8.
Induction and detection of cell differentiation
Osteogenesis
Cells were induced with osteogenic medium, consisting of DMEM-LG supplemented with 10% FBS, 10nM dexamethasone, 0.1mM ascorbic-acid-2-phosphate and 10mM β-glycerophosphate. The medium was changed every four days during osteogenesis. After a 14-day period, the medium was aspirated and the cultures washed twice with PBS, fixed with 10% formaldehyde, washed again with distilled water and incubated with 1% Alizarin Red S (ARS) in dH2O. After incubation, the staining solution was removed and the cultures washed with distilled water to remove excess dye. Stained monolayers were visualized by phase contrast microscopy with an inverted microscope (Zeiss, Thornwood, NY, USA).
Adipogenesis
Cells were induced with adipogenic medium containing DMEM-LG supplemented with 10% FBS, 50µg/ml ascorbic acid, 0.1µM dexamethazone, and 50µg/ml indomethacin. Adipogenesis was determined by Oil Red O (ORO) staining. Briefly, cells were washed with PBS, fixed with 10% formalin for 10 min, washed twice with dH2O and incubated with 60% isopropanol for 5 min. Cells were then stained with ORO solution for 5 min and washed again to remove excess dye. The presence of lipid droplets was visualized by phase contrast microscopy with an inverted microscope.
RT-PCR
RNA was extracted from DFSCs, hASCs, MC3T3-E1 cells and bone tissue using the RNAqueous-4PCR® kit according to manufacturer’s instructions (Ambion, Austin, TX, USA). The RNA was purified with DNase 1 treatment. Reverse transcription was performed with MMLV-Reverse Transcriptase. PCR was performed with Ambion’s RETROscript® kit and rat primers with the sequences listed (forward/reverse [5’ to 3’]): TTGGCATACTGGGAGACGCA/GGCCCAAGATCGTCATCGT (TRPM4; 301bp); CAAGTGTGACATGGTGGCCATCTT/GCTCAGGTGGCTGAGCAGGAT (TRPM5; 600bp); GCAAATGACTCCACTCTC/GATTCTCTCTTCACTCCCAG (TRPM7; 422bp). Rat GAPDH (420bp) and ultrapure water were used as positive and negative controls, respectively. The human primers (forward/reverse [5’ to 3’]) were: AGCATGGTGCCGGAGAA/GGTGTCTCTATTCCGGACCACA (TRPM4; 600bp); CAGAACATCACCTCACACCAG/GGTTCTCGCTCTTCTGGTTC (TRPM5; 640 bp); TGAAACGAGTGAGTTCTCTTGCTG/CACAGGTGTAAATGGAATGCTC (TRPM7; 309bp); AACAGCGACACCCACTCCTC/GGAGGGGAGATTCAGTGTGGT (GAPDH; 258bp).
Electrophysiology
Cells were maintained in standard modified Ringer’s solution of the following composition (in mM): NaCl 140, KCl 2.8, CaCl2 1, MgCl2 2, glucose 4, HEPES-NaOH 10, pH 7.2 adjusted with NaOH. The standard internal solution contained (in mM): Cs-glutamate 120, NaCl 8, MgCl2 1, Cs-BAPTA 10, HEPES-CsOH 10, pH 7.2 adjusted with CsOH. The internal solution’s buffered Ca2+ concentration was adjusted as necessary with CaCl2 (calculated with WebMaxC http://www.stanford.edu/~cpatton/webmaxcS.htm). The Na+-free modified Ringer’s solution contained (in mM): choline-Cl 140, KCl 2.8, CaCl2 1, MgCl2 2, glucose 4, HEPES-CsOH 10, pH 7.2 adjusted with CsOH. The osmolarity of the solutions were ~300 mOsm/L. TRPM4 currents were recorded in the tight-seal whole-cell configuration mode at 21–25 °C. High-resolution current recordings were acquired by a computer-based patch-clamp amplifier system (EPC-10, HEKA, Lambrecht, Germany). Patch pipettes had resistances of 4–7 MΩ and were coated in Sigmacote® silicon solution (Sigma-Aldrich, St. Louis, MO, USA). Immediately following establishment of the whole-cell configuration, voltage ramps of 50 ms duration spanning the voltage range of −100 to +100 mV at a rate of 0.5 Hz over a period of 300–600 s. All voltages were corrected for a liquid junction potential of 10 mV between external and internal solutions, calculated with Igor PPT Liquid Junction Potential software (Wavemetrics, Portland, OR, USA).
Generation of TRPM4 knockdown cells
Lentivirus plasmids were obtained from Sigma-Aldrich (Saint Louis, MO, USA) in a pLKO.1 backbone and contained either nonspecific control (SHC002) or TRPM4 specific shRNA (SHDNA-NM_175130, TRCN0000068684 and TRCN0000068686) under the control of the U6 promoter, plus the puromycin resistance and GFP reporter genes. Cells were selected in 1µg/mL puromycin for one week, and transduction efficiency determined by FACScan flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) to sort GFP+ cells. Stably transduced cells were used for functional experiments.
Cell proliferation assay
An MTT assay was used to compare the proliferation rates between control and TRPM4 knockdown cells. One week following lentiviral transduction 104 cells/well were seeded into three 96-well plates and cultured in normal growth medium for three days. Cells were then placed in osteogenic or adipogenic medium and analyzed by an MTT-based assay kit (Bioassay Systems, Hayward, CA, USA) at 48, 72, and 96 hrs. MTT solution was added to each well and incubated at 37°C for 4 hrs. Then, solubilization buffer was added to dissolve the insoluble formazan product. The absorbance at 550nm was measured with an Ultramark Microplate Imaging System (BioRad, Hercules, CA, USA). Cell proliferation was expressed as the absorbance of the cells minus the background.
Calcium imaging analysis
Cells were loaded with 5µM Fura-2AM for 30 min at 37°C. A Ca2+-imaging buffer containing (in mM) NaCl 136, KCl 4.8, CaCl2 1.2, MgSO4 1.2, HEPES 10, glucose 4, and 0.1% BSA at a pH of 7.3 was used for Fura-2AM loading and perfusion throughout imaging experiments. Calcium measurements were obtained with a dual excitation fluorometric imaging system (TILL-Photonics, Gräfelfingen, Germany) controlled by TILLvisION software. Fura-2AM loaded cells were excited by wavelengths of 340nm and 380nm. Fluorescence emissions were sampled at a frequency of 1 Hz and computed into relative ratio units of the fluorescence intensity of the different wavelengths (F340/F380).
Alkaline phosphatase activity
Quantification of alkaline phosphatase (ALP) enzyme activity at different time points during osteogenesis was made with an ALP assay kit (BioChain, Newark, CA, USA) according to the manufacturer’s instructions. Differentiation experiments were performed in quadruplicate in a 24-well plate (4 wells/time point) and repeated three times. The cells were washed in PBS and lysed with 0.5 ml 0.2% Triton X-100 in distilled water prior to ALP activity determination. Samples were assayed in duplicate in a 96-well plate and analyzed at 405nm with a microcount plate reader BS10000 (Packard Instrument Co., Downers Grove, IL, USA).
Microarray analysis
Raw/normalized intensity values and the log-Ratios of all possible pair wise comparisons were pre-processed by the microarray manufacturer (PhalanxBio Inc., Belmont, CA, USA). The genes that were associated with osteogenesis or adipogenesis and affected by TRPM4 knockdown, were selected based on the following two criteria: first, genes displaying at least 2-fold up/down-regulation during cell differentiation in either cell population [minimum absolute fold-change ≥ 2 between control cells prior to differentiation (C0) and after 14 days (C14) and between TRPM4 knockdown cells prior to differentiation (KD0) and after 14 days (KD14)]. Second, fold change differences in genes exhibiting differential up/down-regulation between control and knockdown cell populations during cell differentiation (i.e., differences that exist between fold-change values found in control and knockdown cells).
Following gene selection, we identified the biological functions that are most significantly associated (P<0.05) with selected gene-sets via Ingenuity Pathway Analysis (IPA 9.0, Ingenuity Systems, Redwood City, CA, USA). A right-tailed Fisher’s exact test was used to calculate a P-value, determining the probability that each biological function assigned to that gene-set is due to chance alone. Gene networks were created with Ingenuity pathway designer.
Data analysis
Patch-clamp recordings are shown as means + S.E.M. and were plotted with Igor Pro 5 software program (Wavemetrics, Portland, OR, USA). The OD values from control and TRPM4 knockdown groups in the MTT and alkaline phosphatase assays are shown as means + S.E.M. and were compared by a two-tailed, unpaired Student’s t-test for each time point. Statistical significance was established at P<0.05.
Results
Dental follicle stem cells express TRPM4 and differentiate into osteoblasts and adipocytes
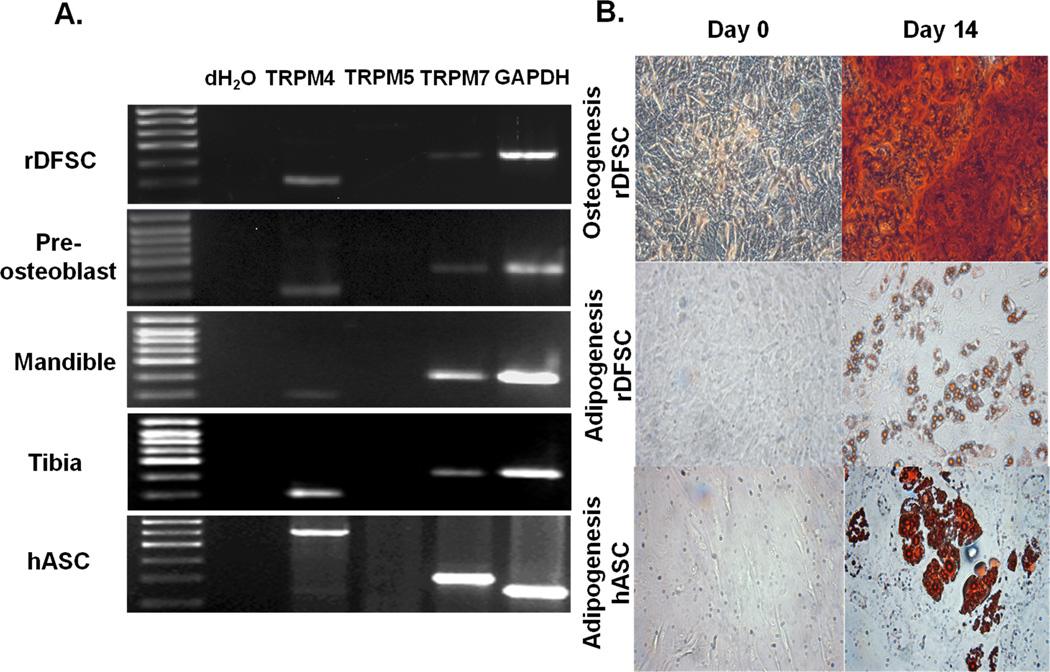
First, we used RT-PCR analysis to determine whether DFSCs expressed the TRPM4 gene. We identified TRPM4 expression with the predicted molecular size (301bp), but not the TRPM5 (600bp), a closely related channel with similar function (Fig. 1A). We also confirmed TRPM4 expression in murine preosteoblast MC3T3-E1cells and rat mandible and tibia bones. Both cells and bone tissues expressed the TRPM7 (422bp), which is required for cell proliferation and viability (8, 24). In order to investigate whether stem cells of human origin expressed TRPM4, we performed RT-PCR with RNA extracted from human adipocyte stem cells (hASCs). We identified similar gene expression pattern as the one observed with DFSCs (Fig. 1A). To demonstrate the multipotency of DFSCs, we tested their differentiation capability into osteoblasts and adipocytes determined by ARS and ORO staining at the end of 14 days (Fig. 1B). These stem cells were capable of differentiation into the respective cell types as indicated by extracellular matrix mineralization and lipid droplet accumulation. The adipogenic potential of hASCs is shown in Figure 1B.
Figure 1. TRPM4 gene expression and multipotency of dental follicle stem cells.
(A) Total RNA isolated from rat DFSCs, mandible and tibia bones, mouse pre-osteoblast MC3T3-E1 cells, human adipocyte stem cells (hASCs) and reverse-transcribed into cDNA. RT-PCR was performed with specific TRPM4 and TRPM5 primers. TRPM7 and GAPDH primers served as positive controls. (B) The multipotency of DFSCs was confirmed after 14 days in osteogenic (upper panel) and adipogenic (lower panel) differentiation medium with Alizarin Red S and Oil Red O staining. The adipogenic potential of hASCs is also shown. Note the presence of mineralization in the extracellular matrix as well as lipid droplet accumulation at 32× magnification.
Dental follicle stem cells have functional TRPM4 channels
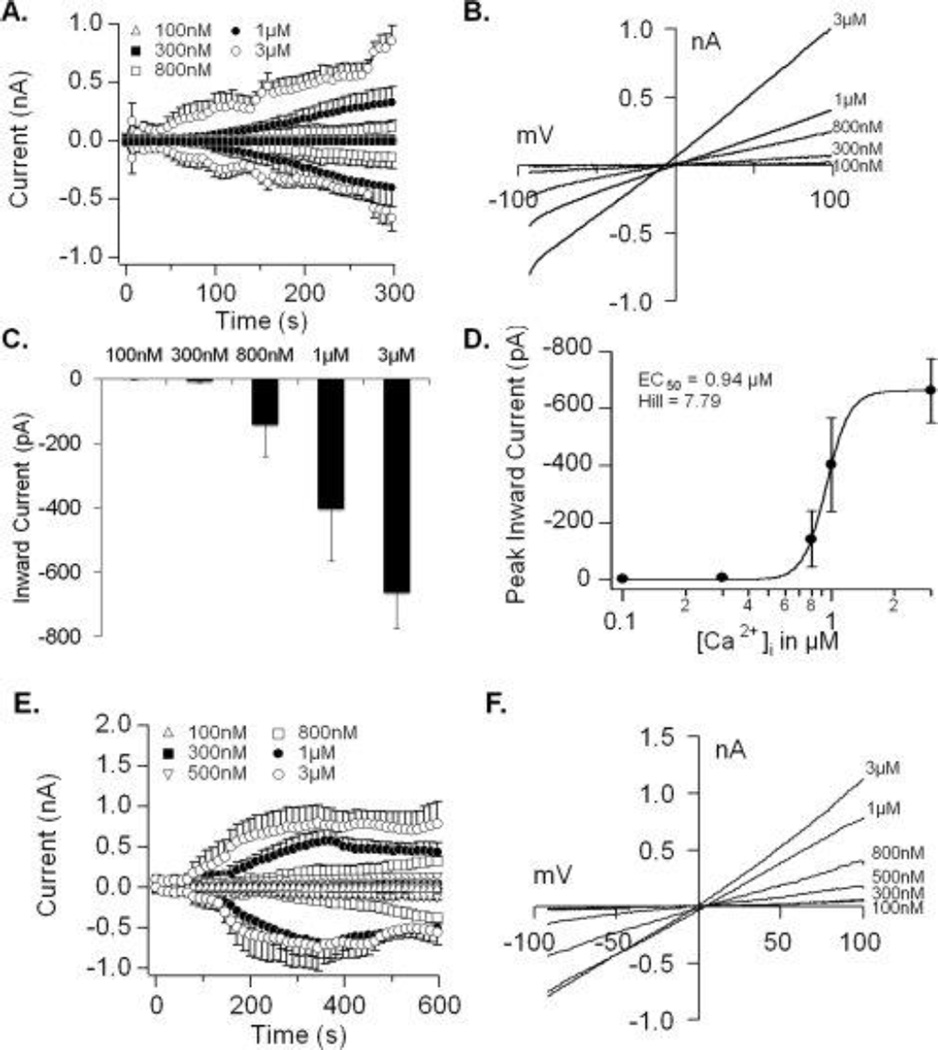
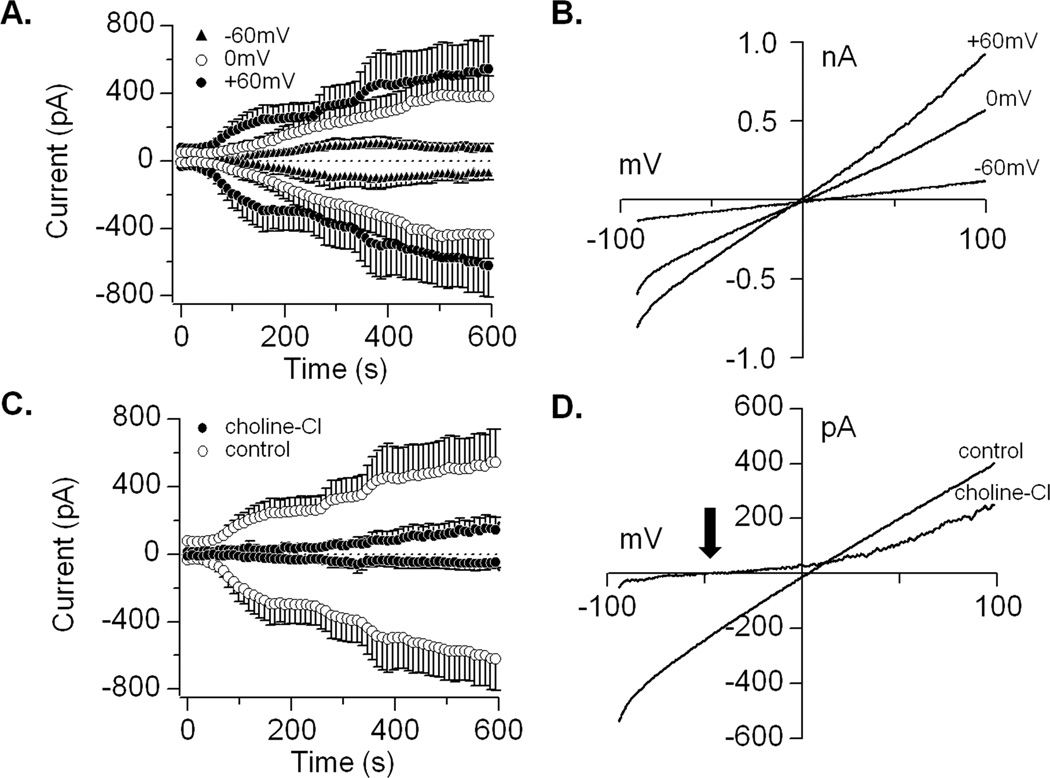
Next, we performed electrophysiological recordings to investigate the biophysical properties of TRPM4, which is activated by increases in [Ca2+]i (5). We performed whole-cell patch-clamp recordings during perfusion of DFSCs with buffered Ca2+ concentrations ranging from 0.1–3µM at 0mV holding potential (Fig. 2A). These experiments resulted in a concentration-dependent activation of TRPM4 currents with an EC50 of 0.94µM and Hill coefficient of 7.79 (Fig. 2C and D). The current-voltage relationships (I/V), that are the signature of an ion channel, are typical of those reported for TRPM4 (Fig. 2B). Perfusion of hASCs with increasing buffered Ca2+ concentrations (0.1–3 µM) also resulted in a concentration-dependent activation of currents and I/V similar to those described for channel (Fig. 2E and F). In addition to Ca2+ activation, TRPM4 is voltage-dependent, where negative potentials inhibit and positive potentials increase its open probability (5, 25). We tested the effect of −60mV, 0mV and +60mV holding potentials on TRPM4 currents with 1µM buffered Ca2+. Patch-clamp recordings at negative potentials suppressed and at positive potentials increased the current amplitude compared to 0mV (Fig. 3A). The effect of voltage on TRPM4 is shown by the I/V from representative cells (Fig. 3B). The opening of TRPM4 results in Na+ entry into cells and depolarization. Hence, we examined channel conductivity by replacing NaCl in the extracellular solution with choline chloride. Under this condition, inward currents were completely abolished compared to cells maintained in NaCl solution during experiments with 1µM buffered Ca2+ and 0mV holding potential (Fig. 3C). The replacement of Na+ caused a noticeable shift in the reversal potential due to hyperpolarization (Fig. 3D; black arrow).
Figure 2. Calcium-dependent activation of TRPM4 currents in dental follicle stem cells.
(A) Average inward and outward currents during perfusion with increasing buffered Ca2+ concentrations. Traces represent the mean + S.E.M. (n=5–8 cells/concentration) recorded at 0mV holding potential. (B) Current-voltage relationship (I/V) under experimental conditions described in (A) taken from representative cells at 300s for each Ca2+ concentration. (C) Average inward currents (mean + S.E.M.) from cells represented in panel A. (D) A dose-response analysis revealed an EC50 of 0.94µM and Hill coefficient of 7.79. (E) Average inward and outward currents during perfusion of hASCs with increasing buffered Ca2+ concentrations. Traces represent the mean + S.E.M. (n=3–6 cells/concentration) recorded at 0mV holding potential. (F) Current-voltage relationship (I/V) under experimental conditions described in (E) taken from representative cells at 600s for each Ca2+ concentration.
Figure 3. Voltage-dependency and ionic conductivity of TRPM4 in dental follicle stem cells.
(A) Average inward and outward currents in response to 1uM Ca2+ at +60mV, 0mV and −60mV holding potentials. Traces are mean + S.E.M. (n=5–6 cells/holding potential). (B) Current-voltage relationship (I/V) under experimental conditions described above at 600s from representative cells. (C) Average inward and outward currents from DFSCs maintained in NaCl solution compared to cells kept in extracellular buffer with choline-Cl replacing NaCl. Traces are mean + S.E.M. (n=6–12 cells). (D) Current-voltage relationship (I/V) taken at 600s from a representative cell. Note the shift in reversal potential (arrow) and hyperpolarization caused by the lack of Na+ entry with choline-Cl substitution.
Molecular suppression of TRPM4 inhibits channel activity and cell proliferation
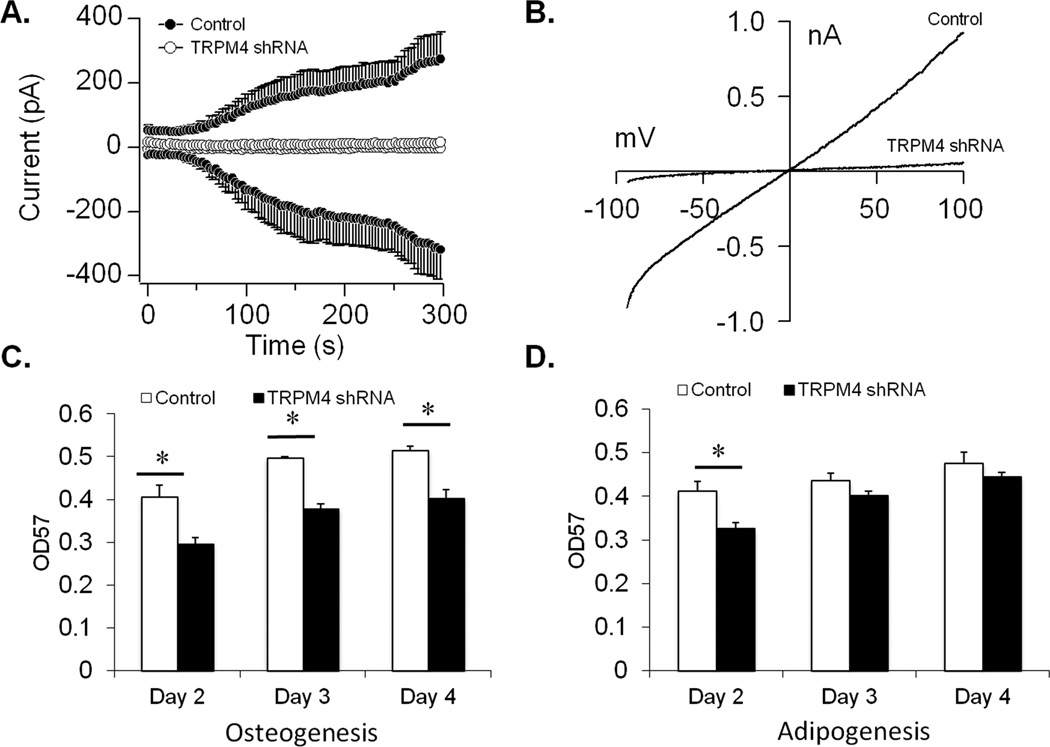
To investigate the functional significance of TRPM4 in DFSCs, we used shRNA and a lentiviral vector to generate stable TRPM4 knockdown cells. We confirmed the effectiveness of the shRNA by performing patch-clamp recordings with 1µM buffered Ca2+ and 0 mV HP to activate the channel. TRPM4 currents were significantly reduced in knockdown cells compared to control shRNA (Fig. 4A), and confirmed by the I/V (Fig. 4B). In stem cells, a decrease in cell proliferation is required prior to differentiation (26, 27). Therefore, we used the MTT assay to examine whether cell proliferation was impacted by TRPM4 during osteogenesis and adipogenesis. Inhibition of TRPM4 significantly decreased cell proliferation compared to control shRNA cells during a 96-hr period when placed in osteogenic medium (Fig. 4C). Under adipogenic conditions, there was a reduction in cell proliferation only during the initial 48-hrs with TRPM4 knockdown (Fig. 4D). Based on these results, we reasoned that if cell proliferation decreased during osteogenesis, there was a possibility that TRPM4 suppression would facilitate osteoblast, but not adipocyte differentiation.
Figure 4. TRPM4 knockdown and its impact on cell proliferation.
(A) Average inward and outward currents from control shRNA (n=9) and TRPM4 shRNA cells (n=6) recorded at +60mV holding potential and 1µM buffered Ca2+. A noticeable reduction in current amplitude is seen with TRPM4 knockdown. Traces represent mean + S.E.M. (B) Current-voltage relationship (I/V) obtained from representative cells at 300s. (C and D) The effect of TRPM4 knockdown on cell proliferation was examined by MTT assay. A significant decrease in cell proliferation was observed in TRPM4 shRNA cells compared to control shRNA at all time points under osteogenic, but not adipogenic conditions. Data are shown as mean + S.E.M. (n=6 wells/time point); *P<0.05.
TRPM4 inhibits osteogenesis but facilitates adipogenesis
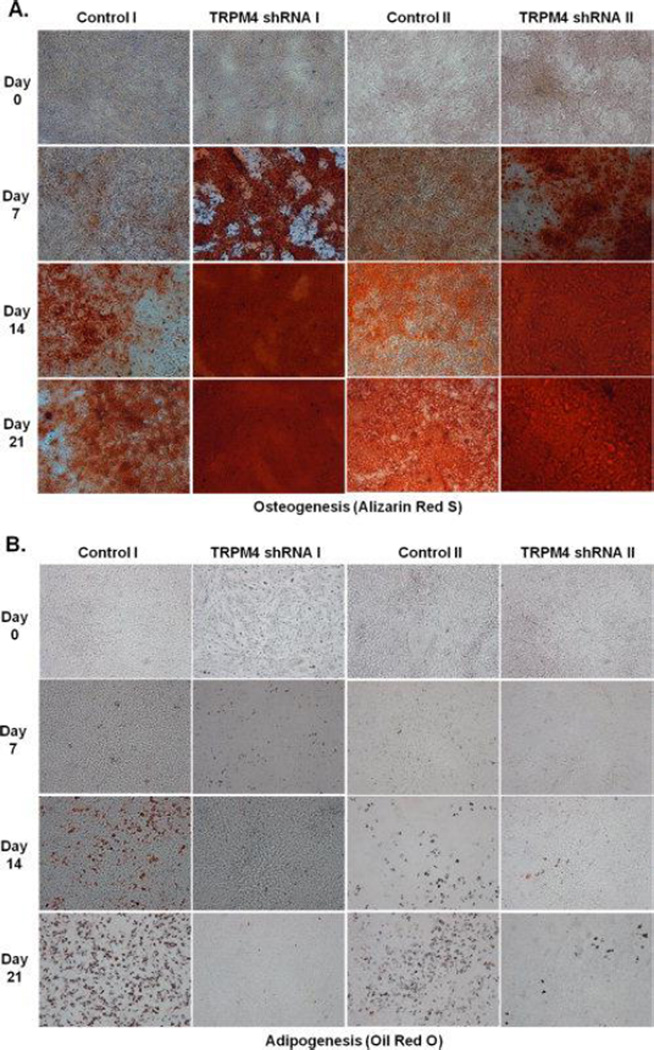
In order to test TRPM4’s function on DFSC differentiation, we cultured cells in osteogenic and adipogenic media over 21-days and performed ARS and ORO staining on days 0, 7, 14 and 21. Inhibition of TRPM4 with two different sets of shRNAs (I and II) enhanced mineralization of DFSCs under osteogenic conditions compared to control shRNA cells, as determined by ARS staining (Fig. 5A). Furthermore, DFSCs cultured under adipogenic conditions failed to differentiate into adipocytes, as indicated by the absence of ORO staining (Fig. 5B). These results suggest that TRPM4 inhibits osteogenesis, but is required for or at least facilitates adipogenesis.
Figure 5. Molecular suppression of TRPM4 enhances osteogenesis but inhibits adipogenesis.
(A) Extracellular matrix mineralization shown by Alizarin Red S staining during a 21-day period in control shRNA and TRPM4 shRNA cells. (B) Lipid droplet accumulation during a 21-day period in control shRNA and TRPM4 shRNA cells after Oil Red O staining. Suppression of TRPM4 enhanced mineralization during osteogenesis, but inhibited lipid droplet accumulation during adipogenesis. Images are representative of three different experiments (10× magnification).
TRPM4 controls Ca2+ signals and alkaline phosphatase enzyme activity
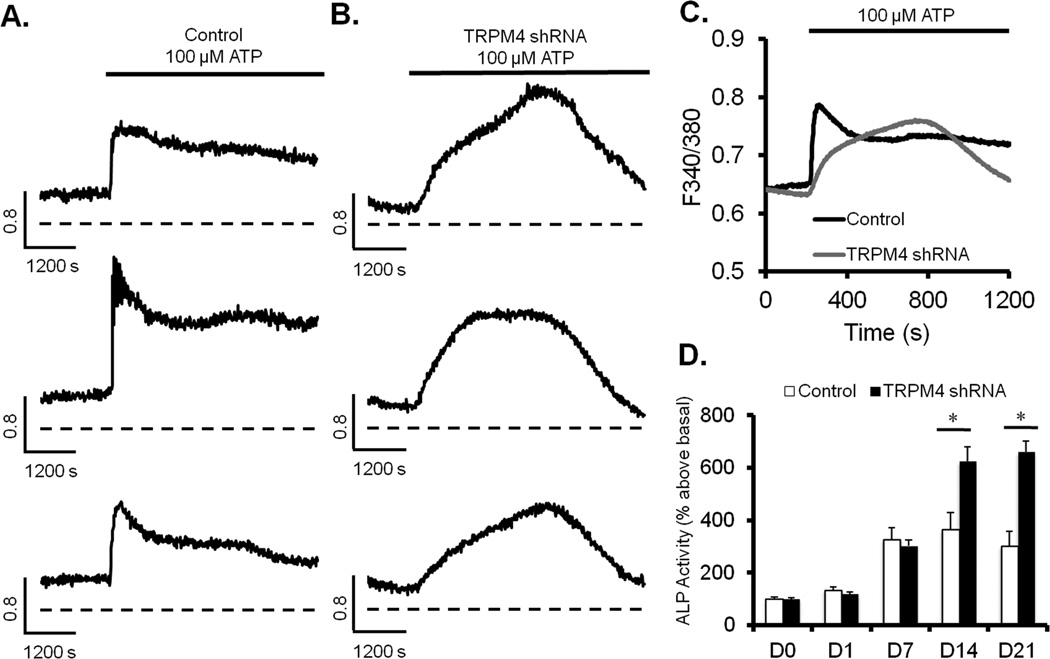
Calcium oscillations are linked to stem cell differentiation and TRPM4 is key regulator of Ca2+ signaling in different cell types (7, 18, 19, 28). Therefore, we performed real-time Ca2+ imaging analysis to determine its impact on Ca2+ signals generated by ATP, which influence stem cell differentiation (29, 30). Stimulation of control shRNA cells with 100µM ATP resulted in the typical increase in [Ca2+]i characterized by a first phase due to Ca2+ release from the ER, followed by a secondary phase due to Ca2+ influx via SOCs (Fig. 6A). TRPM4 knockdown transformed the biphasic Ca2+ pattern into a gradual and sustained increase (Fig. 6B). The average responses of cells from three different cell passages are shown in Figure 6C. The proliferation and osteogenic differentiation assays combined with the Ca2+ signaling data prompted us to investigate whether enhanced osteoblast differentiation and mineralization in knockdown cells were associated with an increase in alkaline phosphatase enzyme activity. This enzyme is one of the hallmarks for osteogenesis (31). We found that there was also a significant increase in alkaline phosphatase enzyme activity on days 14 and 21 of osteoblast differentiation compared to control shRNA cells (Fig. 6D).
Figure 6. TRPM4 regulates Ca2+ signals and increases alkaline phosphatase enzyme activity.
(A) Calcium signals from single control shRNA cells in response to 100µM ATP. (B) Same experiment as in A, except with TRPM4 shRNA cells. (C) Average Ca2+ signals from control shRNA (n=47 cells) and TRPM4 knockdown (n=64 cells) from three different experiments. (D) Alkaline phosphatase enzyme activity during a 21-day period under osteogenic differentiation conditions. Results are presented as mean + S.E.M. from three different experiments (n=4 wells/time point); *P<0.05.
Genes regulated by TRPM4 during osteogenic and adipogenic differentiation
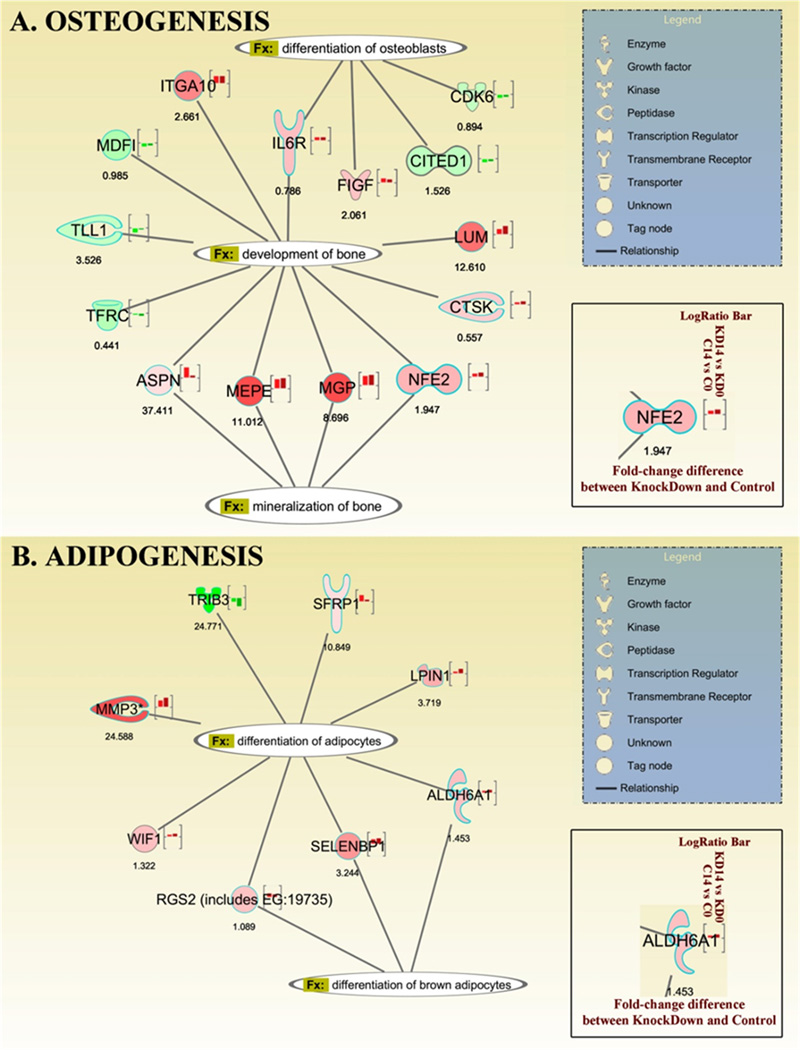
Finally, we used whole genome microarray analysis to investigate potential genes controlled by TRPM4 during 14 days of differentiation. Based on the approach described for microarray analysis, we first examined genes involved in osteogenesis because of the enhancement of mineralization and osteoblast formation with TRPM4 knockdown. We identified four genes involved in bone mineralization (Nuclear factor erythroid-derived 2 (NFE2), Asporin (ASPN), matrix extracellular phosphoglycoprotein (MEPE) and matrix γ-carboxyglutamate protein (MGP)). All four genes were up-regulated (MEPE 11 fold; MGP 8.6 fold; NFE2 1.9 fold) compared to control shRNA cells, while ASPN gene expression was 37.4 fold higher in controls than knockdown cells (Fig. 7A). We also identified four genes involved in osteoblast differentiation (Cbp/p300-interacting transactivator 1 (CITED1), Cyclin-dependent kinase 6 (CDK6), c-fos induced growth factor (FIGF) and Interleukin 6 receptor (IL6R)). Both CITED1 and CDK6 genes were down-regulated 1.5 and 0.9 fold in controls compared to TRPM4 shRNA cells, whereas FIGF and IL6 were up-regulated 2 and 0.7 fold respectively (Fig. 7A). In addition, we found the bone development gene, Lumican (LUM), up-regulated 12.6 fold with TRPM4 suppression. Furthermore, our results indicated that TRPM4 knockdown inhibited adipogenesis. We identified eight genes: Lipin 1 (LPIN1), tribbles homolog 3 (TRIB3), WNT inhibitory factor 1 (WIF1), secreted frizzled-related protein 1 (SFRP1), matrix metallopeptidase 3 (MMP3), selenium binding protein 1 (SELENBP1), aldehyde dehydrogenase 6 family member A1 (ALDH6A1) and regulator of G-protein signaling 2 (RGS2) involved in this process (Fig. 7B). Of the eight, the TRIB3 was the only gene down-regulated, with a 24.7 fold reduction in knockdown compared to control cells. From the up-regulated genes, SFRP1 (10.8 fold) and RGS2 (1 fold) expression were higher in control cells. TRPM4 suppression increased LPIN1 (3.7 fold), WIF1 (1.3 fold), MMP3 (22.9 fold), SELENBP1 (3.2 fold) and ALDH6A1 (1.4 fold) over control cells. The genes involved in brown fat differentiation are shown in the diagram (Fig. 7B).
Figure 7. Whole genome microarray analysis of osteogenic and adipogenic genes impacted by TRPM4.
(A) Gene expression related to osteoblast differentiation, bone development and mineralization in TRPM4 shRNA cells after 14-days in osteogenic conditions. (B) Gene expression related to adipocyte differentiation from TRPM4 shRNA cells after 14-days in adipogenic conditions. RNA was extracted from control shRNA [C] and TRPM4 knockdown cells [KD] prior to differentiation induction [C0 and KD0] and after two weeks of differentiation [C14 and KD14]. The values under each gene represent the difference in fold change between C14 and KD14 groups. Red/Green color indicates Up/Down regulation during differentiation in control (left bars) and knockdown cells (right bars). The biological functions shown in the diagrams have P<0.05.
Discussion
The study describes for the first time the expression and functions of TRPM4 in stem cells. Using rat DFSCs, we identified TRPM4 gene expression and currents typical for the channel, which were Ca2+-dependent, voltage sensitive and conducted mainly Na+. Perfusion of cells with increasing Ca2+ concentrations resulted in the activation of TRPM4 with an EC50 of 0.94µM and Hill coefficient of 7.79. This is consistent with previous findings in pancreatic α and β cells (19); thus it appears that in DFSCs, TRPM4 is more sensitive to changes in [Ca2+]i. The TRPM4 channel also exhibited a strong voltage dependency with currents suppressed at negative, and facilitated at positive holding potentials (5, 25). These currents were mainly due to Na+ entry into cells, since replacement of extracellular NaCl by choline-Cl resulted in almost complete abolishment under elevated Ca2+ concentrations. The absence of Na+ entry resulted in hyperpolarization, which was confirmed by the shift in the reversal potential to more negative values. These findings reveal that DFSCs posses functional TRPM4 channels. We also identified TRPM4 gene expression in mouse pre-osteoblast cells and rat bone tissue from mandible and tibia. This suggests that in addition to osteogenesis, TRPM4 may be important for osteoblast function. Because DFSCs are derived from mesoderm neural crest (23), we tested their differentiation capability into osteoblasts and adipocytes to confirm their multipotency prior to performing functional studies. Under both conditions, we were able to identify mineralization and lipid droplet accumulation after 14 days.
To investigate the role of TRPM4 in DFSC differentiation, we generated stable knockdown cells with shRNA and a lentiviral vector. We selected this approach because of concern over loss of knockdown effect with transient transfection during differentiation. Inhibition of TRPM4 was confirmed with elevated Ca2+ concentration and +60mV holding potential that resulted in a significant reduction in current amplitude. In DFSCs, inhibition of TRPM4 did not cause cell death, which is similar to findings in knockout mice (32, 33), suggesting that TRPM4 is not lethal. With the availability of the knockdown cells, we examined TRPM4’s impact on proliferation and differentiation. A decrease in stem cell proliferation is a pre-requisite for osteogenesis and adipogenesis (26, 27). The fact that TRPM4 suppression decreased DFSCs proliferation during the initial 96-hrs of osteogenesis, but not of adipogenesis, indicated a possible enhancement of osteoblast differentiation. This hypothesis was supported by the increased mineralization and alkaline phosphatase enzyme activity; however, under adipogenic conditions, DFSCs failed to differentiate into adipocytes. These findings suggest that TRPM4 functions as an inhibitor of osteogenesis, while it is required and/or facilitates adipogenesis.
Calcium oscillations are often present during stem cell differentiation and are linked to the differentiation process by activating specific transcription factors (12, 13). To provide insights into the mechanism controlling DFSC differentiation, we investigated their Ca2+ signaling pattern of DFSCs during agonist stimulation. Inhibition of TRPM4 transformed the normal ATP-induced a biphasic Ca2+ pattern, characterized by a sharp increase (ER release - first phase) followed by influx (Ca2+ entry via SOCs - secondary phase) and then a gradual increase followed by a sharp decrease. This was unexpected and different from observations in T-lymphocytes, which also utilize SOCs as the main Ca2+ entry pathway. TRPM4 suppression in immune cells results in a sustained secondary phase without impacting the first phase, and increases IL-2 production via the calcineurin-NFAT pathway (7, 11). It is possible that in DFSCs there could be direct interaction between TRPM4 and the ER to control the rate of Ca2+ release and refill. Such relationship between ER proteins (e.g. Stromal interaction molecule 1 (STIM1)) and ion channels (e.g. SOCs) are essential for Ca2+ signaling in non-excitable cells (34, 35). Another possibility is that of TRPM4 multimerization with SOCs to control the rate of Ca2+ influx. Several TRP members are capable of homo- and heteromultimerization (36–39). However, the gradual increase in [Ca2+]i with TRPM4 suppression, suggests a regulatory mechanism between TRPM4 and the ER. Further studies will be required to elucidate the mechanism underlying the control of Ca2+ signals by TRPM4 in DFSCs.
Functional studies in different cell types provide a direct link between TRPM4, Ca2+ signaling and cellular responses. Together, they control dendritic cell migration and cytokine production in T-lymphocytes (7, 11, 21). They also regulate in insulin and glucagon secretion from pancreatic islet cells and myogenic constriction in cerebral arteries (18–20). Because gene transcription is one of the downstream events regulated by TRPM4, we performed whole genome microarray analysis to identify potential genes responsible for the enhancement of osteogenesis and inhibition of adipogenesis. We identified four genes involved in osteoblast differentiation (CITED1; CDK6; FIGF; IL6R) and four in bone mineralization (NFE2; ASPN; MEPE; MGP). Comparison between osteogenic differentiation genes from control and knockdown cells revealed a maximum of two fold differences, whereas for bone mineralization, the MEPE and MGP genes were up-regulated 11 and 8.6 fold with TRPM4 suppression. These findings are consistent with studies in human dental pulp and murine preosteoblast cells, where MEPE expression enhances osteoblast differentiation and mineralization via the BMP-2 signaling pathway (40, 41). Although the role of MGP in dental tissue is unclear, studies in knockout mice show increased mineralization of pulmonary and renal arteries via the BMP-4 pathway (42). This is contrary to our observation in DFSCs. Perhaps the differences in signaling pathway and tissues affected by MGP expression could account for this disparity. In fact, the BMP-2 pathway is reported to control DFSC differentiation into osteoblasts (43). Furthermore, TRPM4 knockdown inhibited ASPN expression 39.4 fold over control cells. This is relevant to DFSCs because this gene is predominantly expressed in the periodontal ligament (PDL) and in dental follicle cells that give rise to the ligament (44). Since ASPN expression inhibits differentiation and mineralization of the PDL (44), its suppression by TRPM4 knockdown may have contributed to the enhanced osteogenesis. Interestingly, we found the LUM gene up-regulated 12.6 fold over control cells. Its roles in collagen formation, epithelial cell migration and tissue repair suggest a possible role for TRPM4 on bone development (45, 46).
When TRPM4 knockdown cells were placed in adipogenic medium, they failed to differentiate into adipocytes. Of the genes related to this process, the TRB3 was down-regulated 24.7 fold compared to control cells. This finding is consistent with reports in type 2 diabetic rats, where silencing of the TRB3 decreases adipocyte formation (47). Two other notable genes, SFRP1 and MMP3, were up-regulated in both groups. TRPM4 knockdown inhibited SFRP1 expression 10.8 fold compared to control cells and SFRP1 is reported to be a regulator of the Wnt/beta-catenin pathway. Its expression enhances adipocyte differentiation in humans and mice (48). The analysis also revealed a 22.9 fold increase in MMP3 expression compared to control cells. The knockout of this gene also enhances adipogenesis during mammary gland involution in mice, along with adipocyte hypertrophy (49). The effects of TRPM4 knockdown on the TRB3, SFRP1 and MMP3 genes are expected to inhibit adipocyte differentiation based on their roles during adipogenesis.
Conclusion
TRPM4 is a CAN channel present in DFSCs. These stem cells not only express the TRPM4 gene, but have currents typical for the channel. Functional experiments suggested that TRPM4 acts as an inhibitor of osteogenesis, since channel suppression increased mineralization and ALP enzyme activity. However, it appears to be required and/or facilitates adipogenesis due to the absence of adipocyte differentiation in TRPM4 knockdown cells. The effect of TRPM4 on osteogenesis and adipogenesis seems to be linked to the Ca2+ signals, which could be a key factor controlling expression of certain genes.
Acknowledgments
This work was supported by the National Institute of Health/National Institute of Dental and Craniofacial Research grant DE018871 and by an LSU SVM CORP grant to HC.
Footnotes
Piper Nelson: Collection and/or assembly of data, Data analysis and interpretation
Tran Doan Ngoc Tran: Collection and/or assembly of data, Data analysis and interpretation
Hanjie Zhang: Collection and/or assembly of data
Olga Zolochevska: Provision of study material or patients
Marxa Figueiredo: Conception and design, Provision of study material or patients
Ji-Ming Feng: Collection and/or assembly of data, Data analysis and interpretation
Dina L. Gutierrez: Collection and/or assembly of data
Rui Xiao: Data analysis and interpretation
Shaomian Yao: Conception and design, Data analysis and interpretation
Arthur Penn: Data analysis and interpretation
Li-Jun Yang: Provision of study material or patients
Henrique Cheng: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
References
- 1.Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127(8):1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 2.Cho MI, Garant PR. Development and general structure of the periodontium. Periodontol. 2000;24:9–27. doi: 10.1034/j.1600-0757.2000.2240102.x. 2000. [DOI] [PubMed] [Google Scholar]
- 3.Honda MJ, Imaizumi M, Tsuchiya S, et al. Dental follicle stem cells and tissue engineering. J. Oral Sci. 2010;52(4):541–552. doi: 10.2334/josnusd.52.541. [DOI] [PubMed] [Google Scholar]
- 4.Nelson PL, Beck A, Cheng H. Transient receptor proteins illuminated: current views on TRPs and disease. Vet. J. 2011;187(2):153–164. doi: 10.1016/j.tvjl.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Launay P, Fleig A, Perraud AL, et al. TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell. 2002;109(3):397–407. doi: 10.1016/s0092-8674(02)00719-5. [DOI] [PubMed] [Google Scholar]
- 6.Ullrich ND, Voets T, Prenen J, et al. Comparison of functional properties of the Ca2+-activated cation channels TRPM4 and TRPM5 from mice. Cell Calcium. 2005;37(3):267–278. doi: 10.1016/j.ceca.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Launay P, Cheng H, Srivatsan S, et al. TRPM4 regulates calcium oscillations after T cell activation. Science. 2004;306(5700):1374–1377. doi: 10.1126/science.1098845. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H, Feng JM, Figueiredo ML, et al. Transient receptor potential melastatin type 7 channel is critical for the survival of bone marrow derived mesenchymal stem cells. Stem Cells Dev. 2010;19(9):1393–1403. doi: 10.1089/scd.2009.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin J, Wu LJ, Jun J, et al. The channel kinase, TRPM7, is required for early embryonic development. Proc. Natl. Acad. Sci. U. S. A. 2012;109(5):E225–E233. doi: 10.1073/pnas.1120033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S, Liu Y, Lipsky S, et al. Physical manipulation of calcium oscillations facilitates osteodifferentiation of human mesenchymal stem cells. Faseb J. 2007 doi: 10.1096/fj.06-7153com. [DOI] [PubMed] [Google Scholar]
- 11.Weber KS, Hildner K, Murphy KM, et al. Trpm4 differentially regulates Th1 and Th2 function by altering calcium signaling and NFAT localization. J. Immunol. 2010;185(5):2836–2846. doi: 10.4049/jimmunol.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawano S, Otsu K, Kuruma A, et al. ATP autocrine/paracrine signaling induces calcium oscillations and NFAT activation in human mesenchymal stem cells. Cell Calcium. 2006;39(4):313–324. doi: 10.1016/j.ceca.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Kapur N, Mignery G, Banach K. Cell Cycle Dependent Calcium Oscillations in Mouse Embryonic Stem Cells. Am. J. Physiol Cell Physiol. 2006 doi: 10.1152/ajpcell.00181.2006. [DOI] [PubMed] [Google Scholar]
- 14.Nilius B, Prenen J, Voets T, et al. Intracellular nucleotides and polyamines inhibit the Ca2+-activated cation channel TRPM4b. Pflugers Arch. 2004;448(1):70–75. doi: 10.1007/s00424-003-1221-x. [DOI] [PubMed] [Google Scholar]
- 15.Kawano S, Shoji S, Ichinose S, et al. Characterization of Ca(2+) signaling pathways in human mesenchymal stem cells. Cell Calcium. 2002;32(4):165–174. doi: 10.1016/s0143416002001240. [DOI] [PubMed] [Google Scholar]
- 16.Yanagida E, Shoji S, Hirayama Y, et al. Functional expression of Ca2+ signaling pathways in mouse embryonic stem cells. Cell Calcium. 2004;36(2):135–146. doi: 10.1016/j.ceca.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Beck A, Launay P, et al. TRPM4 controls insulin secretion in pancreatic beta-cells. Cell Calcium. 2006 doi: 10.1016/j.ceca.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marigo V, Courville K, Hsu WH, et al. TRPM4 impacts on Ca2+ signals during agonist-induced insulin secretion in pancreatic beta-cells. Mol. Cell Endocrinol. 2009;299(2):194–203. doi: 10.1016/j.mce.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Nelson PL, Zolochevska O, Figueiredo ML, et al. Regulation of Ca(2+)-entry in pancreatic alpha-cell line by transient receptor potential melastatin 4 plays a vital role in glucagon release. Mol. Cell Endocrinol. 2011;335(2):126–134. doi: 10.1016/j.mce.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Earley S, Straub SV, Brayden JE. Protein kinase C regulates vascular myogenic tone through activation of TRPM4. Am. J. Physiol Heart Circ. Physiol. 2007;292(6):H2613–H2622. doi: 10.1152/ajpheart.01286.2006. [DOI] [PubMed] [Google Scholar]
- 21.Barbet G, Demion M, Moura IC, et al. The calcium-activated nonselective cation channel TRPM4 is essential for the migration but not the maturation of dendritic cells. Nat. Immunol. 2008;9(10):1148–1156. doi: 10.1038/ni.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guinamard R, Demion M, Magaud C, et al. Functional expression of the TRPM4 cationic current in ventricular cardiomyocytes from spontaneously hypertensive rats. Hypertension. 2006;48(4):587–594. doi: 10.1161/01.HYP.0000237864.65019.a5. [DOI] [PubMed] [Google Scholar]
- 23.Yao S, Pan F, Prpic V, et al. Differentiation of stem cells in the dental follicle. J. Dent. Res. 2008;87(8):767–771. doi: 10.1177/154405910808700801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz C, Perraud AL, Johnson CO, et al. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114(2):191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- 25.Nilius B, Prenen J, Droogmans G, et al. Voltage dependence of the Ca2+-activated cation channel TRPM4. J. Biol. Chem. 2003;278(33):30813–30820. doi: 10.1074/jbc.M305127200. [DOI] [PubMed] [Google Scholar]
- 26.Reichert M, Eick D. Analysis of cell cycle arrest in adipocyte differentiation. Oncogene. 1999;18(2):459–466. doi: 10.1038/sj.onc.1202308. [DOI] [PubMed] [Google Scholar]
- 27.Owen TA, Aronow M, Shalhoub V, et al. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell Physiol. 1990;143(3):420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 28.Weber KS, Hildner K, Murphy KM, et al. Trpm4 differentially regulates Th1 and Th2 function by altering calcium signaling and NFAT localization. J. Immunol. 2010;185(5):2836–2846. doi: 10.4049/jimmunol.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zippel N, Limbach CA, Ratajski N, et al. Purinergic receptors influence the differentiation of human mesenchymal stem cells. Stem Cells Dev. 2012;21(6):884–900. doi: 10.1089/scd.2010.0576. [DOI] [PubMed] [Google Scholar]
- 30.Coppi E, Pugliese AM, Urbani S, et al. ATP modulates cell proliferation and elicits two different electrophysiological responses in human mesenchymal stem cells. Stem Cells. 2007;25(7):1840–1849. doi: 10.1634/stemcells.2006-0669. [DOI] [PubMed] [Google Scholar]
- 31.Morsczeck C. Gene expression of runx2, Osterix, c-fos, DLX-3, DLX-5, and MSX-2 in dental follicle cells during osteogenic differentiation in vitro. Calcif. Tissue Int. 2006;78(2):98–102. doi: 10.1007/s00223-005-0146-0. [DOI] [PubMed] [Google Scholar]
- 32.Mathar I, Vennekens R, Meissner M, et al. Increased catecholamine secretion contributes to hypertension in TRPM4-deficient mice. J. Clin. Invest. 2010;120(9):3267–3279. doi: 10.1172/JCI41348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerzanich V, Woo SK, Vennekens R, et al. De novo expression of Trpm4 initiates secondary hemorrhage in spinal cord injury. Nat. Med. 2009;15(2):185–191. doi: 10.1038/nm.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang GN, Zeng W, Kim JY, et al. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat. Cell Biol. 2006;8(9):1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 35.Peinelt C, Vig M, Koomoa DL, et al. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat. Cell Biol. 2006;8(7):771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poteser M, Graziani A, Rosker C, et al. TRPC3 and TRPC4 associate to form a redox-sensitive cation channel. Evidence for expression of native TRPC3-TRPC4 heteromeric channels in endothelial cells. J. Biol. Chem. 2006;281(19):13588–13595. doi: 10.1074/jbc.M512205200. [DOI] [PubMed] [Google Scholar]
- 37.Rosker C, Graziani A, Lukas M, et al. Ca(2+) signaling by TRPC3 involves Na(+) entry and local coupling to the Na(+)/Ca(2+) exchanger. J. Biol. Chem. 2004;279(14):13696–13704. doi: 10.1074/jbc.M308108200. [DOI] [PubMed] [Google Scholar]
- 38.Hellwig N, Albrecht N, Harteneck C, et al. Homo- and heteromeric assembly of TRPV channel subunits. J. Cell Sci. 2005;118(Pt 5):917–928. doi: 10.1242/jcs.01675. [DOI] [PubMed] [Google Scholar]
- 39.Murakami M, Xu F, Miyoshi I, et al. Identification and characterization of the murine TRPM4 channel. Biochem. Biophys. Res. Commun. 2003;307(3):522–528. doi: 10.1016/s0006-291x(03)01186-0. [DOI] [PubMed] [Google Scholar]
- 40.Cho YD, Kim WJ, Yoon WJ, et al. Wnt3a stimulates Mepe, matrix extracellular phosphoglycoprotein, expression directly by the activation of the canonical Wnt signaling pathway and indirectly through the stimulation of autocrine Bmp-2 expression. J. Cell Physiol. 2012;227(6):2287–2296. doi: 10.1002/jcp.24038. [DOI] [PubMed] [Google Scholar]
- 41.Wei X, Ling J, Wu L, et al. Expression of mineralization markers in dental pulp cells. J. Endod. 2007;33(6):703–708. doi: 10.1016/j.joen.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Yao Y, Jumabay M, Wang A, et al. Matrix Gla protein deficiency causes arteriovenous malformations in mice. J. Clin. Invest. 2011;121(8):2993–3004. doi: 10.1172/JCI57567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverio KG, Davidson KC, James RG, et al. Wnt/beta-catenin pathway regulates bone morphogenetic protein (BMP2)-mediated differentiation of dental follicle cells. J. Periodontal Res. 2012;47(3):309–319. doi: 10.1111/j.1600-0765.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada S, Tomoeda M, Ozawa Y, et al. PLAP-1/asporin, a novel negative regulator of periodontal ligament mineralization. J. Biol. Chem. 2007;282(32):23070–23080. doi: 10.1074/jbc.M611181200. [DOI] [PubMed] [Google Scholar]
- 45.Ishiwata T, Yamamoto T, Kawahara K, et al. Enhanced expression of lumican inhibited the attachment and growth of human embryonic kidney 293 cells. Exp. Mol. Pathol. 2010;88(3):363–370. doi: 10.1016/j.yexmp.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Kiga N, Tojyo I, Matsumoto T, et al. Expression of lumican and fibromodulin following interleukin-1 beta stimulation of disc cells of the human temporomandibular joint. Eur. J. Histochem. 2011;55(2):e11. doi: 10.4081/ejh.2011.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ti Y, Xie GL, Wang ZH, et al. TRB3 gene silencing alleviates diabetic cardiomyopathy in a type 2 diabetic rat model. Diabetes. 2011;60(11):2963–2974. doi: 10.2337/db11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagathu C, Christodoulides C, Tan CY, et al. Secreted frizzled-related protein 1 regulates adipose tissue expansion and is dysregulated in severe obesity. Int. J. Obes. (Lond) 2010;34(12):1695–1705. doi: 10.1038/ijo.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maquoi E, Demeulemeester D, Voros G, et al. Enhanced nutritionally induced adipose tissue development in mice with stromelysin-1 gene inactivation. Thromb. Haemost. 2003;89(4):696–704. [PubMed] [Google Scholar]