Abstract
Epigenetic changes play important roles in carcinogenesis and influence initial steps in neoplastic transformation by altering genome stability and regulating gene expression. To characterize epigenomic changes during the transformation of normal plasma cells to myeloma, we modified the HELP assay to work with small numbers of purified primary marrow plasma cells. The nano-HELP assay was used to analyze the methylome of CD138+ cells from 56 subjects representing premalignant (MGUS), early and advanced stages of myeloma as well as healthy controls. Plasma cells from premalignant and early stages of myeloma were characterized by striking, widespread hypomethylation. Gene specific hypermethylation was seen to occur in the advanced stages and cell lines representative of relapsed cases were found to be sensitive to decitabine. Aberrant demethylation in MGUS occurred primarily in CpG islands while differentially methylated loci in cases of myeloma occured predominantly outside of CpG islands and affected distinct sets of gene pathways, demonstrating qualitative epigenetic differences between premalignant and malignant stages. Examination of the methylation machinery revealed that the methyltransferase, DNMT3A, was aberrantly hypermethylated and underexpressed, but not mutated in myeloma. DNMT3A underexpression was also associated with adverse overall survival in a large cohort of patients, providing insights into genesis of hypomethylation in myeloma. These results demonstrate widespread, stage specific epigenetic changes during myelomagenesis and suggest that early demethylation can be a potential contributor to genome instability seen in myeloma. We also identify DNMT3A expression as a novel prognostic biomarker and suggest that relapsed cases can be therapeutically targeted by hypomethylating agents.
Introduction
Despite therapeutic advances, multiple myeloma remains incurable and needs newer insights into the pathogenic mechanisms that cause this disease. Gene expression profiling has been used extensively in myeloma and has led to the development of a risk model that remains robust even in the age of newer therapies.(1) The use of gene expression profiling by different groups has also led to the identification of distinct molecular subgroups that show defined clinical features (2, 3). However pathogenic mechanisms driving these differences in gene expression have not been well described, especially for early stages in myelomagenesis (4).
Recent studies of the epigenome and in particular the methylome have shown that cancer is characterized by widespread epigenetic changes. These changes lead to altered gene expression that can result in activation of oncogenic pathways. Furthermore, methylome profiling has shown greater prognostic ability than gene expression profiling in AML and has led to identification of newer molecular subgroups with differences in overall survival (5). We have shown that methylome profiling is able to identify changes that occur early during carcinogenesis in solid tumors such as esophageal cancer (6). These studies have been conducted with the use of the HELP assay, which is a genome wide assay that provides a reproducible analysis of the methylome that is not biased towards CpG islands (7). Analyzing the methylome of myeloma can help identifying changes that can define disease subsets and lead to identification of newer therapeutic targets. Recent sequencing studies in myeloma have also revealed mutations in enzymes that are involved in epigenetic machinery, again reinforcing the need to study the epigenetic alterations in this disease (8). We have conducted a genome wide analysis of changes in DNA methylation in MGUS, newly diagnosed MM as well as relapsed MM and compared them to normal plasma cell controls. We used a modification of the HELP assay (nano-HELP) that was developed to work with low amounts of DNA to interrogate the methylome of sorted CD138+ cells from patient samples. We report that widespread alterations in DNA methylation are seen in myeloma and have the power to discriminate between MGUS and new / relapsed cases. We report that hypomethylation is the predominant early change during myelomagenesis that is gradually transformed to hypermethylation in relapsed cases, thus providing the epigenetic basis for the differentiation between these different stages of myeloma.
Material and Methods
Patient samples
Bone marrow aspirates were obtained with signed informed consent from 11 patients with monoclonal gammopathy of uncertain significance (MGUS), 4 patients with smoldering myeloma (SMM), 13 patients with newly diagnosed myeloma (NEWMM), 16 patients with relapsed myeloma (REL), including 2 patients with serial samples, and 2 patients in clinical complete remission (REM) who are followed at the myeloma clinic at the Robert H. Lurie Cancer Center at Northwestern University. All specimen underwent CD138+ microbead selection (Miltenyi Biotec). Purity was confirmed by flow cytometry staining for CD38 and CD45. Samples with a purity less than 90% by flow cytometry were not used for this study. 8 samples from normal donors without known malignancies were obtained commercially from AllCells, LLC (Emeryville, CA). All normal donor samples had a purity of ≥85%.
DNA methylation analysis using the nano-HELP assay
Genomic DNA was extracted using the Gentra PureGene kit using 1 × 10e5 to 5 × 10e5 cells.
DNA methylation analysis was done using the HpaII tiny fragment Enrichment by Ligation mediated PCR (HELP) assay, as described previously (7, 9).
In brief, 100ng of genomic DNA (nano-HELP modification) was digested overnight using the two isoschizomers HpaII and MspI. The digestion products were then purified once with phenol-chloroform. After overnight adapter ligation the HpaII – and MspI-products were amplified in a 2-step protocol as previously described (7). HpaII- and MspI-generated genomic fragments between 200 and 2000 base pairs (Bp) in length were labeled with either Cy3- or Cy5-labeled random primers and then cohybridized onto a human (HG17) custom designed oligonucleotide array 50-mers) covering 25 626 HpaII amplifiable fragments (HAFs) annotated to 14 214 gene promoters. HAFs are defined as genomic sequences contained between 2 flanking HpaII sites found 200 to 2000 bp apart. Each HAF on the array is represented by a probe set consisting of 14 to 15 individual probes, randomly distributed across the microarray slide. Methylation data presented in this manuscript have been deposited in the NIH Gene Expression Omnibus (GEO, National Center for Biotechnology Information [NCBI], http://www.ncbi.nlm.nih.gov/geo/) under accession number ### (currently awaiting assignment of the accession number).
HELP data analysis and quality control
All microarray hybridizations were subjected to extensive quality control using the following strategies. First, uniformity of hybridization was evaluated using a modified version of a previously published algorithm (10, 11) adapted for the NimbleGen platform, and any hybridization with strong regional artifacts was discarded and repeated. Second, normalized signal intensities from each array were compared against a 20% trimmed mean of signal intensities across all arrays in that experiment, and any arrays displaying a significant intensity bias that could not be explained by the biology of the sample were excluded. Signal intensities at each HpaII amplifiable fragment were calculated as a robust (25% trimmed) mean of their component probe-level signal intensities. Any fragments found within the level of background MspI signal intensity, measured as 2.5 mean-absolute-differences (MAD) above the median of random probe signals, were categorized as “failed.” These “failed” loci therefore represent the population of fragments that did not amplify by PCR, whatever the biological (e.g. genomic deletions and other sequence errors) or experimental cause. On the other hand, “methylated” loci were so designated when the level of HpaII signal intensity was similarly indistinguishable from background. PCR-amplifying fragments (those not flagged as either “methylated” or “failed”) were normalized using an intra-array quantile approach wherein HpaII/MspI ratios are aligned across density-dependent sliding windows of fragment size-sorted data. The log2(HpaII/MspI) was used as a representative for methylation and analyzed as a continuous variable. For most loci, each fragment was categorized as either methylated, if the centered log HpaII/MspI ratio was generally less than zero, or hypomethylated if on the other hand the log ratio was greater than zero.
Quantitative DNA methylation analysis by MassArray Epityping
Validation of the findings of the HELP assay was carried out by MALDI-TOF mass spectrometry using EpiTyper by MassArray (Sequenom, CA) on bisulphite converted DNA as previously described (6). Validation was done on all samples with sufficient available DNA. MassARRAY primers were designed to cover the flanking HpaII sites for a given HAF, as well as for any other HpaII sites found up to 2000 bp upstream of the downstream site and up to 2000 bp downstream of the upstream site, to cover all possible alternative sites of digestion within our range of polymerase chain reaction amplification.
Microarray data analysis
Unsupervised clustering of HELP data by hierarchical clustering was performed using the statistical software R version 2.6.2. A two-sample T-test was used for each gene to summarize methylation differences between groups. Genes were ranked on the basis of this test statistic and a set of top differentially methylated genes with an observed log fold change of >1 between group means was identified. Genes were further grouped according to the direction of the methylation change (hypomethylated versus hypermethylated in MDS), and the relative frequencies of these changes were computed among the top candidates to explore global methylation patterns. Validation with MassArray showed good correlation with the data generated by the HELP assay. MassArray analysis validated significant quantitative differences in methylation for differentially methylated genes selected by our approach
Sequencing of DNMT3A
Screening for mutations in the DNMT3A catalytic domain was carried out using direct genomic sequencing. PCR primers were designed to amplify and sequence coding exons 18–23 (available upon request). For each polymerase chain reaction (PCR), 20 ng genomic DNA was used for PCR amplification followed by purification using Montage Cleanup kit (Millipore, Billerica, MA). Sequencing was performed using ABI 3730×l DNA analyzer (Applied Biosystems, Foster City, CA). PCR was carried out using: 94°C, 4 min, followed by 35 cycles of 94°C, 30 sec, specific annealing temperature for each primer, 30 sec, and extension 72°C, 30 sec, final 72°C, 5 min. Results were compared to reference (NM_175629) and SNP databases (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP). If chromatograms suggested a possible mutation, bidirectional resequencing was performed.
Pathway analysis and Transcription Factor binding site analysis
The Ingenuity Pathway Analysis software (IPA) (Redwood City, CA) was used to determine biological pathways associated with differentially methylated genes as performed previously (6, 10). Enrichment of genes associated with specific canonical pathways was determined relative to the Ingenuity knowledge database for each of the individual platforms and the integrated analysis at a significance level of p<0.01. Biological networks captured by the microarray platforms were generated using IPA and scored based on the relationship between the total number of genes in the specific network and the total number of genes identified by the microarray analysis. The list of differentially methylated genes was examined for enrichment of conserved gene-associated transcription factor binding sites using IPA as well as other published gene sets available through the Molecular Signatures Database (MSigDB) (12).
Meta-Analysis of Myeloma Gene Expression studies
We obtained gene expression data from the Arkansas datasets GSE5900 and GSE2658 (4) from NCBI’s Gene Expression Omnibus (GEO) database. The datasets were quantile normalized to ensure cross-study comparability, based on our previous approach (13, 14). Analyses were performed using SAS (SAS Institute, Cary, NC) and the R language (http://www.r-project.org). The final database had 22 Normal controls, 44 MGUS and 559 new MM samples.
Cell lines and culture conditions
Cell lines U266, RPMI8226 were purchased from ATCC (Manassas, VA). Cell line Cell line MM1.S and MM1.R were kindly provided by Dr. S. Rosen. All cell lines were cultured in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum and penicillin (100 U/mL), streptomycin (100 mg/mL) and 4 mM glutamine. Cells were maintained at 37°C and humidified with 95% air and 5% CO2 for cell culture. For the viability assays the cells were cultured in 0.5, 1 and 5 µM Decitabine (Sigma) 5 days. Decitabine was added to the culture daily, DMSO was served as control. Viability was measured on day 6 using MTS (Promega) according to the manufacturer’s instruction.
Results
Genome wide analysis of DNA methylation reveals epigenetic alterations in plasma cells from patients with Myeloma and MGUS
Extensive study of gene expression profiling of myeloma cells has led to several molecular models that allow the classification of myeloma in different risk categories, which has led to significant improvements in treatment strategies and outcomes (1, 15). Epigenetic alterations including aberrant DNA methylation can regulate gene expression and can also be used as better prognostic markers in tumor models (5). To determine whether there was aberrant differential methylation in different stages of multiple myeloma, we used the nano-HELP assay (7) on CD138+ selected bone marrow (BM) cells from 11 patients with monoclonal gammopathy of uncertain significance (MGUS), 4 patients with smoldering myeloma (SMM), 13 patients with newly diagnosed myeloma (NEWMM) and 16 patients with relapsed myeloma (REL), which also included 2 patients with serial samples. We also included 2 myeloma patients in complete remission (REM). CD138+ selected BM plasma cells from 8 healthy donors (normal, NL) served as controls. Clinical characteristics of the 48 patients with plasma cell dyscrasias (PD) are listed in supplemental table S1.
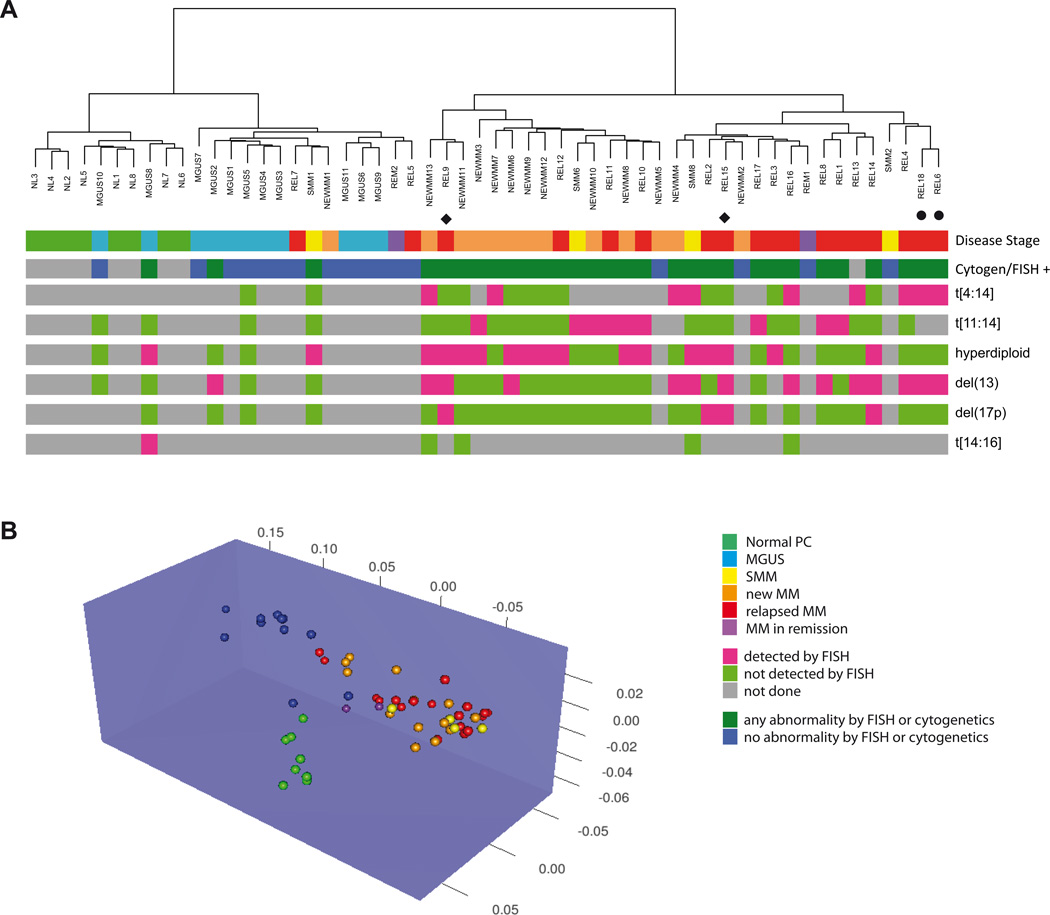
At first we performed an unsupervised analysis of the generated methylation profiles with hierarchical clustering and the nearest shrunken centroid algorithm. Both methods showed that the healthy controls formed a tight cluster that was distinct from samples with abnormal plasma cells, demonstrating epigenetic dissimilarity between these groups (Figures 1A and B). Interestingly, MGUS samples also clustered in a distinct group, suggesting definite alterations in DNA methylation that occur early during myelomagenesis. The new (NEW) and relapsed (REL) myeloma cases demonstrated great epigenetic dissimilarity to the NL and MGUS samples and separated in two subgroups with the majority of NEWMM in one and the majority of REL samples in the other group (Fig 1A). These findings show that methylation profiling can differentiate between NL, MGUS, NEWMM and REL, in an unsupervised manner. Included in our analysis were two sets of consecutive samples taken from the same patient at different time points. In the first case, the initial sample (REL15) clusters with the group of relapsed myeloma and the post-treatment sample with clusters with the NEWMM samples (REL9). The second case included samples taken at distinct time points without any treatments in the interval. Both samples cluster together in the REL group (REL18, REL6) and thus show the biological validity of our analysis.
Figure 1. DNA methylation patterns can differentiate between different stages of plasma cell dyscrasias.
A) Unsupervised hierarchical clustering of using methylation profiles generated by the HELP assay separate the samples in two major clusters containing the majority of NEWMM (orange) and REL (red) samples or the majority of NL (green) and MGUS (light blue) samples respectively. These two clusters were also identified by the presence (dark green) or absence (dark blue) of abnormalities detected by FISH or conventional cytogenetics. The clusters are each further separated into two subgroups resulting in a total of four cohorts representing the majority of NL, MGUS, NEWMM and REL. Green = NL, blue = MGUS, yellow = SMM, orange = NEWMM, red = REL, purple = REM. The bottom 6 lines represent FISH data, green = not detected by FISH, pink = detected by FISH, gray = not done. ◆ or ● indicate paired samples.
B) Unsupervised 3D clustering based on nearest shrunken centroid algorithm using methylation profiles also shows distinction between normal and myeloma samples. Among the myeloma samples, clustering of MGUS samples is distinct from New and Relapsed cases. Green = NL, blue = MGUS, yellow = SMM, orange = NEWMM, red = REL, purple = REM
MGUS and NEWMM show predominant hypomethylation, whereas REL are predominantly hypermethylated
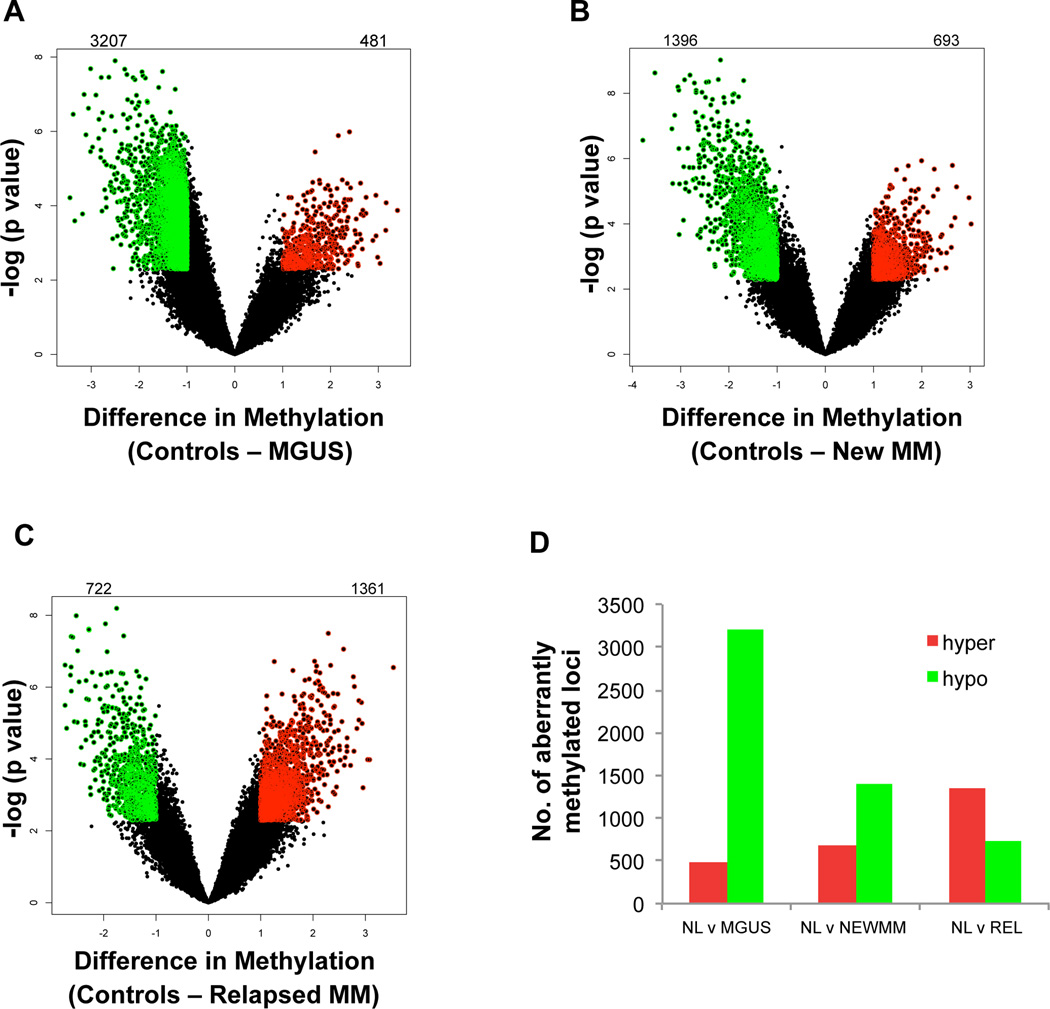
After demonstrating global epigenetic dissimilarity between normal and myelomatous plasma cells, we wanted to determine the specific differences in DNA methylation between the different stages of myeloma. We performed a supervised analysis comparing the MGUS, NEWMM or REL cases versus control samples. Volcano plots comparing the difference of mean methylation of all individual loci were plotted against the significance (log (p value) based on T Test) of the difference (Figure 2). Stringent cut-offs comprising an absolute fold change of ≥2 (= log(HpaII/MspI) > 1) and a p value of <0.005 were used to identify differentially methylated loci. All probes thus identified were significant after multiple testing with Benjamini-Hochberg correction with a FDR of less than 5%. We observed that majority of differentially methylated loci in MGUS were found to be hypomethylated when compared to normal plasma cells. (Figure 2A; Supplemental Table S2A and B). Hypomethylation was the predominant change also seen in NEWMM samples, yet this is was less pronounced than in MGUS (Figure 2B; Supplemental Table S2C and D). Interestingly, in cases of relapsed myeloma, we observed increased hypermethylation demonstrating that there was a trend towards predominant hypomethylation in MGUS to predominant hypermethylation in REL (Figure 2 C,D; Supplemental Table S2E and F). Overall differential methylated genes at the transition between stages are depicted in supplemental figure S1A. When analyzing this differential methylation at the transition from one disease state to the next (i.e. NL to MGUS, MGUS to NEWMM and NEWMM to REL) we found that of the 2963 unique genes that were hypomethylated at the transition from NL to MGUS, 2472 became hypermethylated at the transition of MGUS to NEWMM. Of those 2 were also significantly hypermethylated at the transition from NEWMM to REL, albeit with a q-value > 0.05 (p < 0.05, supplemental Figure S1B). A list of the differentially methylated genes between the different disease states can be found in supplemental table S2A–K.
Figure 2. MGUS and NEWMM show predominant hypomethylation, whereas REL are predominantly hypermethylated.
Volcano Plots showing difference of mean methylation (X Axis) and significance of the difference (Y Axis) demonstrate aberrant hypomethylation in MGUS (A) and both hypo and hypermethylation in New (B) and relapsed (C) cases of myeloma. The number of differentially methylated HpaII amplifiable fragments (HAFs) are indicated above each volcano plot. Numbers to the left indicate hypomethylated HAFs and numbers to the right indicate hypermethylated HAFs. Aberrant hypermethylation is the predominant change in Relapsed cases (D).
The HELP assay has been validated quantitatively in many studies (5, 6) and we also validated our findings by analyzing the methylation status of differentially methylated genes, ARID4B and DNMT3A by bisulfite Massarray analysis. Examination of the promoter regions of these genes demonstrated a strong correlation of quantitative methylation values obtained from MassArray with the findings of HELP assay, demonstrating the validity of our findings (Supplemental Figure S2).
Differentially methylated genes show specific functional and genomic characteristics
We next analyzed the biological pathways that were associated with differentially methylated genes in myeloma and observed that pathways involved in cell proliferation, gene expression, cell cycle and cancer were significantly involved by aberrantly methylated genes in MGUS, New MM as well as relapsed cases of MM (Table 1). The genes affected by aberrant methylation of these pathways included many genes that were hypomethylated (CEBPalpha, Interleukins and their receptors, GFI1 etc.) and hypermethylated (EZH2, various HOX members, SOX and WNT family members) that have not been previously implicated in myelomagenesis.
TABLE 1.
GENES AND PATHWAYS ABERRANTLY METHYLATED IN MYELOMA
| BIOLOGICAL FUNCTIONS | GENES | |
|---|---|---|
| Genes Hypomethylated in MGUS | ||
| 1 | Cell Morphology, Cellular Assembly and Organization, Cellular Function and Maintenance | ADNP, AFF2, ARL15, B4GALT3, BRSK2, CASC3, DIS3L, EBAG9, EXOSC1, EXOSC2, EXOSC8, FBXO9, FLNC, GDI2, IL15, MRPS15, NAGA, NGF, NR3C2, P2RX3, PLCG1, PRKCZ, PRPH, STARD10, STK11, TGFA, TNK1, TSPAN7, TUBE1, UPF3B, WDR6, ZNF74, ZNF83 |
| 2 | Cell Signaling, Molecular Transport, Nucleic Acid Metabolism | FFAR3, FZD1, FZD5, FZD10, GALR1, GPR6, GPR26, GPR44, GPR68, GPR77, GPR84, GPR87, GPR132, GPR137, GPR149, GPR153, GPR161, GPR176, GPRASP2, GRM6, HRH2, LGR5, LPHN1, MC3R, NPY2R, NPY5R, OPRK1, PTGER1, PTGER3, QRFPR, SCTR, TAS1R2, VIPR1 |
| 3 | Cellular Development, Hematological System Development and Function, Hematopoiesis | ALOX5, ALX4, C12orf44, CEBPE, CLEC11A, COPB2, COPE, COPG, COPG2, COPZ1, CXCL5, DDA1, ETS2, FARSB, FLI1, GFI1, GPX1, HMGB2, HSD17B4, IL6R, NAA15, NAA16, S100A9, SACM1L, SIPA1L3, SPI1, SPIB, SRGN, TARS, TDP2, UBA5, UQCRC2, ZXDC |
| 4 | Cell Cycle, Hair and Skin Development and Function, Embryonic Development | BANF1, BLOC1S1, CCNE1, CHEK1, CHMP5, CHMP2A, CUL4A, CUL4B, DCAF11, DCAF16, DDB1, ERCC8, GRP, KAT2A, LIN28B, MED20, NEK9, NUDCD1, PHIP, PKM2, PRPF31, SART3, TADA1, TBL3, USP4, USP5, USP8, USP35, USP36, USP37, USP43, WDR5B, ZNF277 |
| 5 | Amino Acid Metabolism, Cellular Assembly and Organization, Genetic Disorder | ANKRD28, AP1G1, ATP2A3, CCDC47, CCT7, CLN3, CLPX, DDIT4L, DGKZ, DPM1, GAR1, GEMIN4, GEMIN5, HOOK1, LOC100290142/USMG5, NECAP1, NHP2, NUP93, PFDN2, SEC61A1, SH2D3C, SHC1, SLC25A10, SLC25A11, SLC25A22, SMAP1, SNRPG, TNFRSF14, TSC22D1, UNC45A, VBP1, WDR8 |
| Genes Hypermethylated in Relapsed MM | ||
| 1 | Genetic Disorder, Inflammatory Disease, Respiratory Disease | AMH, ATF5, BATF, BTG2, CHPF, CHSY3, CTH, EXOSC1, EXOSC3, EXOSC6, EXOSC10, FERMT3, GDF9, GET4, ILF3, LMO2, LSM3, LSM7, LTBP4, MAGED1, NAA38, NOD1, NOD2, NUDT21, PBK, RIPK2, SCLT1, SCRIB, SGTA, SNAPC5, SUGT1, TRIB3, UNC5A |
| 2 | Gene Expression, Cellular Development, Cellular Growth and Proliferation | ACY1, AKR1B1, BHLHE41, CSMD1, DLX4, ERG, EZH2, FRZB, GNB4, GNB5, HEXIM2, HOXA11, HOXB2, HOXB4, IER5L, KRT6A, LHX2, NPM3, RFTN1, RGS9, SLC29A1, SOX2, SOX4, SOX14, SYTL1, TNF, TNKS2, TNKS1BP1, TTC1, ZBTB11 |
| 3 | Cell Signaling, Molecular Transport, Vitamin and Mineral Metabolism | ADRB1, BAI3, CCR6, CNR2, CXCR1, CYSLTR1, GAB1, GABBR2, GNG7, GPER, GPR1, GPR15, GPR25, GPR44, GPR62, GPR88, GPR125, GPR146, GPR151, GPRC5A, LGR4, LPAR2, MC4R, NPY, PIK3CG, PTGDR, PTGER1, RXFP4, SSTR2 |
| 4 | Genetic Disorder, Neurological Disease, Dermatological Diseases and Conditions | APEX1, CDCA8, CDK16, CDK19, CDK5R1, CORO1C, COX7A2L, EIF6, EIF4B, GDI1, GDI2, GNL1, HOXC13, LZTS1, MSN, NOL3, NUDT5, OSGEP, PAICS, PLEK, RAB5C, RCC2, RPS6, RUFY1, SAP30BP, SCT, SET, TPM2, TUFM, ZNF212 |
| 5 | DNA Replication, Recombination, and Repair, Cell Morphology, Cellular Function and Maintenance | APLF, ARHGEF4, DLX2, DNTT, ERCC4, GATA6, HEYL, IKZF1, KIF5A, LIG4, MECOM, MSX2, PCNA, PDCD6, PECAM1, POLI, RFC4, RPA1, SP8, TAOK3, VPS28, VPS37B, WNT5B, WRN, XPA, XRCC2, XRCC6 |
| Genes Hypomethylated in New MM | ||
| 1 | Cell Signaling, Carbohydrate Metabolism, Lipid Metabolism | ADORA1, BDKRB1, BDKRB2, CHRM2, FFAR1, FFAR3, FPR1, GABBR2, GLP2R, GPR20, GPR32, GPR113, GPR120, GPR139, GPR161, GPR109B, GPRASP2, HBEGF, MC2R, MCHR1, MTNR1A, NMUR1, NPY5R, OPN1LW, OXGR1, RGR, RHO, TAC3, TACR2, TAS1R2, VN1R4 |
| 2 | Neurological Disease, Immunological Disease, Inflammatory Disease | CAPN9, CAPN11, CDH4, CDH7, CDH17, CST3, DNAH1, DNAH5, DNAH10, DNAH17, DNAI2, EDN3, EID1, EIF4EBP2, IL1R1, IL1RAPL1, LIG3, MAPK6, MBP, MYOZ1, NRAP, PDLIM2, PLP1, RNASE2, SIGIRR, STK39, TGM2 |
| 3 | Antigen Presentation, Cell-To-Cell Signaling and Interaction, Hematological System Development and Function | BGN, DOCK1, EFNA3, Hsp70, IFNA1/IFNA13, IgG, IL1, IL25, IL12, IL17A, JPH3, KIR2DL3, KRT2, KRT3, KRT9, KRT14, KRT20, KRT23, LBP, LGALS7/LGALS7B, MYLK2, POTEKP, S100A3, SELP, SERPINB3, SERPINB4, SH3GL2, Tlr, TNFRSF1B, TREM1, TUBA3C/TUBA3D |
| 4 | Cancer, Connective Tissue Disorders, Reproductive System Development and Function | ACR, ACTG2, ADCY, ADCY8, cacn, CACNA1B, Cacng, CACNG1, CACNG3, CACNG5, CNGA3, CNR1, COTL1, GNAO1, GNG2, KIF23, LPO, MYLK3, MYO7A, PCP4, PDE6G, POU2F2, PPP3R2, PSCA, RIT2, SHROOM3, SPTB, TUBB8, VIL1 |
| 5 | Lipid Metabolism, Molecular Transport, Small Molecule Biochemistry | ABCG5, ABCG8, ADAMTS13, Akt, AMPK, APOB, CD22, CEBPA, COL15A1, COL5A3, COL6A3, CYP3A4, CYP4F2, CYP8B1, FCAR, GRB10, HNF4α dimer, KLK8, LIPE, MRC2, NR0B2, NR1H4, PCK1, PTPN3, SCARB1, SLC22A7, STAR, T3-TR-RXR, VWF |
| Genes Hypermethylated in New MM | ||
| 1 | Gene Expression, Amino Acid Metabolism, Post-Translational Modification | ACTR3, ANP32E, BTAF1, CBX5, CDK9, DPYSL2, ERCC4, FUBP1, GTF2H1, HNRNPA0, HNRNPA1, HNRNPUL1, HOXA11, LSM7, MDFIC, MEPCE, NAA38, NNT, PLOD3, POU2AF1, PRMT5, PRUNE, SART3, TAF4, TAF11, TUBB |
| 2 | Genetic Disorder, Inflammatory Disease, Respiratory Disease | AATF, AIMP1, AIMP2, BARD1, EFNA1, EPHA5, FXC1, HNRNPC, HSF1, LMO3, LZTR1, NEDD9, NOD1, NOD2, NR1H3, SIRT1, SS18L1, STMN2, SUGT1, TIMM9, TIMM8A, TOMM40L, TOMM70A, TRIP6, XRCC6, ZNF584 |
| 3 | DNA Replication, Recombination, and Repair, Cell Cycle, Cellular Development | ACACB, AKT1S1, ARFIP2, ATF5, BTG2, CD34, CDKN2A, CDKN2B, CENPK, CHAF1A, CHTF18, GSC, HIST1H2AB/HIST1H2AE, ID3, PCNA, PLEKHA8, POLD1, RFC4, RPS16, SLC3A2, TBX5, TCF19, UBR7, ULK1, WRN |
| 4 | Gastrointestinal Disease, Organismal Injury and Abnormalities, Genetic Disorder | ANKS1A, APOB, BRMS1, C14orf156, DNAJA1, DNAJB11, DNAJC9, EML1, EVX1, FOS, FTL, KCNQ4, KIAA1279, LCP1, PDPK1, PIK3CA, PRKCB, RASSF5, SAP18, SET, SGK3, SLC4A10, SPTBN2, TPM2 |
| 5 | Lymphoid Tissue Structure and Development, Tissue Morphology, Molecular Transport | BIRC2, BPGM, C1orf56, CCDC71, CHAT, DHX15, FAM81A, GGA3, ID2, KCNC4,NDUFS3, NFKBIE, NOLC1, PECI, POGK, POLK, PTEN, PTPN2, SLC9A3R1, SLITRK1, SOX4, STC1, TRAPPC3, ZNF264/ZNF805 |
Aberrant methylation was not distributed randomly across chromosomes. Differentially methylated HpaII fragments showed significant regional differences with positional association with chromosomes 1, 9, 11, 15, 17, 19, 20 and 21. Furthermore, to determine whether these aberrantly methylated regions shared any common DNA elements, we performed a search for transcription factor binding sites enriched in these loci. Significant overrepresentation of binding sites for transcription factors (TFs) that have been implicated (PAX, MEF and MAZ) in myelomagenesis was observed (Table 2). Various TFs such as HNF6, PAX4, STAT3, EVI1 and others were significantly enriched in relapsed cases of MM, including some that have not been implicated in the pathophysiology of MM previously.
Table 2.
Transcription factor binding sites enriched in differentially methylated loci
| Transcription factor | Genes in Overlap | Motif |
|---|---|---|
| MGUS | ||
| ARNT | 202 | NDDNNCACGTGNNNNN |
| ATF6 | 95 | TGACGTGG |
| E47 | 205 | VSNGCAGGTGKNCNN |
| USF | 203 | NNRNCACGTGNYNN |
| LXR | 55 | TGGGGTYACTNNCGGTCA |
| TCF1P | 184 | GKCRGKTT |
| New MM | ||
| PAX2 | 36 | NNNNGTCANGNRTKANNNN |
| GATA | 161 | WGATARN |
| MEF2 | 21 | NNTGTTACTAAAAATAGAAMNN |
| E2F | 56 | TWSGCGCGAAAAYKR |
| CACCC BINDING FACTOR | 205 | CANCCNNWGGGTGDGG |
| Relapsed MM | ||
| STAT3 | 113 | NNNTTCCN |
| PAX4 | 193 | NAAWAATTANS |
| EGR3 | 56 | NTGCGTGGGCGK |
| EVI1 | 44 | AGATAAGATAA |
| HNF6 | 189 | HWAAATCAATAW |
| MAZ | 135 | GGGGAGGG |
| SREBP1 | 136 | NATCACGTGAY |
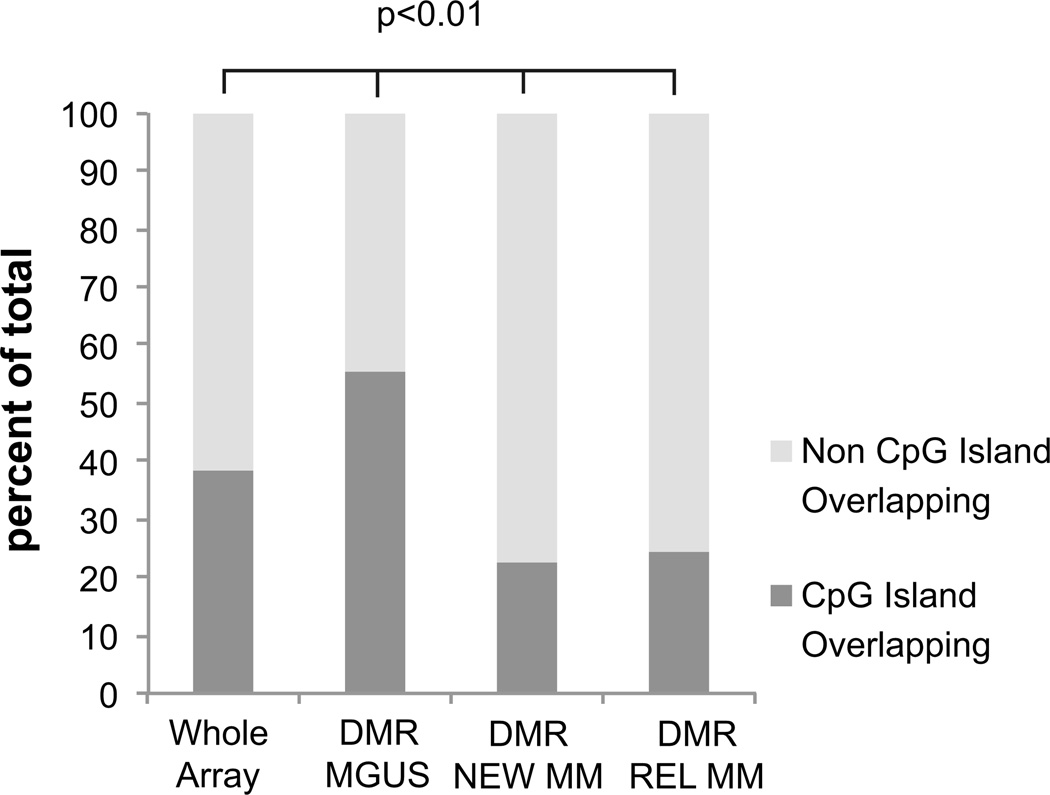
Finally, we also sought to determine if CpG islands were predominantly affected by differential methylation in myeloma. We observed that aberrant methylation occurring in MGUS was significantly found to occur within CpG islands, while both new and relapsed cases of MM had aberrantly methylated loci located outside of CpG islands (Figure 3). This shows that there are qualitative differences in epigenetic alterations between MGUS and myeloma and are also consistent with recent findings that have highlighted that aberrant methylation in cancer can occur outside of CpG islands.(16)
Figure 3. Differential methylation in MGUS occurs mainly within CpG islands.
The genomic location of differentially methylated regions was mapped to CpG islands for each subcategory of myeloma. DMRs in MGUS were significantly enriched within CPG islands (dark gray) compared to the percentage of probes in the whole array (Proportions Test, P Value<0.01). DMRs in NEWMM and REL were significantly located outside of CpG islands (Proportions Test, P Value<0.01).
Genes involved in DNA methylation machinery are differentially methylated and expressed in myeloma
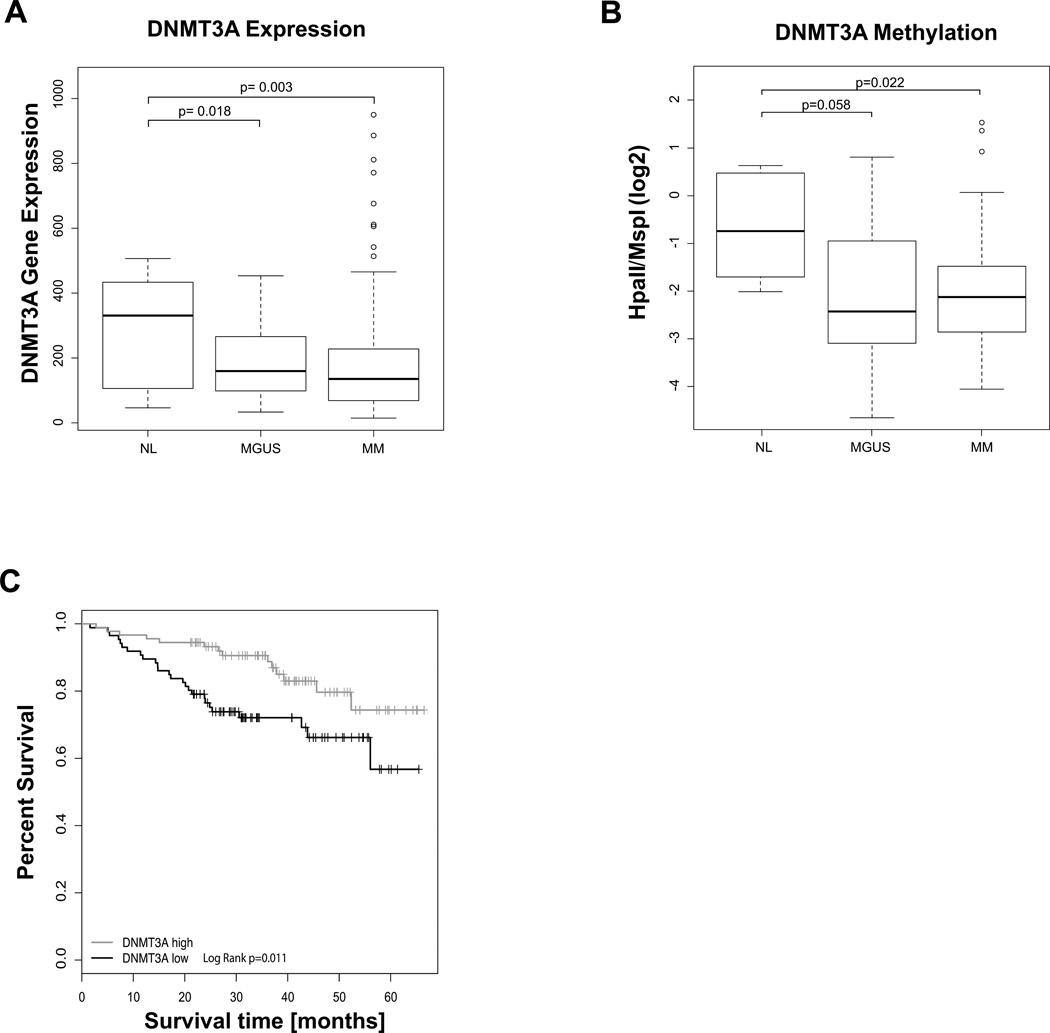
Since we saw an overall decrease in methylation in MGUS we analyzed our dataset for the methylation status of genes involved in this process and also evaluated for the expression of these genes in large gene expression datasets (Arkansas datasets GSE5900 and GSE2658) that have been published previously.(4) Specifically, we analyzed the methyltransferases DNMT1, DNMT3a and DNMT3b and observed that DNMT3a was significantly underexpressed in a large independent cohort of myeloma gene expression profiles (Figure 4A) and had a significant impact on survival. After separating patients into four quartiles according to their DNMT3A expression we found that high DNMT3A expression (Q4) at baseline conveyed a significantly overall survival compared to patients with very low DNMT3A expression (N=140 in each group, Log Rank test, P Value=0.01, Figure 4C). In our cohort of patients we found the promoter of DNMT3A to be aberrantly hypermethylated (Figure 4B). Mutations of the gene encoding DNMT3a have recently been reported for MDS and AML (17) and have not been examined in myeloma. Sequencing of the catalytic domain of DNMT3A in 23 of our CD138 purified plasma cell samples revealed no exonic mutation or SNPs. These results suggest that DNMT3A expression in myeloma is mainly reduced by aberrant hypermethylation.
Figure 4. DNMT3A is underexpressed and hypermethylated in myeloma.
A) Box plots showing gene expression values in CD138+ cells from NL (n=22), MGUS (n= XX) and MM (n= 559) from Arkansas datasets GSE5900 and GSE2658 shows significantly reduced expression levels in myeloma (TTest, P Value< 0.05). The MM samples included in these datasets contained untreated samples. B) Boxplots representing the methylation of the DNMT3A promoter in normal PCs (NL), MGUS and myeloma samples (MM) shows hypermethylation in MM compared to NL (TTest, P Value <0.05). C) Low DNMT3A expression is associated with worse overall survival in TT2. Differences between groups with top and bottom quartile gene expression are shown with Kaplan Meier graphs.
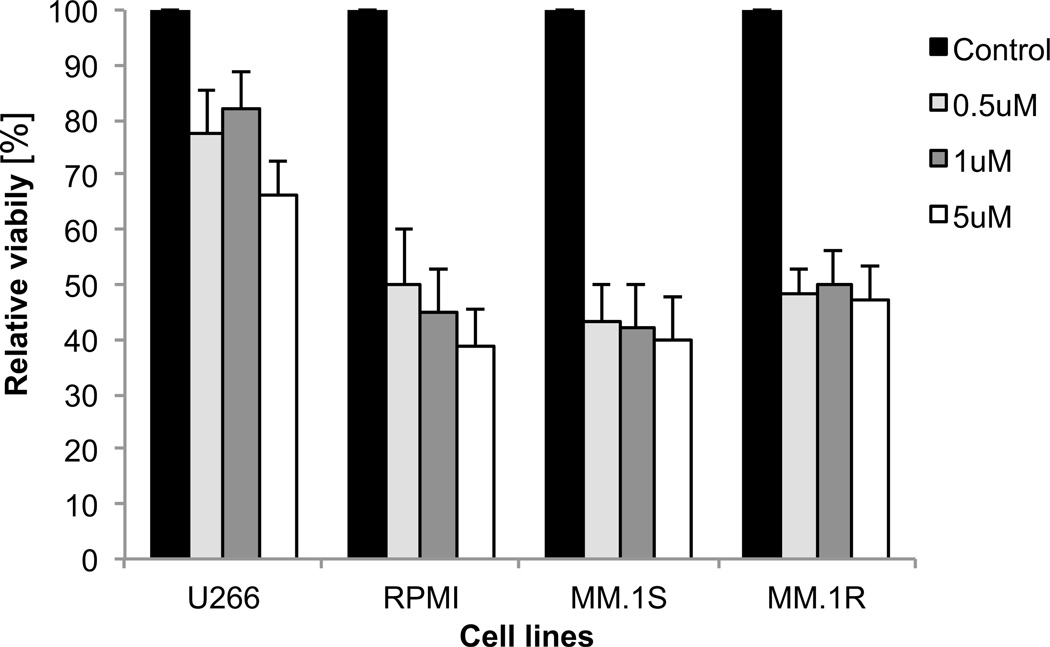
Myeloma cell lines are sensitive to DNMT inhibitors
Lastly, since we observed that relapsed cases of myeloma exhibit increased aberrant methylation, we wanted to determine the sensitivity of these cells to hypomethylating agents. We tested the efficacy of DNMT inhibitor, Decitabine, in long standing myeloma cell lines that exhibit various cytogenetic alterations and are reflective of relapsed cases. Decitabine was able to significantly inhibit the proliferation of these myeloma cell lines at both low and high doses (Figure 5). In fact the dose of 0.5µM has shown to be hypomethylating without causing DNA damage and was significantly able to inhibit myeloma cell proliferation thus suggesting that reversal of hypermethylation can be a potential therapeutic strategy in these cases.
Figure 5. Decitabine treatment leads to growth inhibition in myeloma cell lines.
Cell lines were treated with different doses of decitabine for 5 days and proliferation was assessed by the MTS assay. Significant inhibition of growth was seen after treatment even with low doses of Decitabine (TTest, P Value <0.05). Shown is one representative of three experiments.
Discussion
We show that myeloma is characterized by widespread alterations in DNA methylation that are specific for different stages of the disease. Early stage of MGUS is characterized by predominant hypomethylation that is prevalent enough to distinguish these cells from normal control plasma cells. The later stages acquire progressive hypermethylation with maximum methylation seen in relapsed cases. These data provide a comprehensive epigenomic map of myelomagenesis and also identify important oncogenic gene pathways that are targeted by these aberrant changes.
The observation of global hypomethylation in MGUS builds upon recent two recent studies that noticed loss of methylation in plasma cell neoplasms (18, 19). The first study by Salhia et al. reports hypomethylation as early as in MGUS and noted no difference noted between methylation of new and treated myeloma samples (19). The second study by Walker et al. noticed hypomethylation in myeloma samples, but did not see any significant differences between normal plasma cells and MGUS samples (18). Remethylation was seen in plasma cell leukemia samples in comparison to MM. In contrast to the latter study which interrogated 27578 CpG sites (corresponding to 14495 genes), the former study used an approach looking at a limited set of only 1505 CpG sites (corresponding to 807 genes) thus limiting generalizability of those findings. The study presented here was able to distinguish between normal, MGUS and myeloma (new and relapsed) samples on unsupervised clustering and revealed clear-cut, widespread differences between these groups. We used the HELP assay, a high-resolution assay, that is not biased towards CpG islands and has been able to show stage-specific differences in various tumor models (5, 20). This assay analyses 25626 CpG sites and thus is comparable to the system used by Walker et al. Just like Walker et al. we observe relative hypermethylation in late stages of MM, however we observe hypomethylation much earlier, i.e. already at the level of MGUS an not only MM, similar to the reports of Salhia et al. This discrepancy might be due to the limited number of NL and MGUS samples (3 and 4 respectively) used by Walker et al. Our findings of early hypomethylation in MGUS, which is maintained throughout the early MM stage but then converts to predominant hypermethylation thus represents a novel assessment of myelomagenesis
The finding of hypomethylation in MGUS and myeloma is also different from the previous single locus studies that have focused on hypermethylation of tumor suppressor genes in myeloma (21). The classical cancer associated epigenetic alteration is promoter CpG island hypermethylation. Even though global hypomethylation was reported in the pioneering epigenetic studies in cancer (22) most investigators have subsequently focused on hypermethylation in CpG islands within selected gene promoters. Array based DNA methylation assays which have mainly focused on CpG islands have supported the biased perception that CpG island are uniquely responsible for methyl-DNA induced genomic changes. Newer iterations of assays for the analysis of DNA methylation, which are based on next generation sequencing, have revealed a much more complex picture with DNA methylation occurring further upstream in CpG island shores, in introns as well as extending much further downstream. With this in mind it is difficult to speculate about the true meaning of DMRs in CpG islands and clearly points to the need of further in-depth investigations with these newer methods. Hypomethylation has been hypothesized to lead to carcinogenesis by promoting genomic instability (23, 24) as well as by aberrant activation of oncogenes (23). Since myeloma is characterized by various chromosomal translocations and deletions, the finding of early hypomethylation may be an important pathogenic mechanism that promotes secondary genetic events that lead to the development of full blown disease.
Efforts to distinguish MGUS from MM in an unsupervised manner using Gene expression profiling (GEP) data from large clinical trials have not been successful to date (25, 26). Although MGUS samples can readily be separated from NL samples, they appear identical to MM at a GEP level. Using methylation data we were able to clearly distinguish between the majority of NL, MGUS, MM and REL samples. Gene expression profiling can be affected by large scale changes in just a few transcripts. Since methylation analysis is dependent on evaluation of DNA, it is not globally biased by differences in a few loci and thus is not affected by changes that may occur only in a minority of the analyzed cells. It is therefore more reflective of biology of premalignant conditions, as illustrated by our previous study in esophageal carcinogenesis(27). In the presented study we used CD138+ selection to isolate the myelomatous and normal plasma cells, thus it is possible, that the observed difference between MGUS and MM samples is in part due to “dilution” with normal samples. Strategies such as multi-parameter flow cytometry sorting are being used to ensure clonality in future studies. At this point it should be noted that although two samples included in our study were in clinical remission with less than 3% of PC observed on bone marrow biopsy, they were quite different at the epigenetic level. While REM2 as expected clustered with MGUS samples, REM1 clustered with the relapsed samples, suggesting the presence of epigenetic higher risk features that were not appreciated with conventional methods.
Despite the continued progress and the improvement of treatment with newer drugs, drug resistance remains a big concern. Strategies to use highly active drugs in combination upfront has resulted in a significant improvement of progression free survival however only has had limited effect on overall survival. Multiple studies in other cancers have shown association of hypermethylation phenotypes with resistance to treatment via the inactivation of various cell cycle and other genes involved in chemosensitivity. The presented study identifies a relative hypermethylation in cases of relapsed myelomas compared to normal plasma cells. This difference is further enhanced in the comparison between newly diagnosed MM and relapsed MM (data not shown) and consistent with the observation made by Walker et al. comparing plasma cell leukemia, a very advanced high-risk stage of MM, to newly diagnosed MM. At this point it is not possible to say whether this hypermethylation phenotype is the result of treatment or due to the biology of the disease. However it suggest that inclusion of demethylating agents such as DNMT inhibitors as a viable treatment option. In vitro findings by us an others (28, 29) with representative myeloma cell lines using the DNMT inhibitor decitabine support this assumption and clinical trials testing this hypothesis are currently under way.
In addition to global quantitative differences in methylation, we also found differences in sites of aberrant methylation between MGUS and Myeloma. We observed that even though changes in MGUS involved CpG islands, the later changes seen in myeloma preferentially occurred outside of CpG islands. Recent work has similarly shown that cytosines present outside of CpG islands can be aberrantly methylated/ hypomethylated in cancer, and assays that cover these loci are critical to discovering the full landscape of altered methylome of malignancies (16). Our novel findings thus demonstrate both qualitative and quantitative differences between these subsets of disease.
Finally, we also provide insights into the genesis of these epigenetic changes. We observed that DNMT3A is significantly reduced in myeloma and is aberrantly methylated early in both MGUS and myeloma. DNMT3A is an important methyltransferase which has been shown to be mutated in a large proportion of AML cases (17). We did not find any DNMT3A mutations in our cases and showed that this enzyme can be aberrantly methylated in cancer as well. It is generally accepted that DNMT3A and DNMT3B are responsible for the establishment of methylation whereas DNMT1 ensures maintenance of methylation, thus guaranteeing the faithful transmission of DNA methylation marks from one cell generation to the next. However this longstanding view is being challenged and newer models suggest a role for the de novo enzymes DNMT3A and DNMT3B in methylation maintenance. (30) In fact recent data shows that DNMT3A mutations impart an adverse prognosis in AML (31). Our data shows that dysregulation of this methyltransferase can be also seen in myeloma and adds to the importance of this gene in hematologic malignancies. Analysis of the publicly available data from Walker et al. confirm a significantly higher level of methylation for newly diagnosed MM compared to normal samples and a very high methylation status of MGUS samples with a strong trend towards significance (data not shown). However generalization of this data is limited due to the low number of normal and MGUS samples included in that study. Unfortunately, due to sample limitation, we were not able to obtain RNA for gene expression profiles from the samples used to perform the methylation analysis. While we are not able to directly correlate methylation status and gene expression, the finding of hypermethylation and underexpression (compared to normal plasma cells) seen in two different datasets respectively, as well as the DNMT3A expression effect on survival suggest a role for DNMT3A in myelomagenesis that warrants further investigation. If DNMT3A expression and methylation plays a role in the progression of MGUS to MM requiring treatment is the subject of an ongoing investigation.
In contrast to early MM relapsed MM shows predominant hypermethylation. Although we cannot show a clear reason for this, others have shown that exposure to chemotherapy can lead to large scale genetic and epigenetic changes in tumor samples (reviewed in (32–34)). The relapsed cases examined in our cohort were heavily pretreated patients who have been exposed multiple MM agents including Bortezomib. It is plausible that our relapsed population is enriched with patients who have a higher overall degree of methylation as hypermethyalted phenotype have been associated with chemotherapy resistance in multiple cancers (35–37) and has been linked with resistance to Bortezomib (38). In the present study we have shown that low dose decitabine significantly reduces viability of several MM cell lines as models of late stage disease. Low doses of decitabine have been shown to significantly reduce DNMT levels without the additional DNA damage activity (39, 40). Ongoing longitudinal studies following patients from diagnosis to relapse will hopefully answer the question whether hypermethylation can be detected early on and whether it is associated with survival. If hypermethylation were to be detected early and to predict for short progression free survival, it would be rational to add a demethylating drug such as decitabine to the initial therapy.
Supplementary Material
Acknowledgments
Supported by NIH Immunooncology Training Program T32 CA009173, Gabrielle’s Angel Foundation, Leukemia and Lymphoma Society, American Cancer Society, Department of Defense and Partnership for Cures grant.
Footnotes
The authors have no conflicts of interest to declare
Bibliography
- 1.Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, Dhodapkar M, Nevins J, Barlogie B, Shaughnessy JD., Jr Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27:4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Zangari M, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–1700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez H, Opalinska J, Zhou L, Sohal D, Fazzari MJ, Yu Y, Montagna C, Montgomery EA, Canto M, Dunbar KB, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Opalinska J, Sohal D, Yu Y, Mo Y, Bhagat T, Abdel-Wahab O, Fazzari M, Figueroa M, Alencar C, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286:25211–25223. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, Zheng X, Kim K, Greally JM. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24:1161–1167. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohal D, Yeatts A, Ye K, Pellagatti A, Zhou L, Pahanish P, Mo Y, Bhagat T, Mariadason J, Boultwood J, et al. Meta-analysis of microarray studies reveals a novel hematopoietic progenitor cell signature and demonstrates feasibility of inter-platform data integration. PLoS ONE. 2008;3:e2965. doi: 10.1371/journal.pone.0002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, Nguyen AN, Sohal D, Ying Ma J, Pahanish P, Gundabolu K, Hayman J, Chubak A, Mo Y, Bhagat TD, et al. Inhibition of the TGF-beta receptor I kinase promotes hematopoiesis in MDS. Blood. 2008;112:3434–3443. doi: 10.1182/blood-2008-02-139824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair B, Shaughnessy JD, Jr, Zhou Y, Astrid-Cartron M, Qu P, van Rhee F, Anaissie E, Alsayed Y, Waheed S, Hollmig K, et al. Gene expression profiling of plasma cells at myeloma relapse from tandem transplantation trial Total Therapy 2 predicts subsequent survival. Blood. 2009;113:6572–6575. doi: 10.1182/blood-2009-02-207803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, Davies FE, Ross FM, Morgan GJ. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117:553–562. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 19.Salhia B, Baker A, Ahmann G, Auclair D, Fonseca R, Carpten J. DNA methylation analysis determines the high frequency of genic hypomethylation and low frequency of hypermethylation events in plasma cell tumors. Cancer Res. 2010;70:6934–6944. doi: 10.1158/0008-5472.CAN-10-0282. [DOI] [PubMed] [Google Scholar]
- 20.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A, Heuck CJ, Fazzari MJ, Mehta J, Singhal S, Greally JM, Verma A. DNA methylation alterations in multiple myeloma as a model for epigenetic changes in cancer. Wiley Interdiscip Rev Syst Biol Med. 2010;2:654–669. doi: 10.1002/wsbm.89. [DOI] [PubMed] [Google Scholar]
- 22.Cravo M, Pinto R, Fidalgo P, Chaves P, Gloria L, Nobre-Leitao C, Costa Mira F. Global DNA hypomethylation occurs in the early stages of intestinal type gastric carcinoma. Gut. 1996;39:434–438. doi: 10.1136/gut.39.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003;532:39–49. doi: 10.1007/978-1-4615-0081-0_5. [DOI] [PubMed] [Google Scholar]
- 25.Hardin J, Waddell M, Page CD, Zhan F, Barlogie B, Shaughnessy J, Crowley JJ. Evaluation of multiple models to distinguish closely related forms of disease using DNA microarray data: an application to multiple myeloma. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1018. Article10. [DOI] [PubMed] [Google Scholar]
- 26.Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, Dhodapkar M, Nevins J, Barlogie B, Shaughnessy JD., Jr Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez H, Opalinska J, Zhou L, Sohal D, Fazzari MJ, Yu Y, Montagna C, Montgomery EA, Canto M, Dunbar KB, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS genetics. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilop S, van Gemmeren TB, Lentjes MH, van Engeland M, Herman JG, Brummendorf TH, Jost E, Galm O. Methylation-associated dysregulation of the suppressor of cytokine signaling-3 gene in multiple myeloma. Epigenetics : official journal of the DNA Methylation Society. 2011;6:1047–1052. doi: 10.4161/epi.6.8.16167. [DOI] [PubMed] [Google Scholar]
- 29.Nojima M, Maruyama R, Yasui H, Suzuki H, Maruyama Y, Tarasawa I, Sasaki Y, Asaoku H, Sakai H, Hayashi T, et al. Genomic screening for genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:4356–4364. doi: 10.1158/1078-0432.CCR-08-3336. [DOI] [PubMed] [Google Scholar]
- 30.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature reviews. Genetics. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, Yun H, Gohring G, Schlegelberger B, Hoelzer D, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 32.Kubota M, Lin YW, Hamahata K, Sawada M, Koishi S, Hirota H, Wakazono Y. Cancer chemotherapy and somatic cell mutation. Mutation research. 2000;470:93–102. doi: 10.1016/s1383-5742(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 33.Karp JE, Smith MA. The molecular pathogenesis of treatment-induced (secondary) leukemias: foundations for treatment and prevention. Seminars in oncology. 1997;24:103–113. [PubMed] [Google Scholar]
- 34.Lewandowska J, Bartoszek A. DNA methylation in cancer development, diagnosis and therapy--multiple opportunities for genotoxic agents to act as methylome disruptors or remediators. Mutagenesis. 2011;26:475–487. doi: 10.1093/mutage/ger019. [DOI] [PubMed] [Google Scholar]
- 35.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA, Brown R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–4576. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Rao X, House MG, Nephew KP, Cullen KJ, Guo Z. GPx3 promoter hypermethylation is a frequent event in human cancer and is associated with tumorigenesis and chemotherapy response. Cancer letters. 2011;309:37–45. doi: 10.1016/j.canlet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Cheetham S, Tang MJ, Mesak F, Kennecke H, Owen D, Tai IT. SPARC promoter hypermethylation in colorectal cancers can be reversed by 5-Aza-2'deoxycytidine to increase SPARC expression and improve therapy response. British journal of cancer. 2008;98:1810–1819. doi: 10.1038/sj.bjc.6604377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leshchenko V, Kuo P-Y, Ulahannan N, Braunschweig E, Yu Y, Suh S, Weniger M, Goy A, Wymdham W, Wiestner A, et al. High-Resolution Sequencing Identifies NOXA1 De-Methylation As a Novel Strategy to Overcome Bortezomib Resistance in Mantle Cell Lymphoma. Blood. 2011;118 [Google Scholar]
- 39.Negrotto S, Ng KP, Jankowska AM, Bodo J, Gopalan B, Guinta K, Mulloy JC, Hsi E, Maciejewski J, Saunthararajah Y. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2012;26:244–254. doi: 10.1038/leu.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.