Abstract
Introduction
The use of antiretroviral therapy (ART) has led to a significant decrease in morbidity and mortality in HIV-infected individuals. Nevertheless gene-based therapies represent a promising therapeutic paradigm for HIV-1, as they have the potential for sustained viral inhibition and reduced treatment interventions. One new method amendable to a gene-based therapy is the clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 gene editing system.
Areas covered
CRISPR/Cas9 can be engineered to successfully modulate an array of disease-causing genetic elements. We discuss the diverse roles that CRISPR/Cas9 may play in targeting HIV and eradicating infection. The Cas9 nuclease coupled with one or more small guide RNAs (sgRNAs) can target the provirus to mediate excision of the integrated viral genome. Moreover, a modified nuclease deficient Cas9 fused to transcription activating domains may induce targeted activation of proviral gene expression allowing for the purging of the latent reservoirs. These technologies can also be exploited to target host dependency factors such as the co-receptor CCR5, thus preventing cellular entry of the virus.
Expert opinion
The diversity of the CRISPR/Cas9 technologies hold great promise for targeting different stages of the viral life cycle, and have the capacity for mediating an effective and sustained genetic therapy against HIV.
Keywords: CRISPR, HIV-1, gene excision, gene editing, gene activation, latency, RNA therapy
1. Introduction
HIV/AIDS persists as a global health problem with little hope in the near future for an efficacious vaccine. Despite this, ART has reduced the morbidity, mortality and transmission of HIV-related illness 1 resulting in a slow turning of the tide, and newfound optimism, in the fight against this chronic viral disease. However, current treatment regimens do have significant limitations. These include drug toxicity, resistance to antiretrovirals (ARVs) and the inability to eradicate latent viral infection. Moreover, the daily medication burden has made it difficult to ensure adequate patient compliance with treatment, and the costs associated with life-long therapy and monitoring remains problematic in those countries where HIV/AIDS is endemic. Consequently, alternative strategies to inhibit HIV-1 replication in the absence of viral resistance, and which do not require long-term administration of ARVs, is an attractive goal, and in theory could result in a “functional cure”; whereby individuals can control their ongoing HIV-1 infection, and prevent associated pathologies. This has prompted the search for a different treatment paradigm, whereby the focus lies more with single-intervention or long-lasting combination therapies aimed at blocking replicating virus, preventing drug resistance, and eliminating latent viral reservoirs. One such approach is gene therapy.
The last three decades have seen tremendous progress in the development of gene therapy approaches for HIV with many different technologies having shown elimination of actively replicating virus. Antisense RNAs, ribozymes, dominant-negative mutants, TAR-decoys and RNAi modalities are currently being tested in pre-clinical and clinical settings 2, 3. Additionally, the use of viral vectors to deliver therapeutic gene payloads to hematopoietic stem and progenitor cells (HSPCs) has paved the way for the generation of HIV-resistant cells, bringing the promise of a single intervention therapy closer to reality. Indeed, new strategies aimed at eliminating or reducing the latent reservoir, are necessary to abrogate the requirement for life-long treatment.
Here we review the function and application of the gene editing and gene modulation technology, CRISPR/Cas9, as a therapy for HIV-1 (Figure 1). Much of our understanding of CRISPR function has been built on the foundation of customized DNA-binding and editing proteins, such as transcription activator-like nucleases (TALENs) and zinc finger nucleases (ZFNs), whose function and therapeutic utility are touched on briefly, but are mostly explored elsewhere 4, 5.
Figure 1. The life cycle of HIV with targeted intervention points.
HIV binds to the cell surface via the CD4+ receptor and the CCR5/CXCR4 co-receptors. Following fusion with the cell membrane the viral particle enters the cell and its genome is transcribed from RNA to DNA. It is the DNA that is integrated into the host cell genome, and that provides the template to drive transcription of HIV RNA, producing progeny virions that bud off from the cell, completing the infectious cycle. Nuclease-directed disruption of the HIV life cycle could occur at any of the following stages: A. Targeting and preventing the integration of proviral DNA into the genome. B. Proviral DNA, once integrated into the genome, is a target for excision, or deactivation by mutagenic disruption. C. Cellular factors necessary for the HIV life cycle present further targets, and include the co-receptor CCR5 or other host dependency factors.
2. The emergence of CRISPR/Cas9: a facile tool for gene editing and transcriptional modulation
CRISPR, in its native function, provides adaptive immunity in bacteria by introducing targeted DNA mutations in pathogenic viruses and plasmids 6, 7. A breakthrough came in 2012 with the development of modified CRISPR components comprising a short chimeric single guide RNA (sgRNA) and a Cas9 nuclease from Streptococcus pyogenes (Figure 2A) 8. It now appears that this simplified system can be adapted to target any DNA sequence from virtually any organism 9-11, thereby greatly expanding its function and utility.
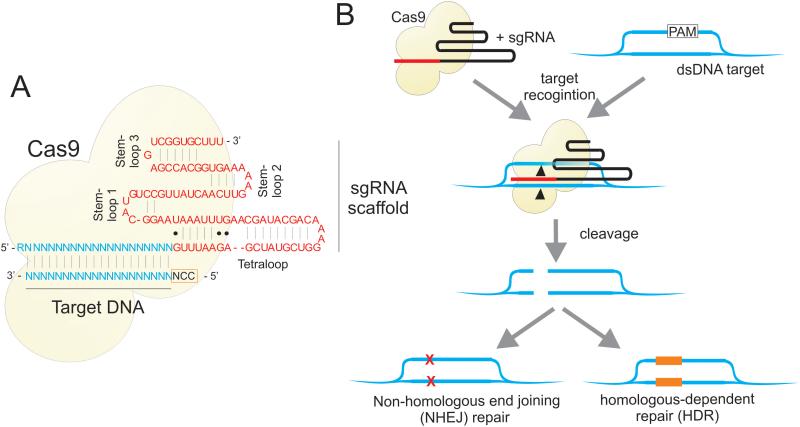
Figure 2. Mechanisms of CRISPR/Cas9-directed cleavage.
The Cas9 protein forms a complex with a sgRNA, which guides the nuclease to a specific genomic address for cleavage. A. Cas9 catalyzed DNA cleavage is guided by a 17-20 nucleotide sequence within the sgRNA. B. The Cas9 protein “scans” genomic DNA for regions of homology with the guide sequence 77, where it unwinds the DNA and its nuclease domain directs site specific cleavage 78. This results in deletions generated by non-homologous end joining (NHEJ) at the binding site, or in homologous-dependent repair (HDR) (reviewed in 79).
Cas9 catalyzed DNA cleavage is guided by a 17-20 nucleotide sgRNA, resulting in deletions generated by non-homologous end joining (NHEJ) at the binding site, or alternatively, in homologous-dependent repair (HDR) (Figure 2B). Mutations in the two Cas9 endonuclease domains, or deactivated Cas9 (dCas9), have also helped develop programmable RNA-dependent DNA-binding proteins 12, 13. Deactivated Cas9 has been successfully fused to accessory proteins, such as transcription activator and repressor domains, to provide RNA-guided locus-specific targeted interactions 12-17. CRISPR has the advantage over other approaches (e.g. TALENs and ZFNs) in that the sgRNA component is easily “programmable”, remains physically separate from Cas9/dCas9 expression, and that many sgRNAs can target multiple DNA sites when expressed simultaneously with the same Cas9. Here we explore the application of CRISPR/Cas9 as a therapeutic antiviral gene editing and gene-modulating tool.
3. CRISPR/Cas9 mediated excision and elimination of the integrated latent provirus
The most significant limitation of ART is its inability to purge HIV from the reservoirs in which it remains latent; which means that the virus persists even under life-long treatment. Latent viral reservoirs, which mostly lie within resting memory CD4+ T cells, are extremely long-lived and can persist for as long as 60 years in patients receiving ART 18. Novel approaches are therefore urgently needed to target the latent provirus specifically for excision/elimination and/or activation.
In order to achieve the objective of a functional cure for HIV/AIDS, strategies that specifically target integrated HIV-1 proviral DNA allowing for deactivation or elimination remains an attractive option (Figure 3). Influential early work by the Hauber and Buchholz labs made use of Tre-recombinases 19, 20. These modified nucleases were developed using directed in vitro evolution to specifically target the long terminal repeat (LTR) regions of the virus, resulting in excision of the provirus. Furthemore, Tre-recombinases were effectively delivered into the cell when fused to a cell permeable translocation motif derived from the PreS2 surface antigen of Hepatitis B virus (HBV) 21. HIV-1 proviral DNA cleavage was observed in a dose-dependent manner, and did not appear to have any cytotoxic effects. An LTR-targeting Tre-recombinase was also delivered ex vivo into CD4+ T cells and CD34+ HSPCs using lentiviral vectors 22. The Tre-recombinase was under the control of a modified TAR sequence, thus limiting transgene expression to Tat-expressing cells – as would be the case during active infection by HIV-1. Engraftment of these cells in humanized mice resulted in significant antiviral effects in vivo, coupled with positive selection of vector-transduced cells 22. While these results are promising, it is uncertain if latent cells express sufficient Tat to stimulate Tre-recombinase activity.
Figure 3. Targeting the HIV genome.
The HIV provirus is flanked by identical viral long terminal repeat (LTR) sequences. Therefore CRISPR/Cas9 targeted to the LTR could cleave at both ends of the virus. DNA repair of the excised region between the cleavage sites would result in a single LTR “footprint” within the genome providing a reference to identify the position of the HIV proviral DNA 17, 23, 24. Guide RNAs targeting two or more sites within the 5' LTR can result in loss of promoter activity, leading to deactivation of the provirus 17. Guide RNAs can also be targeted to specific viral reading frames, causing indels that affect viral protein function, and concomitant virion production 26.
ZFNs have also been used successfully for the excision of the HIV-1 provirus. Qu et al. directed ZFNs to the TAR region of the HIV-1 LTR by transient transfection of T cells that contained a single stably integrated copy of the virus, resulting in excision of the integrated viral DNA 23. One major drawback of the Tre-recombinase and ZFN approaches is that the enzymes themselves encode the sequence specificity of the cleavage site. To alter the cleavage site, further rounds of in vitro directed evolution, or protein engineering is required. In contrast, the CRISPR/Cas9 cleavage site can be reprogrammed simply by changing the sequence of the sgRNA.
Proof-of-concept that CRISPR/Cas9 gene editing can similarly eradicate proviral DNA using an sgRNA guide targeted to the viral LTRs of a reporter HIV-1 virus was provided by Ebina et al.24. Here, CRISPR/Cas9-mediated proviral excision also prevented viral gene activation following treatment with a combination of the cytokine TNFα, the HDAC inhibitor trichostatin A, and the demethylating agent 5-aza-2'-deoxycytidine. Interestingly, both the Qu and Ebina studies gave similar frequencies for excision of the reporter provirus from stably infected Jurkat T cells of 44.9% and 40% respectively. Further convincing evidence of CRISPR/Cas9-mediated HIV excision was obtained using sgRNAs targeted to conserved sites within the U3 region of the HIV-1 LTR in a number of latently infected microglial, promonocytic and T cell lines 25. The highly efficient editing induced by using two sgRNAs simultaneously prevented viral reactivation and replication in these infected cells. Moreover, cells pretreated with dual sgRNAs and CRISPR/Cas9 were resistant to de novo HIV infection, although there was little evidence to indicate whether the site of activity is inhibiting integration or a result of cleavage of the circular double-LTR proviral DNA in the pre-integration complex. Recently the Belmonte lab also demonstrated the efficacy CRISPR/Cas9 targeted to multiple sites within the HIV genome 26. This study also showed that pre-treatment of cells with CRISPR/Cas9 successfully targeted non-integrating virus, providing much stronger evidence for blocking integration of viral DNA. In concordance with preceding studies, sustained targeting of sites within the LTR proved to be the most effective in patient-derived cells 26. Moreover, Cas9 was permanently introduced into human HPCS and differentiated into monocyte and macrophage lineages without adverse effects.
The studies conducted thus far show that cells tolerate the long-term presence of the CRISPR/Cas9 machinery, and that this system might be able to “immunize” cells against further infection. One confounding issue with CRISPR/Cas9 is the potential for non-specific sequence “off-target” effects. Hu et al. assessed the risk of off-target effects by performing whole-genome sequencing on treated cells 25. Despite an abundance of differential indels in the treated cells, no indels appeared to map to off-target sites. Nevertheless, the possibility for off-target effects, especially when using multiplexed sgRNAs deserves much further scrutiny.
4. The potential for CRISPR/Cas9-mediated latency reactivation of HIV
Until now, the primary strategy to eradicate latent HIV reservoirs has been to purge the pool of latently infected cells in the presence of ARVs by reactivating dormant virus: a strategy known as “shock and kill”. Reactivation of latent HIV purges infected cells directly (via active viral replication), or indirectly via the host immune system; ARVs can then act to prevent new infection from the released virus and thereby extinguish the reservoir 27. A seminal clinical study using the histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (Vorinostat) resulted in viral reactivation, but it remains uncertain whether only partial transcriptional reactivation was induced in memory CD4+ T cells 28. The Siliciano lab has determined that the viral reservoir is larger than originally thought, and that activation from latency is largely driven by stochastic events in both active and resting memory T cells 29. Thus, HDAC inhibitors and cell-reactivation strategies alone are unlikely to reverse the mechanisms of latency for the entire reservoir 29. In addition, there are major safety concerns with this approach due to the risk of widespread and non-specific induction of host gene expression. Novel viral activation agents that function specifically by inducing HIV expression are therefore an attractive prospect, and CRISPR-based technologies provide a potential solution.
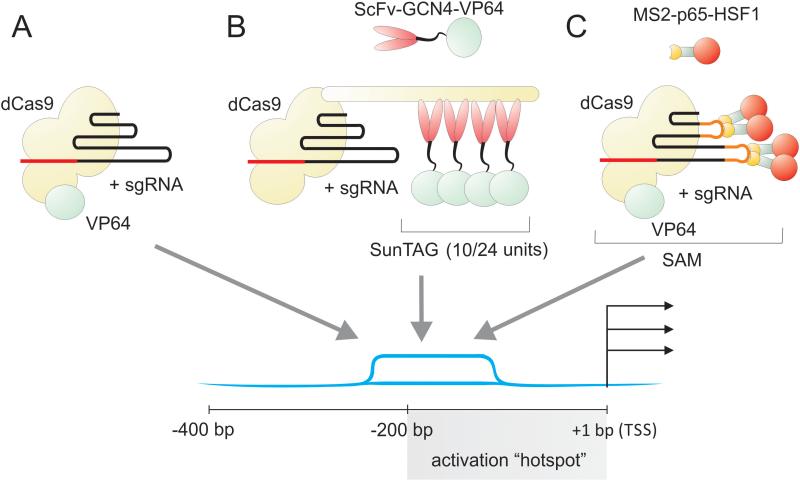
Gene specific transcriptional activation has been achieved using various engineered forms of the CRISPR/Cas9 system. Initial systems made use of a nuclease deficient mutant of Cas9 (dCas9) fused to a C-terminal herpes virus transcriptional activation domain (VP16, or four copies of VP16 = VP64) to modulate endogenous gene expression (Figure 4A). Coupling with single sgRNAs, induced highly specific enhanced expression of targeted genes 14-16, 30. While this represents an exciting new tool, the levels of gene activation have been modest. Activation can be enhanced by using multiple non-overlapping sgRNAs targeted to the same promoter, which in turn recruit multiple copies of the dCas9-VP64 fusion proteins 14-16, 30. Using a different approach, the recruitment of multiple activation domains to a single dCas9 molecule can also enhance RNA-guided transcriptional activation levels 31. This system uses a polypeptide scaffold termed SunTag, which recruits multiple antibody-fusion proteins to link a promoter-localized dCas9 to multiple VP64 domains (Figure 4B). The SunTag platform allows a single sgRNA to mediate robust activation of gene expression, as demonstrated with CXCR4 and the cell cycle inhibitor CDKN1B 31.
Figure 4. Strategies to activate HIV provirus using CRISPR/Cas9.
Gene specific transcriptional activation using engineered forms of CRISPR/Cas9 targeted to the activation “hotspot” within the 200bp upstream of the HIV proviral transcriptional start site (TSS). A. Nuclease deficient mutant of Cas9 (dCas9) fused to a C-terminal VP64 activation domain, which is boosted using multiple sgRNAs are targeted to this region 12. B. A modified dCas9 that uses a polypeptide scaffold termed SunTag, can recruit multiple antibody-fusion proteins resulting in enhanced activation 31. C. Aptamers that selectively bind to the dimerized MS2 bacteriophage coat protein can create a synergistic activation mediator (SAM) system that can recruit multiple activation domains.
Structural analysis of dCas9 co-crystalized with a sgRNA has provided further insights into potential applications of CRISPR/Cas9 32. This work has identified modifiable regions that can interact with RNA aptamers that facilitate the recruitment of effector domains to the dCas9 complex. Experimentally, Konermann et al. used short hairpin aptamers that selectively bind to the dimerized MS2 bacteriophage coat protein, creating a synergistic activation mediator (SAM) system that can recruit multiple activation domains (Figure 4C). For example, MS2 fused to the NF-κB trans-activating subunit, p65, and the activation domain from human heat-shock factor 1 (HSF1), results in enhanced activation at a single sgRNA-targeted dCas9-VP64 bound locus.
RNA-guided transcriptional activation using CRISPR/Cas9 provides an exciting new avenue to potentially target and induce transcription from latent HIV reservoirs, and ongoing studies continue to define rules that enable the design of systems with increased activity. One of the most important factors affecting activation of HIV-1 is the position of the sgRNA target site relative to the transcription start site (TSS); sgRNA ‘hotspots’ tend to reside within -200 to +1 bp upstream of the TSS 32, 33 (Figure 4). Future efforts will focus on developing a combination of sgRNAs that target upstream 5’ LTR promoter sequences, and achieve synergistic high-level activation. In spite of this promising new technology, additional considerationis owed to the complex locations of reservoirs of latent provirus. Whilst it is argued that concomittent treatment with ARVs would limit viral replication and act to prevent new infection from the released virus and thereby extinguish the reservoir 27 important latency resevoirs, such as the brain, are compartmentalized and therefore not readily accesible to ARVs or immune surveillance.
5. CRISPR/Cas9 targeted editing of the CCR5 gene locus
HIV-1 entry into target cells requires CD4, and either CCR5 or CXCR4 as a co-receptor 34 (Figure 1). The targeting of CD4, and possibly even CXCR4, is not advisable since these receptors are vital for a functional immune system 35. CCR5 is a proven target for therapy with the CCR5 agonist Maraviroc approved for clinical use. Individuals harboring a rare homozygous 32-bp deletion in the CCR5 gene (CCR5Δ32) are otherwise healthy, and are naturally resistant to R5-tropic HIV-1 infection 36, 37, albeit living with the risk of a viral switch to the CXCR4 tropism to regain infectivity 38. In one of the most celebrated cases, transplantation of allogeneic donor CCR5Δ32 hematopoietic stem progenitor cells (HSPCs) was successfully applied in the case of Timothy Ray Brown (the HIV-1 positive “Berlin Patient”), giving rise to the first “sterilizing cure” for HIV 39-41. It does however remain to be proven that HIV was eradicated from latent reservoirs. Indeed, the possibility exists that this is simply a “functional cure”, whereby HIV emerging from the latent reservoir is adequately controlled by a functional, albeit genetically modified, immune system. Nevertheless, while this result was encouraging, allogeneic transplantation is unlikely to be widely applied because there are too few homozygous CCR5Δ32 donors (approximately 1% of the Caucasian population) 42, 43. Moreover, finding suitable HLA-matched donors and performing full bone marrow ablation and immune suppression is not feasible at a large scale. Autologous transplantations represent a less toxic alternative, as complete bone marrow ablation or immune suppression is not a prerequisite for successful engraftment. However, the trade-off with such an approach is that latent reservoirs are likely to persist, making a cure difficult to achieve. Nevertheless, recent research has focused on generating similarly disruptive homozygous mutations in CCR5 using a variety of engineered nucleases for ex vivo modification of autologous, patient-derived CD34+ HSPCs or CD4+ T cells.
CCR5-specific ZFNs have been successfully delivered to primary human CD4+ T cells 44 and CD34+ HSPCs 45, 46 which were then transplanted into murine models of HIV infection. In most cases, cells with ΔCCR5 showed long-lasting persistence and evidence of selective survival. In the most significant clinical study to date, Tebas et al. used an adenoviral vector expressing a ZFN targeted to CCR5 in CD4+ T cells isolated from 12 HIV patients 47. After ex vivo expansion, the cells were autologously re-infused. Gene-modified T cells could be detected in all patients in follow-up studies, for up to 42 months, with some evidence of selection of modified cells in patients who underwent ART interruption. Importantly, while HIV DNA had decreased in most patients, the study also suggests that delay in viral recrudescence may be correlated with the degree of biallelic disruption of CCR5.
CRISPR/Cas9 technologies represent a strong alternative to ZFN or TALEN-based approaches for disrupting CCR5 because of its ease of use, the requirement for a single sgRNA to “program” the location of the cleavage site, and the improved levels of on-target specificity. Cho et al. showed that significant gene distruption within CCR5 could be induced when cells were co-transfected with plasmids encoding Cas9 and distinct CCR5-targeting sgRNAs 48. Ye et al. combined TALENs or CRISPR/Cas9 together with a piggyBac transposon donor sequence to seamlessly reproduce the naturally-occurring CCR5Δ32 deletion in induced pluripotent stem cells (iPSCs) 49. Modified iPSCs were differentiated into monocytes/macrophages that were resistant to challenge by HIV-1 49. More recently, lentiviral vectors expressing Cas9 and CCR5-targeted sgRNAs were used in a single round transduction into engineered CD4+ T cells 50. While high frequencies of CCR5 gene disruption could be shown in cell lines, the same system transduced into primary T cells resulted in toxicities 50. It is possible that such deletarious effects may be caused by off-targeting by some CCR5 sgRNAs, as the CCR5 locus is closely related to the CCR2 locus 51, 52. It may also be that the innate immune system of T cells reacts adversely to foreign DNA 53, a problem which may be overcome by administering in vitro transcribed mRNA 54. Recently, Mandal et al. utilized the CRISPR/Cas9 system in human primary CD4+ T cells and CD34+ HSPCs to target CCR5 and the clinically relevant gene B2M 55. Interestingly, the activity of the CRISPR/Cas9 showed little activity in CD4+ T cells. However, by using a dual sgRNA approach to generate deletions rather than indels, improved biallelic CCR5 disruption efficacy could be obtained in both cell types. As with iPSCs, modified CD34+ HSPCs retained multilineage potential in vitro and also in vivo upon transplantation in mice 55.
6. CRISPR/Cas9 modulation of HIV-1 expressed antisense long non-coding RNAs
Long non-coding RNAs (lncRNAs) represent an abundant class of RNAs that have recently been identified in human cells and which are extremely diverse in their function, genomic origin, as well as their mechanism of action 56. Long non-coding RNAs are often expressed as an antisense transcript relative to a protein coding sense transcript and have been observed to act as epigenetic and transcriptional modulators of gene expression 57-60. The integrated HIV-1 provirus is also regulated by an internal antisense lncRNA. This antisense transcript is expressed from the nef gene, which is located at the 3’ end of the viral genome 61-63, and appears functional in modulating the epigenetic and transcriptional state of HIV 64. Over-expression of the nef lncRNA leads to suppression of the virus and inhibition of the nef lncRNA results in increased HIV expression 64
Interestingly, the promoter region of this nef expressed antisense lncRNA 61 lies within a region of nef that is known to be deleted in a cohort of ‘elite controllers’ (patients who appear to control their infection indefinitely) 65 This is the same region that was observed to be susceptible to small RNA directed transcriptional silencing 64. When targeted with small RNAs, transcriptional suppression of the antisense transcript resulted in increased activation of HIV-1 64. While it is not yet clear, it is possible that the nef lncRNA functions to epigenetically modulate the virus and contribute towards the latent state 64. As such, the promoter region in nef might serve as a useful target for CRISPR/Cas9 directed gene excision or gene suppression. CRISPR/Cas9 editing of the promoter elements in nef could essentially recapitulate those deletions found in elite controllers. It is recognized that being an untested strategy, mutations introduced in this region might disrupt viral activation though the production of shortened transcripts and truncated viral proteins. The approach therefore requires further investigation. In any case, this highlights the potential need to identify conserved viral and host lncRNAs that could serve as therapeutic targets of CRISPR/Cas9 editing.
7. Conclusion
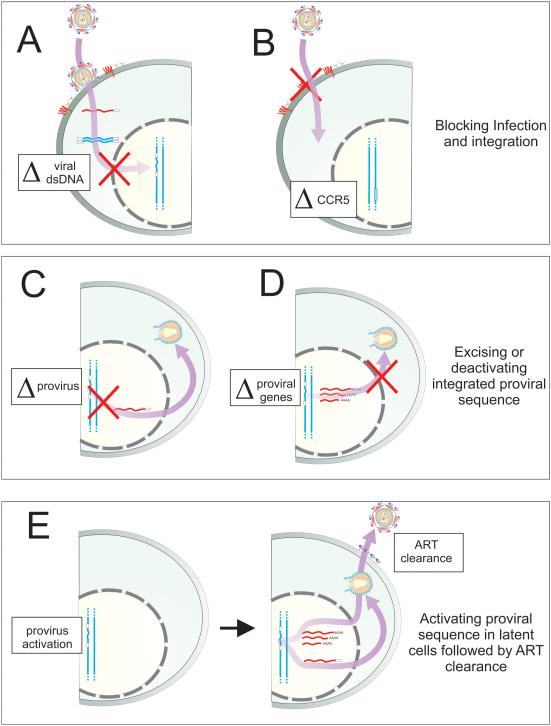
The emergence of CRISPR/Cas9 has had a fundamental impact on our ability to alter the genome and to control gene expression. CRISPR/Cas9 is also an attractive technology for the development of novel therapies. While still in a proof-of-concept stage, CRISPR/Cas9 technologies can be used to block integration and progressive infection (Figure 5A). In addition, integrated HIV-1 proviral sequences can be excised and deactivated, providing important evidence to suggest that one day we may be able to successfully remove latent virus that is hidden away in reservoirs (Figure 5C&D). Secondly, CRISPR/Cas9-based activators represent a promising approach for specific and targetable reactivation of the latently infected viral reservoir, and for subsequent purging of replicating virus whereby ART is used in combination to “shock and kill” (Figure 5E). Finally, CRISPR/Cas9 editing can generate deletion variants of CCR5 that recapitulate the naturally occurring homozygous CCR5Δ32 mutant, which provides important evidence to suggest that permanent cellular immunization from infection and a functional cure is a real possibility (Figure 5B). While many hurdles remain, including concerns over safety and technical issues about the delivery of the expression vectors, it is very clear that CRISPR/Cas9 technologies offer significant hope, for the eventual eradication of HIV/AIDS.
Figure 5. The different CRISPR/Cas9 therapeutic approaches against HIV.
New HIV infection can be blocked by directly targeting the (A) pre-integrated proviral dsDNA (and possibly 2-LTR circular DNA) 17, 26 and (B) by disrupting early-stage host dependency factors such as the co-receptor CCR5 48-50, 52. By targeting the integrated provirus for cleavage, downstream viral production can be blocked by (C) excision of the viral genome by targeting the LTRs (or using multiple sgRNAs) 24-26 and (D) by disrupting viral genes thereby preventing viral genome assembly and budding 26. Lastly, by targeting upstream regulatory sequences using CRISPR/Cas9-based activators in latent cells, subsequent viral output can be restricted by ART and immune clearance in a “shock and kill” approach.
8. Expert opinion
In less than three years, CRISPR/Cas9 technology has advanced to the point where it will likely be remembered as a cornerstone of bioscience and translational research. Heralded as a method to edit any gene in any organism, we have seen its utility expand from the basic manipulation of cells in vitro, to an ability to rapidly engineer new transgenic animal models. It is becoming increasingly clear that a long-lasting solution to HIV/AIDS will require more than just additions to the current growing arsenal of ARVs - which themselves will continue to battle with drug resistance, side effects, cost, and patient adherence. And despite the success of ART, and our ability to achieve viral suppression, there will be the growing future burden of treating an aging population of HIV-infected individuals suffering additional HIV-related illness. Recent reports indicate that elite controllers who maintain viral levels below detectable limits in the absence of ART, have a 2-fold increased level of hospitalization66 and add a further confounder to our understanding of the long term effects of HIV infection. Gene therapy using CRISPR/Cas9 presents new opportunities to address some of the fundamental barriers to progress.
On the one hand, we have discussed how CRISPR/Cas9 might complement the presently available ARV treatment regimens, providing therapeutic approaches against targets that include the HIV provirus, the cellular co-receptor CCR5, and possibly the nef antisense RNA. Indeed, its innate strength as an easily programmable weapon, that can hit multiple targets using the most currently available gene therapy vectors, offers not only a promising tool, but the potential to minimize the risk of the development of viral resistance, and to address many of the disadvantages of alternative gene editing technologies such as ZFNs, TALENs and other nucleases/recombinases.
On the other hand, perhaps the single greatest hurdle that any cure must overcome is the ability to totally eradicate latent virus. With proof of principle using in vitro cell models that CRISPR/Cas9 can excise provirus from “latently” infected cells being the first stage in realizing clinical potential, one might consider that we are close to finding a cure. However, there remain significant questions to be answered. The first relates to safety.
A major safety concern with all nucleases, but specifically with CRISPR/Cas9, is the potential off-target activity. Whilst it is true that low-to-absent levels of Cas9-directed cleavage has been detected within the genome at sites of close homology to the intended target, this can be misleading. Significant off-target cleavage does occur with surprising regularity, even for sgRNAs that have 6 or more off-target mismatches 67. Of course, any single one of these events has the potential to cause cellular transformation. Much effort has been placed on reducing these non-specific effects, from truncating sgRNAs 68 to generating Cas9 “nickases” 69, 70 or dCas9-FokI obligate heterodimers 71. However, these approaches remain less efficient and require further optimization. The ability to switch on, and then off, the CRISPR/Cas9 system could provide one solution. While there are many inducible gene expression systems, all suffer from being stringently controlled. One elegant alternative to inducible expression systems includes the use of protein-destabilizing domains (DD) which result in the rapid degradation of the fusion protein in the absence of a DD-specific small molecule inhibitor 72-74. DD-Cas9 fusions would allow for temporal and dose-dependent control of Cas9 activity by modulating the concentration and the timing of treatment intervention with the specific DD inhibitor, thereby avoiding potential off-target effects that may otherwise result from constitutive Cas9 activity. The ability to control Cas9 expression may also have the added advantage of addressing the concern of long-term clearance of cells expressing exogenous non-human proteins by the immune system.
The second major challenge faced is the identification and characterization of the cellular targets that harbor latent HIV provirus. Cell models containing stable viral integrants are in flux between cellular quiescence, methylation status, and histone compaction. Recent studies indicate that each of these cell models differs in the biological interpretation of latency that they offer, and in particular, the agents required for reversal of viral latency 75. Significantly, an ex vivo model from the Siliciano lab indicates that none of the candidate latency reversal agents tested could adequately demonstrate reversal of viral latency in the absence of cellular activation 76 Therefore, the use of generalized activation agents for therapy remains problematic. While CRISPR/Cas9-mediated HIV activation may be a much more specific and a more promising technology than activating agents in vivo, it remains important to be cautious about the challenges that lie ahead as these novel approaches remain untested in robust in vivo models of infection and latency. Also, while we have a good understanding of circulating memory T-cells, which appear to be the major harbor of HIV provirus, very little is known about the populations of memory T-cells that remain embedded in hidden tissue compartments. Studies planned to dissect these “sanctuary” sites, and analyze the cellular environment in which latent provirus is maintained, will play a pivotal role to resolving this question. Perhaps a new generation of viral or non-viral vectors will provide the means to deliver CRISPR/Cas9 precisely to the sites where it is needed.
Lastly, there are significant barriers to therapy. Even with a potent and specific CRISPR/Cas9 therapeutic candidate, it remains uncertain whether necessary regulatory approvals will be forthcoming for the gene therapy vectors required to deliver these new agents. Moreover, a significant hurdle to targeting latent viral pools is the presence of inaccessible reservoir sites (such as the brain). One possible solution may be the use of smaller Cas9 orthologs and vectors which are capable of being administered safely to the brain, such as Adeno-associated virus (AAV).
There is little doubt of the transformative power of CRISPR/Cas9 technologies for gene editing and transcriptional activation. All things considered, it is still very early to determine the long-term value of this new strategy as a therapeutic against HIV. However, with no immediate prospects of a vaccine, and with over 34 million people infected, the advent of any new path towards a functional cure deserves serious consideration.
Article highlights.
CRISPR/Cas9 as an emerging tool for gene editing and transcriptional modulation of HIV.
CRISPR/Cas9 mediated excision of integrated HIV.
CRISPR/Cas9 mediated activation of HIV gene expression.
CRISPR/Cas9 mediated modulation of HIV host dependency factors.
CRISPR/Cas9 mediated modulation of HIV-associated long non-coding RNAs.
The limitations of the CRISPR/Cas9 technology as a novel therapeutic for HIV.
Acknowledgements
We thank members of the Weinberg and Morris laboratories for helpful discussion.
MSW is supported by funding from the South African Medical Research Council (MRC), and National Research Foundation (NRF). He has received grants for materials/supplies/salaries from the NIH. KVM is supported by NIAID PO1 AI099783-01 and has received grants for materials/supplies/salaries from the NIH. SA is supported on a student fellowship from the MRC of South Africa. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial and competing interests
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
Bibliography
- 1.Palella FJ, Jr., Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. The New England journal of medicine. 1998 Mar 26;338(13):853–60. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 2.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Science translational medicine. 2010 Jun 16;2(36):36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung J, Scherer LJ, Gu A, Gardner AM, Torres-Coronado M, Epps EW, et al. Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Molecular therapy : the journal of the American Society of Gene Therapy. 2014 May;22(5):952–63. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013 Jul;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai M, Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Current gene therapy. 2014 Feb;14(1):2–9. doi: 10.2174/156652321402140318165450. [DOI] [PubMed] [Google Scholar]
- 6.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009 Mar;155(Pt 3):733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. 2005 Feb;60(2):174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 15;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]; * (11) A seminal paper (And one of the first, along with 9 and 10 amongst others) describing the first applications of CRISPR/Cas9 for editing in mammalian cells.
- 12.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013 Jul 18;154(2):442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013 Oct;10(10):977–9. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013 Oct;10(10):973–6. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell research. 2013 Oct;23(10):1163–71. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Lei Y, Wong WK, Liu S, Lee KC, He X, et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic acids research. 2014 Feb 5; doi: 10.1093/nar/gku109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature medicine. 2003 Jun;9(6):727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007 Jun 29;316(5833):1912–5. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 20.Buchholz F, Hauber J. In vitro evolution and analysis of HIV-1 LTR-specific recombinases. Methods. 2011 Jan;53(1):102–9. doi: 10.1016/j.ymeth.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Mariyanna L, Priyadarshini P, Hofmann-Sieber H, Krepstakies M, Walz N, Grundhoff A, et al. Excision of HIV-1 proviral DNA by recombinant cell permeable tre-recombinase. PloS one. 2012;7(2):e31576. doi: 10.1371/journal.pone.0031576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauber I, Hofmann-Sieber H, Chemnitz J, Dubrau D, Chusainow J, Stucka R, et al. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS pathogens. 2013;9(9):e1003587. doi: 10.1371/journal.ppat.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]; * (22) Provided one of the first insights how nucleases could be adapted to target HIV. Led to the directed evolution of recombinases that show activity in humanized mouse models for HIV-1.
- 23.Qu X, Wang P, Ding D, Li L, Wang H, Ma L, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic acids research. 2013 Sep;41(16):7771–82. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific reports. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014 Jul 21; doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature communications. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 27.Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends in microbiology. 2013 Jun;21(6):277–85. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012 Jul 26;487(7408):482–5. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]; **(28) Provided what is perhaps the most compelling demonstration that CRISPR/Cas9 can be used to excise the HIV-1 provirus from latently infected cell lines.
- 29.Ho Y-C, Shan L, Hosmane Nina N, Wang J, Laskey Sarah B, Rosenbloom Daniel IS, et al. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell. 2013;155(3):540–51. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013 Sep;31(9):833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014 Oct 23;159(3):635–46. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; * (31) A vastly improved CRISPR/Cas-mediated activation complex. The rules for effective transcriptional activation provided which allow for multiple genes to be activated simultaneously.
- 32.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014 Dec 10; doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014 Oct 23;159(3):647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995 Dec 15;270(5243):1811–5. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 35.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996 Aug 15;382(6592):635–8. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 36.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nature medicine. 1997 Mar;3(3):252–3. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 37.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996 Aug 22;382(6593):722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 38.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nature medicine. 1997 Nov;3(11):1259–65. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 39.Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011 Mar 10;117(10):2791–9. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 40.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. The New England journal of medicine. 2009 Feb 12;360(7):692–8. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 41.Hutter G, Thiel E. Allogeneic transplantation of CCR5-deficient progenitor cells in a patient with HIV infection: an update after 3 years and the search for patient no. 2. Aids. 2011 Jan 14;25(2):273–4. doi: 10.1097/QAD.0b013e328340fe28. [DOI] [PubMed] [Google Scholar]
- 42.Michael NL, Chang G, Louie LG, Mascola JR, Dondero D, Birx DL, et al. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nature medicine. 1997 Mar;3(3):338–40. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 43.Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997 Jan;3(1):23–36. [PMC free article] [PubMed] [Google Scholar]
- 44.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nature biotechnology. 2008 Jul;26(7):808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nature biotechnology. 2010 Aug;28(8):839–47. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schleifman EB, Bindra R, Leif J, del Campo J, Rogers FA, Uchil P, et al. Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chemistry & biology. 2011 Sep 23;18(9):1189–98. doi: 10.1016/j.chembiol.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014 Mar 6;370(10):901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013 Mar;31(3):230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 49.Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2014 Jul 1;111(26):9591–6. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 Gene Disruption via Lentiviral Vectors Expressing Cas9 and Single Guided RNA Renders Cells Resistant to HIV-1 Infection. PloS one. 2014;9(12):e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014 Jan;24(1):132–41. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic acids research. 2013 Nov 1;41(20):9584–92. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014 Jan 24;343(6169):428–32. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truong L, Wood T, Henley J, Ya-Li L, Kim K, Zhou Y, et al. Autologous Hematopoietic Stem/Progenitor Cell (HSPC) Therapy For Monogenic Blood Disorders: Scalable, cGMP-Compliant Process For Generating Highly Efficient Genome Edited HSPC. Blood. 2013;122(21):4213–13. [Google Scholar]
- 55.Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell. 2014 Nov 6;15(5):643–52. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris KV, Mattick JS. The rise of regulatory RNA. Nature reviews Genetics. 2014 Apr 29; doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013 Apr;20(4):440–6. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010 Nov;1(3):165–75. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008 Jan 10;451(7175):202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008 Nov;4(11):e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, et al. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludwig LB, Ambrus JL, Jr., Krawczyk KA, Sharma S, Brooks S, Hsiao CB, et al. Human Immunodeficiency Virus-Type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products. Retrovirology. 2006;3:80. doi: 10.1186/1742-4690-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, et al. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology. 2012 May 8;9(1):38. doi: 10.1186/1742-4690-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]; **(63) CRISPR/Cas9 targeted to CCR5 in primary human CD4+ T cells and CD34+ HSPCs. CRISPR/Cas9 edited HSPCs retained multilineage potential.
- 64.Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, et al. An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol Ther. 2014 Jun;22(6):1164–75. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995 Jan 26;332(4):228–32. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]; **(65) The use of integrase-defective lentiviral vectors (IDLVs) to detect and refine our understanding of CRISPR/Cas9 off-target cleavage effects.
- 66.Crowell TA, Gebo KA, Blankson JN, Korthuis PT, Yehia BR, Rutstein RM, et al. Hospitalization Rates and Reasons among HIV Elite Controllers and Persons With Medically Controlled HIV Infection. The Journal of infectious diseases. 2014 Dec 15; doi: 10.1093/infdis/jiu809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nature biotechnology. 2015 Jan 19; doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 68.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014 Jan 26; doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013 Sep 12;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nature methods. 2014 Mar 2; doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 71.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology. 2014 Jun;32(6):569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sellmyer MA, Chen LC, Egeler EL, Rakhit R, Wandless TJ. Intracellular context affects levels of a chemically dependent destabilizing domain. PloS one. 2012;7(9):e43297. doi: 10.1371/journal.pone.0043297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chemistry & biology. 2010 Sep 24;17(9):981–8. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; * (73) Defining the physiological parameters that define latency are paramount to understanding both the approaches to “shock and kill”, as well as the mechanisms by which HIV becomes dormant, and then reactivates. This work provides a current overview by conducting an extensive examination of the effects of knows stimuli on the activation of HIV in a wide range cell models of HIV latency.
- 74.Tai K, Quintino L, Isaksson C, Gussing F, Lundberg C. Destabilizing domains mediate reversible transgene expression in the brain. PloS one. 2012;7(9):e46269. doi: 10.1371/journal.pone.0046269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS pathogens. 2013;9(12):e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nature medicine. 2014 Apr;20(4):425–9. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014 Jan 29; doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014 Sep 25;513(7519):569–73. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014 Nov 28;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]