Abstract
Ataxia oculomotor apraxia type 2 (AOA2) is a rare autosomal recessive cerebellar ataxia. Recent evidence suggests that the protein defective in this syndrome, senataxin (SETX), functions in RNA processing to protect the integrity of the genome. To date, only patient-derived lymphoblastoid cells, fibroblasts and SETX knockdown cells were available to investigate AOA2. Recent disruption of the Setx gene in mice did not lead to neurobehavioral defects or neurodegeneration, making it difficult to study the etiology of AOA2. To develop a more relevant neuronal model to study neurodegeneration in AOA2, we derived neural progenitors from a patient with AOA2 and a control by induced pluripotent stem cell (iPSC) reprogramming of fibroblasts. AOA2 iPSC and neural progenitors exhibit increased levels of oxidative damage, DNA double-strand breaks, increased DNA damage-induced cell death and R-loop accumulation. Genome-wide expression and weighted gene co-expression network analysis in these neural progenitors identified both previously reported and novel affected genes and cellular pathways associated with senataxin dysfunction and the pathophysiology of AOA2, providing further insight into the role of senataxin in regulating gene expression on a genome-wide scale. These data show that iPSCs can be generated from patients with the autosomal recessive ataxia, AOA2, differentiated into neurons, and that both cell types recapitulate the AOA2 cellular phenotype. This represents a novel and appropriate model system to investigate neurodegeneration in this syndrome.
Introduction
Ataxia oculomotor apraxia type 2 (AOA2) was first described 15 years ago and subsequently mapped to chromosome 9 (1). This disorder is characterized by progressive cerebellar atrophy, peripheral neuropathy, oculomotor apraxia in ∼50% of the patients and elevated α-fetoprotein levels with an age of onset between 10 and 20 years (2). The gene defective in AOA2 was identified as SETX coding for senataxin, a 2667 amino acids protein that contains a highly conserved C-terminal seven-motif domain of the superfamily 1 of DNA/RNA helicases and an N-terminal domain important for protein–protein interactions (3).
Using lymphoblastoid cells and fibroblasts from AOA2 patients and SETX-RNAi-depleted cells as model systems, roles for senataxin have been uncovered that encompass (i) protection against DNA damage (3), (ii) transcription regulation including transcription termination, splicing efficiency of specific mRNAs and alternate splice site selection (4) and R-loop resolution (5) and (iii) localization at the interface of transcription and replication (6). R-loops are RNA/DNA hybrids that form over transcription pause sites by interaction with an ssDNA template behind an elongating RNA Pol II complex, which are potentially harmful and can cause genomic instability if left unresolved (7). A role for senataxin in transcription elongation and termination is further supported by a report, showing that cells with senataxin knockdown display an increase in RNA readthrough and RNA Pol II density downstream of the Poly(A) site and also exhibit increased levels of R-loop formation (5). Senataxin was found to localize to distinct nuclear foci in S/G2 phase cells, and the number of these foci increased in response to impaired replication, suggesting that senataxin localizes to collision sites between the transcription apparatus and components of the replisome (6). In addition, a SUMO-dependent interaction between senataxin and Rrp45, a core component of the exosome, was also shown to co-localize in nuclear foci corresponding to sites of R-loops, suggesting that senataxin connects transcription, DNA damage and RNA surveillance (8).
More recently, we generated a mouse model of AOA2 revealing that senataxin is involved in homologous recombination and gene silencing (9). Disruption of the Setx gene caused accumulation of R-loops, leading to the persistence of DNA double-strand breaks (DSB) and a failure of crossing-over. Senataxin localized to the XY body and persistence of RNA Pol II activity, altered ubH2A distribution and abnormal XY-linked gene expression in Setx−/− demonstrated an essential role for senataxin in meiotic sex chromosome inactivation (MSCI) (9). These data support key roles for senataxin in coordinating meiotic crossing-over with transcription and in gene silencing to protect the integrity of the genome. Furthermore, mutation of SETX has been shown to lead to disease-specific alterations of gene expression in patients that are conserved across cell type and species, including the cerebellar neurons of Setx−/− mice (10). Unfortunately, the Setx−/− mouse did not exhibit neurobehavioral defects or neurodegeneration and thus was not an appropriate model to study the neurodegenerative changes in AOA2 (9,11).
Given the current lack of a neuronal model system to study neurodegeneration in AOA2, we decided to reprogram AOA2 patient fibroblasts into induced pluripotent stem cells (iPSCs), which have the potential to be further differentiated into mature neurons and glial cells. Relevant to developing a neuronal model system to study neurodegeneration in AOA2, Muguruma et al. (12) recently reported the successful generation of polarized cerebellar structure from three-dimensional (3D) human embryonic stem cell (hESC) cultures in which the self-organized neuroepithelium differentiated into functional Purkinje cells (12). Given that cerebellar atrophy and loss of Purkinje cells are key features of AOA2 (2), the development of AOA2 iPSCs represents a first step toward the generation of cerebellar progenitors and the study of cerebellar development in AOA2. Here we report the generation of bona fide footprint-free AOA2 iPSCs that recapitulate the AOA2 cellular phenotype, the differentiation of AOA2 iPSCs into neural progenitors and neurons that exhibit signs of oxidative stress, sensitivity to DNA-damaging agents, R-loop accumulation and genome instability, providing evidence of the suitability of this model system to investigate neurodegeneration in AOA2. Furthermore, differential gene expression, pathway analysis and system analysis of gene co-expression in AOA2 neural progenitors are consistent with findings from AOA2 patients and provide novel insights into the role of senataxin in gene regulation and neurodegeneration.
Results
Generation and characterization of AOA2 iPSC
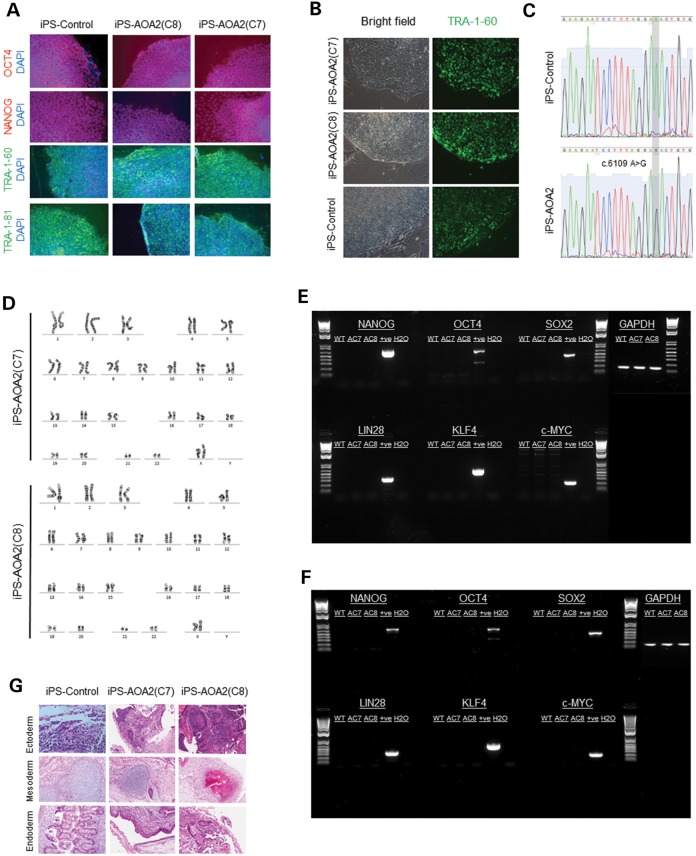
In order to optimize conditions and to reduce the risk of chromosomal instability, we used early passage (P < 5) fibroblasts for reprogramming. Following transfection with pEP4EO2SCK2MEN2L and pEP4EO2SET2K episomal plasmids, we stepwise adapted the cells to knockout serum replacement (KOSR) medium over the first 4–5 days of iPSC generation, as direct replacement with the KOSR medium was found to lead to extensive death of the AOA2 fibroblasts. After 2 weeks, transduced AOA2 patient and matched control fibroblasts gave rise to colonies of small round cells with a high nucleus-to-cytoplasm ratio typical of pluripotent human stem cells (Fig. 1A). Although our data show that it was possible to reprogram AOA2 fibroblasts, the efficiency was somewhat reduced compared with that of controls. Thirteen colonies from the AOA2 homozygote patient who expressed the TRA-1-60 stem cell surface marker (13) (Fig. 1B) were expanded for further analysis. Two of these AOA2 clones, AOA2(C7) and AOA2(C8), were selected for further analysis as these clones displayed robust expression of the pluripotency markers TRA-1-60, TRA-1-81, Nanog and Oct4 (Fig. 1A). Control clones were screened similarly as described earlier and conformed to the same criteria. The presence of the c.6109 A>G homozygous missense mutation in AOA2 iPSC was confirmed at the mRNA levels by sequencing (Fig. 1C), and a normal karyotype was observed for AOA2(C7) and AOA2(C8) iPS clones (Fig. 1D). OCT4, NANOG and SOX2 expression in the AOA2 iPSC was not derived from integrated or persistent reprogramming plasmids as (i) polymerase chain reaction (PCR) analysis of genomic DNA revealed no amplicons following 36 rounds of PCR using IRES-anchored primers designed to amplify the reprogramming genes OCT4, SOX2, LIN28, KLF4 and c-MYC (plasmids used as positive controls) (Fig. 1E) and (ii) reverse transcriptase (RT)–PCR analysis of RNA isolated from the AOA2 and control iPSC showed an absence of transgene expression (human fibroblasts transiently transfected with the reprogramming plasmids used as a positive control) (Fig. 1F). iPSC from both AOA2 patients and controls formed teratomas when injected into SCID mice comprising tissue types from all three germ layers (endoderm, mesoderm and ectoderm), indicating pluripotent tri-lineage differentiation (Fig. 1G). We conclude from these analyses that the phenotype of AOA2 iPSC is consistent with a fully reprogrammed iPSC phenotype.
Figure 1.
Characterization of AOA2 iPS cells. (A) Expression of pluripotency markers Oct4, Nanog, TRA-1-60 and TRA-1-81 in both control and AOA2 iPSC colonies. (B) Expression TRA-1-60 stem surface marker in control and AOA2 iPSCs. (C) mRNA sequencing confirmed the c.6109 A>G homozygous missense mutation in AOA2 iPSC. Chromatograms showing the wild-type (c.6109A) and mutated (c.6109G) sequences in controls and AOA2 iPSC lines, respectively (based on NCBI reference sequence NM_015046.5). (D) Normal karyotype was observed for both AOA2(C7) and AOA2(C8) iPS clones. (E) PCR analysis of genomic DNA revealed no amplicons following 36 rounds of PCR using IRES-anchored primers designed to amplify the reprogramming genes OCT4, SOX2, LIN28, KLF4 and c-MYC (plasmids used as positive controls). (F) RT–PCR analysis of RNA isolated from the AOA2 and control iPSC showed an absence of transgene expression (human fibroblasts transiently transfected with the reprogramming plasmids used as a positive control). (G) Teratoma formation in SCID mice following control and AOA2 iPSC injections. Tissue types from all three germ layers (endoderm, mesoderm and ectoderm) were observed indicating pluripotent tri-lineage differentiation.
AOA2 iPSC displays an oxidative stress phenotype and R-loop accumulation
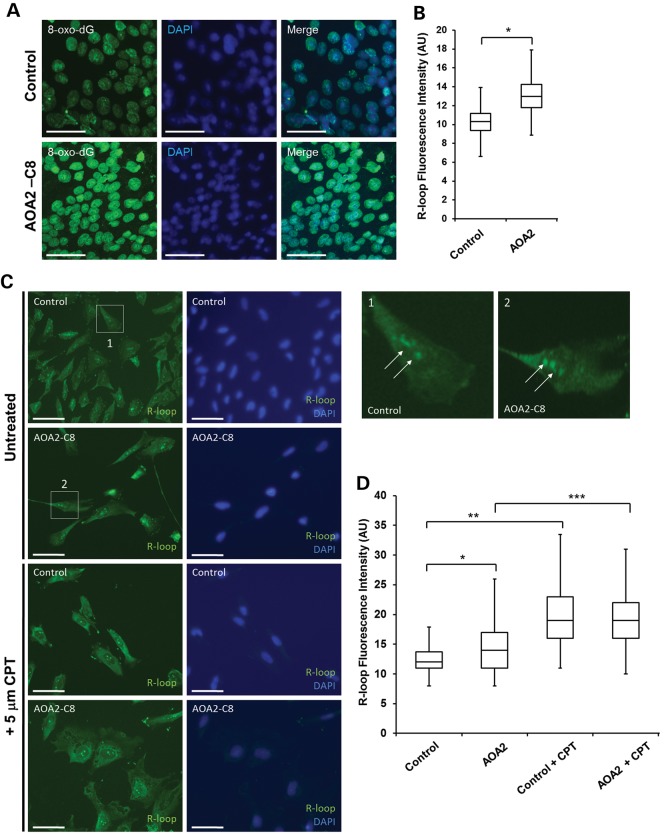
As AOA2 cells were previously shown to exhibit oxidative stress (3), we first wished to examine the oxidative stress status of AOA2 iPSC. Immunostaining for the oxidative DNA damage marker 8-oxo-dG (Fig. 2A) revealed a small but significant increase in the average 8-oxo-dG fluorescence intensity in AOA2 iPSCs, compared with controls (Fig. 2B). Senataxin was recently shown to be involved in the resolution of R-loops (5,9). In order to determine the level of R-loop accumulation in AOA2 iPSCs, we performed immunostaining using the well-characterized anti-RNA/DNA (S9.6) antibody (14). Extranuclear and nucleolar signals for R-loops were observed, in agreement with previous findings showing that R-loops occur readily in the nucleolus and mitochondria (Fig. 2C) (15,16). A small but significant increase in R-loop fluorescence intensity was observed in AOA2 iPSCs compared with controls (Fig. 2D). To verify these data, we exposed the cells to camptothecin (CPT), a topoisomerase poison that blocks topoisomerase I activity, which acts to relax supercoils generated during transcription and DNA replication and therefore readily induces nucleolar R-loop formation (11,15,16). Treatment of control and AOA2 iPSCs with 5 µm CPT led to additional increases in R-loop formation in both cell types as expected (Fig. 2D).
Figure 2.
Oxidative stress and R-loop formation in AOA2 iPSC. (A) Increased levels of oxidative DNA damage in AOA2 iPSC revealed by anti-8-oxo-dG immunostaining. Nuclei were stained with DAPI. Scale bar, 50 µm. (B) Quantitation of 8-oxo-dG fluorescence intensity (AU, arbitrary units) in both control and AOA2 iPSC. Fluorescence intensity was measured for 100 cells per experiment from three technical replicates from one experiment (*P < 0.05, Student's t-test). (C) Detection of R-loop in control and AOA2 iPSC using the S9.6 antibody in untreated (basal levels) and CPT-treated cells. DAPI counterstained nuclei. Scale bar, 20 µm. (D) Quantitation of average R-loop fluorescence intensity (AU) in untreated and CPT-treated iPSc. *, ** and *** indicate P < 0.001, one-way ANOVA.
Differentiation of AOA2 iPSC into neural progenitors
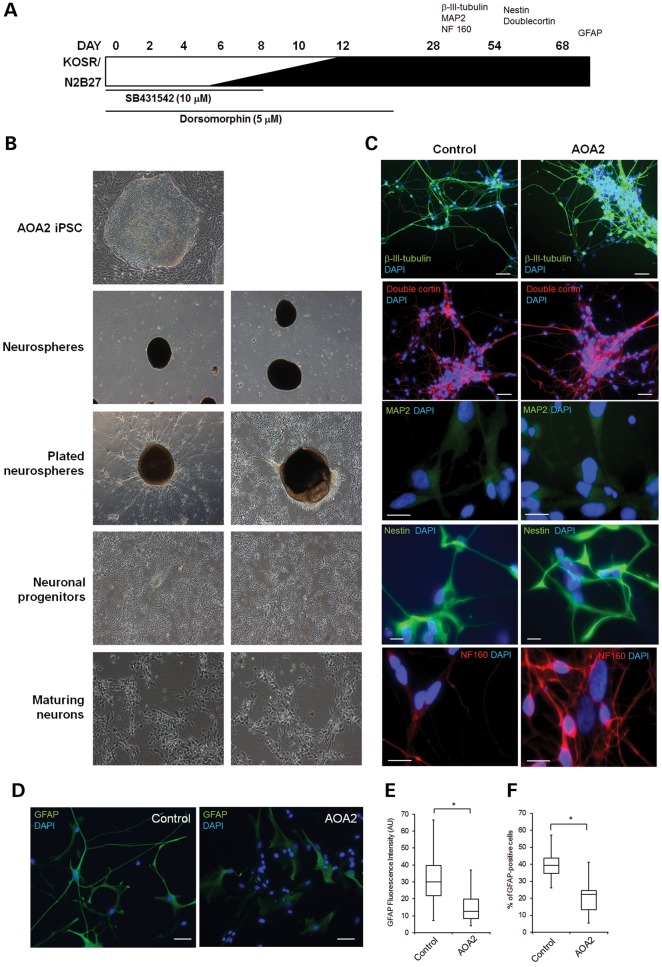
We next differentiated control and AOA2 iPSCs into neural progenitors using a modified version of the dual SMAD inhibition protocol (Fig. 3A) described previously (17). After 6 days, the neuralized cultures were detached and cultured as neurospheres (Fig. 3B). Following plating on Matrigel-coated surfaces, cells with neural projections migrated out of these neurospheres. Staining for neuronal makers β-III tubulin, MAP2, medium neurofilament (NF160), nestin and doublecortin confirmed the presence of neuronally restricted progenitors and differentiated post-mitotic neuronal cells from both control and AOA2 iPSCs (Fig. 3C). Immunoreactivity to glial fibrillary acid protein (GFAP) confirmed the presence of glial-restricted progenitors in both control and AOA2 iPSC cultures differentiated into the neural lineage (Fig. 3D). However, reductions in GFAP fluorescence intensity (Fig. 3E) and the number of GFAP-positive cells (Fig. 3F) were observed for AOA2, suggesting a delay and/or defect in differentiation into type 1 astrocytes compared with controls.
Figure 3.
Directed differentiation of AOA2 iPSCs into neural progenitors. (A) Schematic representation of neural induction protocol involving stepwise addition of N2B27 neurobasal medium and small molecules SB431542 and dorsomorphin for the first 6 and 12 days, respectively. Neurospheres were generated on day 6 of induction and plated after day 12, giving rise to colonies with neuronal projections and morphologies that were assessed for a range of neuronal makers, including β-III tubulin, MAP2, nestin, doublecortin, NF160 and GFAP. (B) Bright field images showing the differentiation of AOA2 iPSC into neural progenitors and maturing neurons. (C) Immunostaining for neuronal markers as indicated in the time line in (A). DAPI counterstained the nuclei. For β-III tubulin, doublecortin and nestin, scale bar represents 50 μm. For NF160 and MAP2, scale bar corresponds to 20 μm. (D) GFAP immunostaining of control and AOA2 neural progenitors. DAPI counterstained nuclei. Scale bar, 50 μm. (E) Reduced GFAP staining intensity in AOA2 compared with controls. More than 100 GFAP-immunoreactive cells for each population were used to quantify the average fluorescence intensity (AU) (*P < 0.05, Student's t-test). (F) Percentage of GFAP-positive cells was determined after counting manually more than 500 cells from each cell population (*P < 0.05, Student's t-test).
DNA damage, oxidative stress and R-loop accumulation in AOA2 neural progenitors
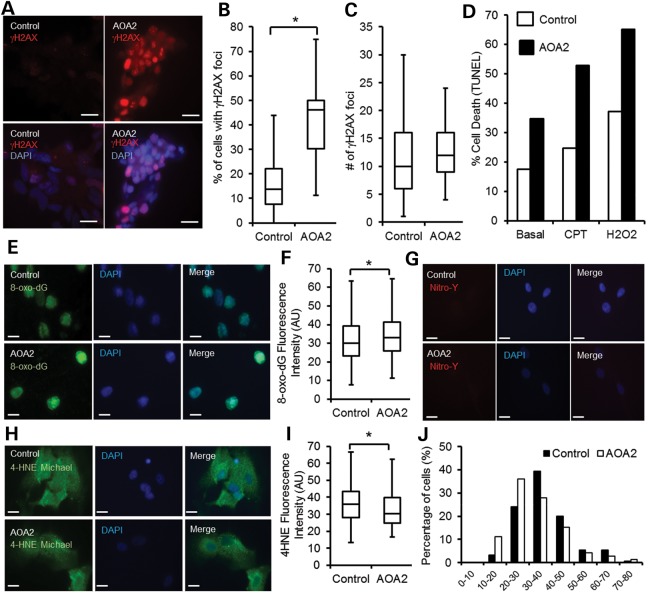
In order to further characterize AOA2 neural progenitors, we next assessed DNA damage, sensitivity to DNA-damaging agents, oxidative stress and accumulation of R-loops in these cells (3,9). Basal levels of DNA DSB were assessed using histone H2AX Ser 139 phosphorylation (γH2AX), a well-characterized marker for DNA DSB (18). Our data show that both control and AOA2 neural progenitors formed nuclear γH2AX foci under normal culture conditions, but that an increased number of γH2AX foci-containing cells was observed for AOA2 (Fig. 4A and B), suggesting a greater degree of genomic instability in the AOA2 neural progenitors population compared with the control one. The number of individual foci per cell did, however, not differ between control and AOA2 neural progenitors (Fig. 4C). We next investigated the sensitivity of AOA2 neural progenitors to DNA-damaging agents by assaying DNA damage-induced apoptosis (Fig. 4D). Interestingly, AOA2 neural progenitors showed an increased basal number of apoptotic cells compared with controls under normal culture conditions, and this was further enhanced after treatment with CPT, a topoisomerase I inhibitor, and hydrogen peroxide (Fig. 4D). These findings agree with the reported sensitivity of other AOA2 genetic models to DNA-damaging agents (3,19–22). In addition, similar to AOA2 iPSCs, AOA2 neural progenitors displayed increased levels of 8-oxo-dG compared with controls (Fig. 4E and F), with 27.46% of AOA2 cells with a fluorescence intensity >40 AU versus 22.36% for controls. Both protein oxidation (nitrotyrosine) (Fig. 4G) and lipid peroxidation (4-HNE Michael adducts) showed similar background levels in AOA2 and controls (Fig. 4H–J).
Figure 4.
DNA damage, oxidative stress and R-loops in AOA2 neural progenitors. (A) Control and AOA2 neural progenitors were immunostained for DNA damage using anti-γH2AX antibody. Scale bar, 20 μm. (B) Greater than 2-fold increase of γH2AX-positive cells in AOA2 neural progenitors compared with controls. About 200–400 cells were manually scored for the presence of γH2AX foci for control and AOA2 populations (*P < 0.05, Student's t-test). (C) Similar number of γH2AX foci per cell in both control and AOA2 neural progenitors (P = 0.34, Student's t-test). (D) DNA-damage-induced cell death after CPT (25 μm) and hydrogen peroxide (H2O2, 0.5 mm) treatments revealed a marked sensitivity of AOA2 neural progenitors to DNA-damaging agents and increased basal levels of apoptotic cells under normal growing conditions. (E) Immunostaining of control and AOA2 neural cultures with anti-8-oxo-dG antibody. DAPI stained nuclei. Scale bar, 20 µm. (F) A small but significant increase in oxidative stress as determined by 8-oxo-dG fluorescence intensity (AU) quantitation. More than 500 individual cells for each population were used for quantitation (*P < 0.05, Student's t-test). (G) Nitrotyrosine (Nitro-Y) staining of control and AOA2 neural progenitors. DAPI stained nuclei. Scale bar, 20 μm. (H) 4-HNE Michael adducts staining of control and AOA2 neural progenitors. DAPI stained nuclei. Scale bar, 20 μm. (I) 4-HNE fluorescence intensity quantitation for control and AOA2 neural progenitors (*P < 0.05, Student's t-test). (J) 4-HNE-Michael adducts fluorescence intensity distribution (x-axis) in control and AOA2 neural progenitors.
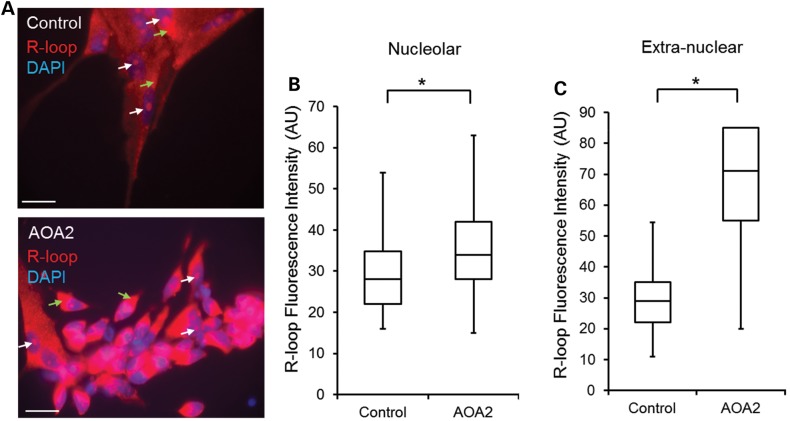
Similar to undifferentiated iPSCs, an increase in both nucleolar and extranuclear mitochondrial R-loop staining was observed in AOA2 neural progenitors (32.43% cells with a fluorescence intensity >40 AU) (Fig. 5A) when compared with control neural progenitor cultures (13.73% cells with a nucleolar R-loop fluorescence intensity >40 AU) (Fig. 5B). For extranuclear mitochondrial R-loops, 18.95% of the control cells had a fluorescence intensity >40 AU, whereas the vast majority of AOA2 (89.1%) had a fluorescence intensity >40 AU (Fig. 5C). Collectively, these data demonstrate that neurally differentiated AOA2 cells recapitulate the cellular features of current AOA2 genetic models as shown by increased cell sensitivity to CPT and hydrogen peroxide and increased oxidative DNA damage and provide evidence for a role for senataxin in resolving R-loops in neural progenitors.
Figure 5.
Accumulation of R-loops in AOA2 neural progenitors. (A) Detection of nucleolar and extranuclear R-loops in control and AOA2 neural progenitors using S9.6 antibody (14). DAPI stained nuclei. Scale, 20 µm. White arrows indicate nucleolar R-loops and green arrows indicate extranuclear R-loops. (B and C) Quantitation of nucleolar and extranuclear R-loop fluorescence intensity (AU) in neural progenitors (*P < 0.05, Student's t-test). A significant increase of R-loop formation in nucleoli and mitochondria was observed in AOA2 neural progenitors compared with controls (*P < 0.05, Student's t-test).
Gene expression profiling identifies AOA2-relevant pathways and co-expression gene networks
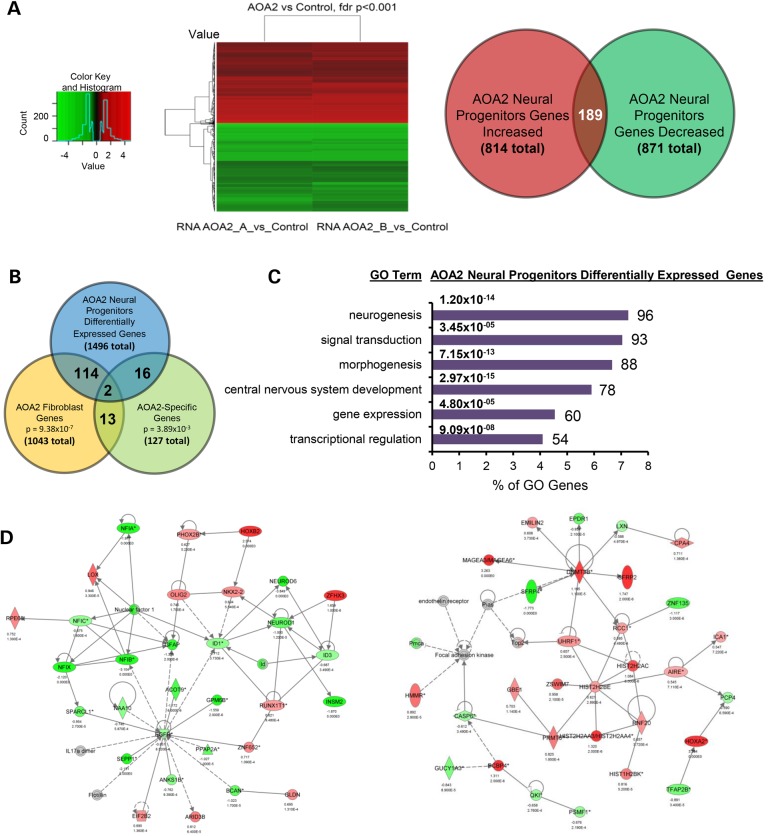
Mounting evidence indicates a role for senataxin in gene expression regulation (4,5,9,10,23). To further characterize our new AOA2 neural model, we used microarray analysis to assess genome-wide expression changes in AOA2 neural progenitors. Gene expression data were analyzed using (i) differential gene expression and pathway analysis using hierarchical clustering to identify individual genes and pathways relevant to AOA2 pathology and senataxin function, along with a systems approach, and (ii) weighted gene co-expression network analysis (WGCNA) to identify biologically meaningful and preserved AOA2-specific transcriptional networks. Overall, differential gene expression between the AOA2 cells and a control cell line derived from an unrelated individual identified 1707 probes (corresponding to 1496 unique genes) whose expression was significantly altered (false discovery rate 0.1%; Fig. 6A and Supplementary Material, File S1). A number of these genes were also differentially expressed in AOA2 fibroblasts derived from a separate AOA2 patient (116 genes, 7.75%, P = 9.38 × 10−7, hypergeometric test) (10) (Fig. 6B and Supplementary Material, File S1), and a marginally significant overlap was also seen with AOA2-specific genes identified in transfected fibroblast cell lines (18 genes, 1.2%, P = 3.89 × 10−3, hypergeometric test) (10) (Fig. 6B and Supplementary Material, File S1). Gene ontology (GO) analysis showed overrepresentation of pathways functionally relevant to the AOA2 neurological phenotype such as neurogenesis, central nervous system development, morphogenesis, signal transduction, gene expression and transcriptional regulation (Fig. 6C). Further analysis using ingenuity pathway analysis (IPA) identified multiple gene sets related to nervous system development and function, neurological and psychological disease, regulation of gene expression and reproductive system disease. Examples of two key pathways related to central nervous system development and function are shown (Fig. 6D and Supplementary Material, File S2). Microarray results were validated by quantitative PCR (Supplementary Material, Fig. S1).
Figure 6.
Identification of functionally relevant biological and molecular pathways related to senataxin function and disease in AOA2 neural progenitors. (A) Heat map for hierarchical clustering of differential gene expression in neural progenitors. Comparison of AOA2 versus control (P < 0.001, adjusted for false discovery rate) and number of significant genes up- and downregulated in AOA2 neural progenitors. (B) Overlap of the genes differentially expressed in neural progenitors with those previously published in AOA2 patient fibroblasts and fibroblasts transfected with mutant SETX (AOA2-specific genes) (10). (C) GO analysis of the AOA2 neural progenitor differentially expressed genes using DAVID (see Materials and Methods). (D) Examples of IPA-derived networks related to the central nervous system development and function and cellular growth and proliferation based on differential gene expression.
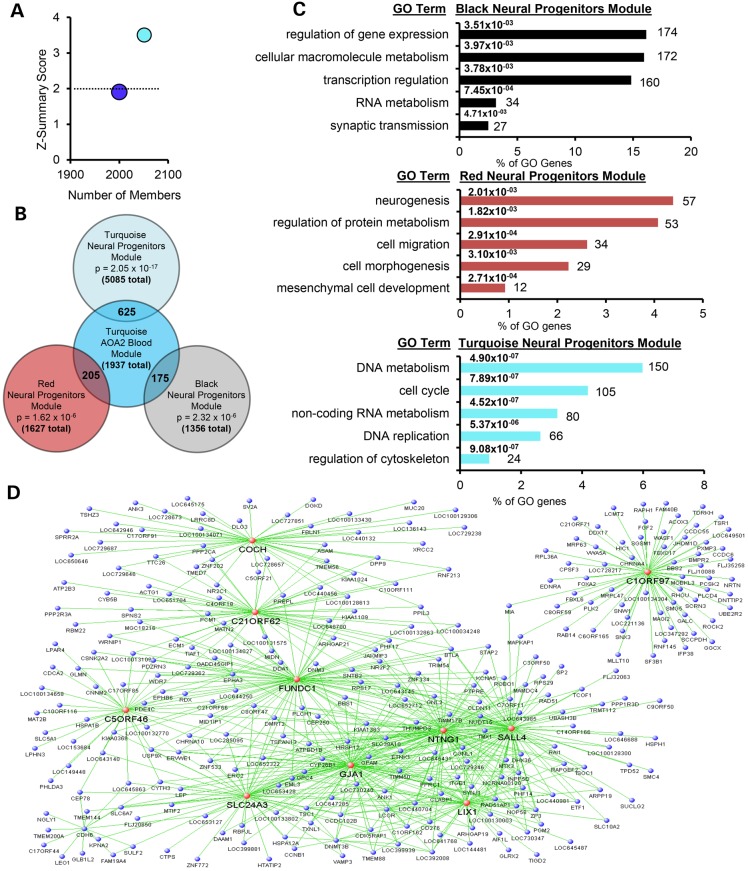
Network analysis is commonly used in systems biology to decipher correlation patterns among genes and to identify related gene sets (or modules) that reflect biologically meaningful relationships to investigate affected molecular pathways, select candidate biomarkers or identify potential therapeutic targets (24,25). WGCNA has been widely applied in various biological contexts of neurological diseases (26–29). Recently, a comprehensive genome-wide expression study and WGCNA using peripheral blood cells of patients from a cohort of 12 AOA2 families revealed that mutation of senataxin alters disease-specific transcriptional networks (10), identifying an AOA2-specific transcriptional signature thought to underlie the AOA2 phenotype (10). A key gene module (Turquoise_AOA2_blood) was found to be related to SETX function in multiple cell types derived from AOA2 patients and across species, including Setx mouse cerebellar neurons (10). To determine whether our AOA2 neural model recapitulated these findings from AOA2 patients, we performed a limited WGCNA. Consistent with findings in other AOA2 and Setx-deficient tissues, the SETX functional module (Turquoise_AOA2_blood) (10) was found to also be preserved in the AOA2 neural progenitors (Fig. 7A). Although it was not possible to generate and interpret a complete WGCNA in this data set due to the small sample size, eight modules were identified that correlated with the disease status and three of them (black, red and turquoise) showed significant overlap with the Turquoise_AOA2_blood SETX functional module (10) (Fig. 7B and Supplementary Material, File S3). GO indicated that these modules were associated with RNA metabolism and transcription regulation, central nervous system development and function and neurogenesis (Fig. 7C and Supplementary Material, File S3). The Turquoise module identified by WGCNA in the neural progenitors, which correlates most closely with the Turquoise_AOA2_blood SETX functional module in patients, is shown in Figure 7D. Highly connected hubs from the black, red and turquoise modules relevant to AOA2 and senataxin function are listed in Table 1 (a complete list of key gene features is listed in Supplementary Material, File S4). Taken together, these findings support our AOA2 neural model as reflecting the key molecular genetic events seen in the cells of AOA2 patients (10).
Figure 7.
WGCNA in AOA2 neural progenitors shows preservation of SETX functional networks and pathways from AOA2 patient blood. (A) Preservation of the key WGCNA SETX modules from AOA2 patient peripheral blood (10) in the AOA2 neural progenitors. Results are plotted by number of module members versus preservation score (Z-summary) calculated using the preservation module function of the WGCNA software package. Z-summary score between 2 and 10 shows moderate preservation and scores less than 2 (dotted line) show weak or no preservation. Moderate preservation of the Turquoise_AOA2_blood module (representing SETX function) from AOA2 patient peripheral blood was observed in the AOA2 neural progenitors. The Blue_AOA2_blood module, representing the AOA2 transcriptional signature, is also weakly preserved. (B) Overlap of the genes in the key AOA2 neural progenitor modules (black, red and turquoise) with those in the Turquoise_AOA2_blood module is shown. (C) GO analysis of the AOA2 neural progenitors black, red and turquoise WGNCA modules. A list of key genes relevant to AOA2 and senataxin function from each module is shown in Supplementary Material, File S5. (D) The turquoise module identified by WGCNA in the AOA2 neural progenitors, which most closely correlates with the Turquoise_AOA2_blood module, is shown. For clarity, only the most highly connected module members are shown. Genes with the highest connectivity (i.e. hubs) are indicated in red.
Table 1.
Module hubs relevant to AOA2 and senataxin function
| Module | Gene | Protein | Function |
|---|---|---|---|
| Black | MIR513A2 | microRNA 513a-2 | Co-regulator of reproductive hormones |
| Black | PPP1R1B | Protein phosphatase 1 | Synapse function |
| Black | DUSP4 | Dual specificity phosphatase 4 | Cellular proliferation and differentiation |
| Black | NMNAT3 | Nicotinamide nucleotide adenylyltransferase 3 | Formation of NAD+ and axonal protection |
| Black | BAIAP2 | BAI1-associated protein 2 | Nerve growth-cone guidance |
| Black | C1ORF151 | Mitochondrial inner membrane organizing system | Mitochondrial morphogenesis |
| Black | BLVRB | Biliverdin reductase B | Heme catabolism |
| Black | LOC653344 | Uncharacterized protein | Uncharacterized protein |
| Black | ZNF177 | Zinc finger protein 177 | DNA transcription regulation |
| Red | SLN | Sarcolipin | Uncharacterized protein |
| Red | LOC646576 | HHIP antisense RNA 1 | Non-coding RNA gene |
| Red | MARVELD3 | MARVEL domain containing 3 | Transmembrane tight junction |
| Red | SPINT1 | Serine peptidase inhibitor, Kunitz type 1 | Neural progenitor cell proliferation |
| Red | RIT1 | Ras-like without CAAX 1 | Regulation of neuronal differentiation |
| Red | ACVR1 | Activin A receptor, type I | Receptor for bone morphogenetic protein |
| Red | EIF4E3 | Eukaryotic translation initiation factor 4E | Initiation and regulation of translation |
| Turquoise | COCH | Cochlin | Unknown function expressed in inner ear |
| Turquoise | C21ORF62 | Chromosome 21 open reading frame 62 | Uncharacterized protein |
| Turquoise | C1ORF97 | Long intergenic non-protein coding RNA 467 | Enhances neuroblastoma cell survival |
| Turquoise | FUNDC1 | FUN14 domain containing 1 | Hypoxia response in mitochondria |
| Turquoise | C5ORF46 | Chromosome 5 open reading frame 46 | Uncharacterized extracellular protein |
| Turquoise | SLC24A3 | Solute carrier family 24, member 3 | K-dependent Na/Ca ion transporter |
| Turquoise | GJA1 | Gap junction protein, alpha 1, 43 kDa | Ion transporter |
| Turquoise | NTNG1 | Netrin G1 | Neuronal circuit formation |
| Turquoise | SALL4 | Spalt-like transcription factor 4 | Transcription regulator |
| Turquoise | LIX1 | Limb expression 1 | RNA metabolism (predicted) |
Discussion
AOA2 is one of a small number of autosomal recessive ataxias in which the gene product involved plays a role in protecting the integrity of the genome. The accumulation of R-loops in AOA2 cells, Setx−/− mouse mutants and cells in which senataxin is knocked down is consistent with senataxin having helicase activity (5,6,9,11,23). All these studies have been carried out in actively replicating cells in which such structures can collide with replication forks or cause genome instability by inducing recombination events if left unresolved (7).
As an approach to employing cells more relevant to the neurological phenotype in AOA2, we have shown here for the first time that fibroblasts from patients with AOA2 can be reprogrammed to pluripotency and differentiated into neural progenitors. As with other AOA2 cells, there was evidence of increased cell death following hydrogen peroxide treatment, elevated levels of 8-oxo-dG in the AOA2 iPSC supporting the presence of oxidative damage. Despite the increase in 8-oxo-dG levels in AOA2 compared with controls being small, it is a significant feature found in other AOA2 genetic models (Table 2). However, this was not a generalized oxidative stress as there was no evidence of oxidative damage to lipids or proteins. Perhaps not unexpectedly, there was accumulation of R-loops in AOA2 iPSCs in keeping with loss of senataxin activity (5,6,9,11), indicating that iPSCs behave like other proliferating cells in this respect (Table 2).
Table 2.
Cellular and molecular features of AOA2 genetic models
| AOA2 model | Sensitivity to DNA-damaging agents | Genomic instability | Oxidative DNA damage | Presence of R-loopsa | References |
|---|---|---|---|---|---|
| AOA2-derived iPSC | ND | ND | Increased levels of 8-oxo-dG | Elevated levels of R-loops with and without CPT treatment | This study |
| AOA2-derived neural progenitors | H2O2, CPT | Increased number of γH2AX foci per cell | Increased levels of 8-oxo-dG | Elevated basal levels of R-loops | This study |
| SETX RNAi knockdown cells | ND | ND | Increased levels of 8-oxo-dG | ND | Suraweera et al. (4) |
| ND | Elevated basal levels of R-loops at the β-actin locus | Skourti-Stathaki et al. (5) | |||
| Elevated levels of R-loops on induction with CPT | Yeo et al. (11) | ||||
| Elevated basal levels of R-loops at the β-actin locus | Hatchi et al. (59) | ||||
| AOA2 lymphoblastoid cells | H2O2, MMC, CPT | H2O2-induced DSB repair defect | ND | ND | Suraweera et al. (3) |
| ND | ND | ND | Airoldi et al. (20) Vantaggiato et al. (60) |
||
| AOA2 fibroblasts | DSB repair defect after H2O2 treatment | Increased levels of 8-oxo-dG | ND | Suraweera et al. (3) | |
| ND | ND | ND | Roda et al. (22) | ||
| Setx−/− mouse | ND | Meiotic DSB repair defect | ND | Elevated levels of R-loops in spermatocytes | Becherel et al. (9) |
| ND | Increased levels of 8-oxo-dG in Setx−/− MEFs | Elevated levels of R-loops in the nucleolus of Setx−/− MEFs | Becherel (unpublished data) | ||
| TPT (analog of CPT) | ND | ND | Elevated levels of R-loops in proliferating cells Elevated levels of R-loops at XY genes in germ cells |
Yeo et al. (11) | |
| ND | ND | Increased levels of 8-oxo-dG in testes of aged male adults | Elevated levels of R-loops at XY genes in germ cells | Yeo (unpublished data) |
aS9.6 antibody employed to detect R-loops. Common features across all AOA2 genetic models are shaded.
Differentiation of AOA2 iPSCs into neuron-restricted progenitors expressing doublecortin, MAP2, nestin, β-III tubulin and immunoreactivity to NF160 indicated the presence of mature post-mitotic neuronal cells in each population. Although both control and AOA2 neural progenitors expressed GFAP, a reduction in GFAP expression and the proportion of GFAP-expressing cells was observed for AOA2, suggesting a delay and/or impairment to differentiate into mature astrocytes. Neural progenitors showed evidence of oxidative stress and DNA damage and R-loop accumulation, indicating that they behaved very much like proliferating AOA2 cells. Although R-loops are readily detected in proliferating cells at sites of collision with replication forks or during meiotic recombination (5,6,9), it has not been possible to detect these in the brain or cerebellum of Setx−/− mice (11,30). The presence of R-loops in neural progenitors is mostly visible in nucleoli which represent sites of active rRNA transcription and in extranuclear sites corresponding to the mitochondria. Loss of DNA topoisomerase I activity or its inhibition leads to stabilization of hybrid structures and R-loop-mediated transcriptional block during ribosomal RNA synthesis (15,16). This is confirmed here as both control and AOA2 iPSC exhibited increased levels of nucleolar R-loops after CPT treatment. R-loops in the mitochondria have been previously described and result from mitochondrial DNA (mtDNA) replication at the leading-strand origin, which is coupled to transcription through the formation of an RNA/DNA hybrid (31). The presence of high levels of R-loops in the mitochondria of AOA2 neural progenitors could affect mtDNA integrity. mtDNA damage is found in affected neurons in every major neurodegenerative disorder and is associated with increased reactive oxygen species production, mitochondrial dysfunction and dysregulation of cellular calcium homeostasis (32). Further studies will be required to investigate mitochondrial function in these cells.
It is not clear what role senataxin plays in the nervous system, but our data with neural progenitors show that in its absence neural progenitors undergo markedly increased cell death under normal culture conditions, are sensitive to DNA-damaging agents and exhibit an accumulation of oxidative DNA damage, DNA DSB and R-loops. Although a direct link between the regulation of R-loops and disease etiology remains to be established, possible scenarios include the following: (i) post-mitotic neurons are sensitive to low levels of R-loops; (ii) unscheduled accumulation of R-loops impacts the functionality of glial cells, a population of cycling cells that interacts with neurons and whose dysfunction contributes to neurodegeneration (33); (iii) R-loop formation in other tissues could indirectly impact neuron function and (iv) uncontrolled R-loop accumulation during neurogenesis could have long-term effects on genome integrity and gene expression in nature neurons. We and others have provided evidence for a broader role for senataxin in transcription and other cellular processes, which may or may not be related to R-loop resolution (4,5,10,19,23). Indeed, we showed that senataxin attenuates the activity of RNA polymerase II at genes stimulated after viral infection and thus controls the magnitude of the host response to pathogens and the biogenesis of various RNA viruses, suggesting that AOA2 patients with defective senataxin may exhibit heightened susceptibility to infection (23). Recent findings suggest that DNA:RNA hybrids containing viral- or bacterial-derived sequences can stimulate the innate immune system response (34–36), which, if uncontrolled, could trigger chronic inflammation and have implications on the onset of neurodegenerative disorders. Further investigation will be required to determine whether the increased cell death observed in AOA2 neural progenitors under normal culture conditions and their increased sensitivity to DNA-damaging agents may be related to innate immune system-mediated inflammation.
A role for senataxin in neuronal differentiation through fibroblast growth factor 8 (FGF8) signaling has been reported (19). FGF8 is one of several growth factors that regulate neurogenesis, neuronal differentiation, survival and synaptic plasticity, both during development and in adulthood. Our data do not support a general defect in differentiation as AOA2 iPSC showed much the same capacity to differentiate into neural progenitors as control iPSC. Thus, it is possible that the defect is more subtle and affects more specific aspects of differentiation, as reported previously (19). Indeed, we observed reductions in the number of GFAP-expressing cells and a downregulation of transcription factors NFIA and NFIX and of GFAP in AOA2 neural progenitors, suggesting a possible defect in the differentiation of glial-restricted progenitors into mature astrocytes.
Biological and molecular pathway analysis from AOA2 neural progenitor gene expression data revealed that key transcription factors of the nuclear factor 1 (NFI) family involved in axon guidance and outgrowth, neuronal migration and glial and neuronal differentiation (37) were downregulated in AOA2 neural progenitors. The downregulation of NeuroD6, a transcription factor that confers tolerance to oxidative stress by triggering an antioxidant response and sustaining mitochondrial biomass (38), is in agreement with the elevated levels of oxidative DNA damage and the sensitivity to H2O2 observed in AOA2 neural progenitors. Genes involved in chromatin modification/histones methylation (DMNT3B, UHRF1 and PRMT6) (39), chromatin condensation (TOP2) (40) and RNA processing (QKI) (41) were upregulated in the AOA2 neural progenitors. DNMT3B mediates large-scale methylation patterns in hESCs and deficiency alters the timing of neuronal differentiation and maturation (42). UHRF1 has been implicated in DNA methylation and histone modification and likely coordinates a macromolecular complex able to serially catalyze histone deacetylation (via HDCA1), histone methylation (via G9) and DNA methylation (via DMNT1), thus playing a key role in the epigenetic code inheritance (43). The relevance of this gene network to senataxin function is supported by recent evidence for an essential role for senataxin in gene silencing and MSCI (9) (44). Finally, QKI encodes a member of the highly conserved STAR/GSG family of RNA-binding proteins expressed in neuronal progenitors and mature glial cells that functions as master developmental regulators of glial cell fate (45). In addition, highly connected hubs from black, red and turquoise modules in AOA2 neural progenitors included NMNAT3, PPP1R1B and BAIAP2, which have been implicated in NAD synthesis and neuroprotection (46), dopamine metabolism (47) and nerve growth-cone guidance (48). RIT1 encodes a member of a subfamily of Ras-related GTPases involved in regulating p38 MAPK-dependent signaling cascades related to cellular stress and cooperates with nerve growth factor to promote neuronal development and regeneration (49). Finally, NTNG1 from the turquoise module, and also seen in the overlap with data from another AOA2 patient (10) (Supplementary Material, File S3), belongs to a conserved family of proteins that act as axon guidance cues during vertebrate nervous system development, and mutations in NTNG1 have been associated with schizophrenia (50). Detailed functional characterization of the molecular pathways relevant to AOA2 neuropathology identified here will require further investigation.
Mutation of senataxin has been previously reported to alter gene expression in a disease-specific manner in cells derived from patients with AOA2 and in Setx−/− mice, and these changes likely underlie the phenotypic difference between AOA2 and ALS4, a dominant motor neuron disease associated with mutations in SETX, distinct from those that cause AOA2 (10). Genome-wide transcriptome analysis from AOA2 neural progenitors showed wide-spread dysregulation of gene expression with some overlap with what was previously described in AOA2 fibroblasts and fibroblast cell lines transfected with mutant SETX (10), consistent with the derivation of these cells from fibroblasts. GO reflected differential expression in genes associated with the functions ascribed to senataxin and reported previously (10). WGCNA showed preservation of the previously identified genetic network reflecting senataxin function in the AOA2 neural progenitors, with significant overlap between this network and neural progenitor network modules corresponding to AOA2 disease status, reflecting SETX functionality, and enriched for differentially expressed genes (Supplementary Material, File S3) (10). It is important to note that although a fraction of the observed changes could result from genetic and/or technical differences due to small sample size and comparison between the AOA2 and unrelated control cell lines, the overall findings in these neuronal cells are consistent with the gene dysregulation previously observed in AOA2 and in Setx−/− cerebellar neurons (10). Interestingly, a previous WGCNA of transcriptional responses to type I interferons identified senataxin in one of the two representative modules of IFN response (51), a relationship recently confirmed by Miller et al. (23). They showed that senataxin suppresses the expression of early infection-induced IFN-β and other antiviral mediators important for the resolution of infection by binding to TAF4, which acts as a binding platform for the recruitment of positive and negative effectors of transcriptional activity (23). In addition, a recent study showed that DNA damage can induce the interferon I response through the transcription factor IRF3 and that this factor localized to sites of DNA DSB (52). Given that cells from AOA2 patients and Setx−/− mice are hyperresponsive to infection, this suggests that chronic inflammation may play a role in AOA2 pathophysiology. The development of this new AOA2 neural model will enable us to test this hypothesis and to ascertain whether chronic inflammation could be one of the triggers of neurodegeneration in AOA2.
Several mouse models have been engineered to study a number of human DNA repair disorders causing neurodegeneration, and so far none has been able to recapitulate the neurodegenerative phenotypes observed in these diseases (53,54), highlighting the difficulty in mimicking progressive DNA damage-induced neurodegeneration in the mouse and therefore calling for the development of more suitable models to study the neuropathology of these disorders. The development of AOA2 iPSC and neural progenitors has confirmed previous gene dysregulation in AOA2 (10) and identified novel AOA2 and SETX functionally relevant gene networks supporting the cellular role of senataxin in the genome-wide regulation of gene expression, RNA processing, chromatin/histone modification and modifying key pathways involved in neurogenesis, differentiation and central nervous system development and function. In spite of limitations regarding the number of control and AOA2 patient samples available for iPSC reprogramming, this study represents an initial characterization of the first AOA2-derived iPSC and neural progenitors and demonstrates the feasibility of reprogramming AOA2 cells that recapitulate the cellular and molecular phenotype. Recent knowledge on the mechanism of cerebellar differentiation has promoted technical advances for in vitro generation of cerebellar neurons from pluripotent stem cells (55). Indeed, Muguruma et al. (12) recently reported the generation of polarized cerebellar tissue in 3D culture of human pluripotent stem cells in which the self-organized neuroepithelium differentiated into functional Purkinje cells. We are now in an excellent position to address cerebellar development and neurodegeneration with the availability of AOA2 iPSC and neural progenitors as they constitute an appropriate model system to investigate the role of senataxin in the nervous system, as shown here. The recent implication of senataxin in the innate antiviral response represents a new avenue of investigation for the role of chronic inflammation in AOA2. Using this resource, we are now set to provide insight in the etiology of a complex human neurological disease that to date has remained a mystery.
Materials and Methods
Generation of iPSC
Primary fibroblasts were established from dermal punch biopsies from a female patient with ataxia oculomotor apraxia type 2 (missense homozygous c.6109A>G; protein amino acid change: p.Asn2037Asp) and control subjects. Fibroblasts were transfected with the oriP/EBNA1-based pCEP4 episomal vectors pEP4EO2SCK2MEN2L and pEP4EO2SET2K from Addgene (Cambridge, MA, USA) and after 2 days of culture in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 0.1 mm non-essential amino acids and 2.0 mm Glutamax, 50 000 fibroblasts were seeded onto mouse embryonic fibroblasts (MEF) feeder plates (36k/cm2) and adapted to KOSR medium containing 100 ng/ml bFGF (all from Life Technologies, Grand Island, NY, USA) over a course of 4 days at 25% per day (56). After the first week of reprogramming, daily medium changes were performed with MEF-conditioned KOSR medium with 100 ng/ml bFGF. After 4–5 weeks, small colonies with a hESC-like morphology emerged. Live staining with TRA-1-60 was performed to identify and manually pick 12–20 putative iPSC colonies that were transferred to organ culture dishes with 36 k/cm2 MEFs in KOSR medium with 100 ng/ml bFGF. AOA2 iPSC cultures were subsequently further expanded using enzymatic passaging as described previously and characterized (56,57). All work was carried out with informed consent from patients under the approval of the Human Research Ethics Committee (HREC/09/QRCH/103).
Cell culture conditions
Human iPS cells were maintained at 37°C at 5% CO2 at high humidity on MEFs feeder layers in DMEM/F12 culture medium supplemented with 20% KOSR, 0.1 mm non-essential amino acids, 1 mm l-glutamine, 0.1 mm β-mercaptoethanol and 100 ng/ml human bFGF (all from Life Technologies), as described previously (57). For experimentation, cells were cultured under feeder-free conditions on Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) in MEF-conditioned hESC media. Cells were passaged either by manual cutting or by treatment with Collagenase IV (Life Technologies) before replating at a seeding ratio of between 1:2 and 1:6. hESC medium was replaced daily and cells were split at ∼80% confluence on days 6–7.
AOA2 mutation confirmation in iPSC and neural progenitors
The presence of the AOA2 homozygous c.6109 A>G mutation was confirmed in the AOA2 iPS lines and the iPSC-derived neural progenitors at the mRNA level by sequencing of the cDNA. Sequencing of the corresponding controls revealed the wild-type sequence at mRNA coding position 6109. Briefly, total RNA was isolated from controls and AOA2 fibroblasts, iPSCs and iPSC-derived neural progenitors using the RNeasy mini kit (Qiagen, USA), according to the manufacturer's protocol. RNA concentrations were determined by UV spectrophotometry using a Nanodrop ND-2000 (Thermo Scientific, USA), and cDNA was made from 1 μg of purified RNA using SuperScript III (Life Technologies, USA), according to the manufacturer's protocol. Sequencing was performed using the BigDye 3.1 Terminator sequencing protocol in the targeted region using the following primer 5′-AGAGGAAGGGGATTCAGACG-3′.
Teratoma formation
iPSCs grown on MEFs were collected by Collagenase IV treatment and injected into hind limb muscles of 6-week-old immune-compromised SCID mice (approximately one well of a six-well plate with 50–80% confluency per mouse). After 8–10 weeks, teratomas were dissected and fixed in 4% paraformaldehyde and examined for the presence of representatives of the three germ layers by an independent pathologist.
Neural differentiation
Undifferentiated iPSC colonies grown under feeder-free conditions were introduced into KOSR medium supplemented with 10 µm SB431542 (Sigma-Aldrich, Sydney, NSW, Australia) and 5 µm dorsomorphin (Stemgent, Cambridge, MA, USA) for 6 and 12 days, respectively. Medium was changed every 2 days. N2B27 media gradually replaced the KOSR at a rate of 25% per feed on days 4, 6, 8 and 10, respectively. On day 6, neurospheres were generated by incubation with 1 mg/ml Collagenase IV (Life Technologies) at 37°C. Neuralized fragments were seeded into Ultra-Low Costar Cluster plates (Sigma-Aldrich). On day 12, the neurospheres were transferred to Matrigel-coated plates in N2B27 media where they attached and neural outgrowth proceeded from the border of these spheres.
Immunostaining and fluorescence quantitation
iPSC and neural progenitors were seeded onto Matrigel-coated glass cover slips or chamber slides from Millipore (Billerica, MA, USA), and immunostaining was performed using TRA-1-60 (1:250), Nanog (1:50), TRA-1-81 (1:250), 4-HNE-Michael Adduct (1:100) and γH2AX (1:1000) from Millipore; Oct 4 (1:50) from Santa Cruz Biotechnology (Dallas, TX, USA); anti-RNA/DNA hybrids (S9.6) (14); nitrotyrosine (1:100), nucleophosmin (1:100) and doublecortin (1:300) from Cell Signalling (Danvers, MA, USA); β-III tubulin (1:1000) from Covance (San Diego, CA, USA); nestin (1:100) and NF160 (1:200) from Abcam (Cambridge, MA, USA); GFAP (1:100) from DAKO (Agilent Technologies, Glostrup, DK, Denmark) and MAP2 (1:100) from Sigma-Aldrich antibodies followed by Alexa Dye-488- or Alexa Dye-594-conjugated secondary antibody (1:250; Life Technologies). Briefly, iPSC and neural progenitors were fixed for 15 min with 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) and permeabilized with PBT (0.5% Triton X-100/PBS) for 10 min. All procedures were carried out at room temperature except when specified. Non-specific sites were blocked using PBFB (10% FBS/2% bovine serum albumin /PBS) for 1 h at room temperature. Immunostaining was performed by incubating the cells with the relevant primary antibody overnight at 4°C in a humidified chamber. After primary antibody incubation, cells were then washed thrice with PBT for 5 min each and then incubated with the appropriate Alexa Dye-488- or Alexa Dye-594-conjugated secondary antibody for 1 h at 37°C in a humidified chamber. Cells were then washed thrice with PBT for 5 min each, rinsed with 1× PBS and nuclei were counterstained with DAPI (1:10 000; Sigma-Aldrich), washed twice with 1× PBS and cover slips were mounted using ProLong Gold Antifade reagent (Life Technologies). Immunostaining for 8-oxo-dG was performed as described previously (3). Images were captured at room temperature using a digital camera (AxioCam Mrm, Carl Zeiss MicroImaging, Germany) attached to a fluorescent microscope (Axioskop 2 mot plus, Carl Zeiss MicroImaging) and the AxioVision 4.8 software (Carl Zeiss MicroImaging). The objective employed was a 63× Zeiss Plan Apochromat 1.4 Oil DIC (Carl Zeiss MicroImaging) or a Zeiss Plan Neofluar ×10/0.30. Images were subsequently assembled in Adobe Photoshop 7 (San Jose, CA, USA), and contrast and brightness were adjusted on the whole image panel at the same time. Fluorescence intensity was quantitated on the RAW images using NIH ImageJ software version 1.42 (Bethesda, MD, USA). Statistical analysis was carried out using one-way analysis of variance (ANOVA) or Student's t-test.
DNA damage-induced cell death
Control and AOA2 neural progenitors were grown on Matrigel-coated Lab-Tek II chamber slide (Thermo Fisher Scientific Inc., Waltham, MA, USA). Cells were then treated or not with CPT (25 μm; Sigma-Aldrich) in culture media for 2 h at 37°C/5% CO2 and hydrogen peroxide (0.5 mm; Sigma-Aldrich) in culture media for 30 min at 37°C/5% CO2. After the treatment period, drug-containing media were removed, and cells were washed with fresh media and further incubated at 37°C/5% CO2. At 8 h post-DNA damage exposure, cells were washed with 1× PBS and fixed with 4% PFA for 15 min at room temperature and apoptotic cells were detected using In Situ Cell Death Detection Kit (Roche, Indianapolis, IN, USA), following the manufacturer's instructions. Nuclei were counterstained with DAPI (Sigma-Aldrich) and images were captured as described earlier. The ratio of TUNEL-positive cells over the total number of cells was used to calculate the percentage of cell death for each condition.
Gene expression and pathway analysis
Total RNA samples from control and AOA2 neural progenitors were extracted using RNeasy Kit (Qiagen, Dusseldorf, Germany) and analyzed using HumanHT-12 v4.0 Beadchip expression arrays according to the manufacturer's instruction (Illumina, San Diego, CA, USA). Data were filtered for batch effects, and biological replicate counts were pooled for gene expression analysis. Statistical comparisons for differential gene expression were made using the limma software package (58), and the false discovery rate was set at 0.1%. Validation was performed on a set of genes using quantitative RT (qRT)–PCR following standard procedures with Power SYBR Green PCR Master Mix (Life Technologies) and StepOnePlus thermocycler (Applied Biosystems, Foster City, CA, USA). Primer pairs used for qRT–PCR validation are indicated in Supplementary Material, Table S1. GOs were determined using the DAVID Bioinformatic resources (NIAID, NIH). Statistical comparisons to other gene lists were made using hypergeometric probability. Genetic network and pathway analysis were performed using WGCNA as described previously (10) and QIAGEN's IPA (Qiagen, Redwood City, CA, USA). All microarray data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Australian Research Council (ARC) and the ARC Centre of Excellence ‘Stem Cells Australia’, the National Health and Medical Research Council (NHMRC; GNT 0631407) and the Australian Research Council (ARC; DP 130100389). B.L.F. was supported by the National Institute for Neurological Disorders and Stroke (R01NS082094) and we thank Bryan White and Xizhe Wang for technical assistance. We also acknowledge the support of NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691to G.C.).
Supplementary Material
References
- 1.Moreira M.C., Klur S., Watanabe M., Nemeth A.H., Le Ber I., Moniz J.C., Tranchant C., Aubourg P., Tazir M., Schols L. et al. (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet., 36, 225–227. [DOI] [PubMed] [Google Scholar]
- 2.Anheim M., Monga B., Fleury M., Charles P., Barbot C., Salih M., Delaunoy J.P., Fritsch M., Arning L., Synofzik M. et al. (2009) Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain, 132, 2688–2698. [DOI] [PubMed] [Google Scholar]
- 3.Suraweera A., Becherel O.J., Chen P., Rundle N., Woods R., Nakamura J., Gatei M., Criscuolo C., Filla A., Chessa L. et al. (2007) Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J. Cell Biol., 177, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suraweera A., Lim Y., Woods R., Birrell G.W., Nasim T., Becherel O.J., Lavin M.F. (2009) Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet., 18, 3384–3396. [DOI] [PubMed] [Google Scholar]
- 5.Skourti-Stathaki K., Proudfoot N.J., Gromak N. (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell, 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuce O., West S.C. (2013) Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol., 33, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groh M., Gromak N. (2014) Out of balance: R-loops in human disease. PLoS Genet., 10, e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richard P., Feng S., Manley J.L. (2013) A SUMO-dependent interaction between senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev., 27, 2227–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becherel O.J., Yeo A.J., Stellati A., Heng E.Y., Luff J., Suraweera A.M., Woods R., Fleming J., Carrie D., McKinney K. et al. (2013) Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet., 9, e1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogel B.L., Cho E., Wahnich A., Gao F., Becherel O.J., Wang X., Fike F., Chen L., Criscuolo C., De Michele G. et al. (2014) Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum. Mol. Genet., 23, 4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo A.J., Becherel O.J., Luff J.E., Cullen J.K., Wongsurawat T., Jenjaroenpoon P., Kuznetsov V.A., McKinnon P.J., Lavin M.F. (2014) R-loops in proliferating cells but not in the brain: implications for AOA2 and other autosomal recessive ataxias. PloS ONE, 9, e90219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muguruma K., Nishiyama A., Kawakami H., Hashimoto K., Sasai Y. (2015) Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep., 10, 537–550. [DOI] [PubMed] [Google Scholar]
- 13.Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A. et al. (2009) Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol., 27, 1033–1037. [DOI] [PubMed] [Google Scholar]
- 14.Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J. (1986) Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Meth., 89, 123–130. [DOI] [PubMed] [Google Scholar]
- 15.El Hage A., French S.L., Beyer A.L., Tollervey D. (2010) Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev., 24, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marinello J., Chillemi G., Bueno S., Manzo S.G., Capranico G. (2013) Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I–DNA cleavage complex and R-loop formation. Nucleic Acids Res., 41, 10110–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayler S., Gatei M., Kozlov S., Gatti R., Mar J.C., Wells C.A., Lavin M., Wolvetang E. (2012) Induced pluripotent stem cells from ataxia-telangiectasia recapitulate the cellular phenotype. Stem Cells Trans. Med., 1, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem., 273, 5858–5868. [DOI] [PubMed] [Google Scholar]
- 19.Vantaggiato C., Bondioni S., Airoldi G., Bozzato A., Borsani G., Rugarli E.I., Bresolin N., Clementi E., Bassi M.T. (2011) Senataxin modulates neurite growth through fibroblast growth factor 8 signalling. Brain, 134, 1808–1828. [DOI] [PubMed] [Google Scholar]
- 20.Airoldi G., Guidarelli A., Cantoni O., Panzeri C., Vantaggiato C., Bonato S., Grazia D'Angelo M., Falcone S., De Palma C., Tonelli A. et al. (2010) Characterization of two novel SETX mutations in AOA2 patients reveals aspects of the pathophysiological role of senataxin. Neurogenetics, 11, 91–100. [DOI] [PubMed] [Google Scholar]
- 21.De Amicis A., Piane M., Ferrari F., Fanciulli M., Delia D., Chessa L. (2011) Role of senataxin in DNA damage and telomeric stability. DNA Repair (Amst.), 10, 199–209. [DOI] [PubMed] [Google Scholar]
- 22.Roda R.H., Rinaldi C., Singh R., Schindler A.B., Blackstone C. (2014) Ataxia with oculomotor apraxia type 2 fibroblasts exhibit increased susceptibility to oxidative DNA damage. J. Clin. Neurosci., 21, 1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M.S., Rialdi A., Ho J.S., Tilove M., Martinez-Gil L., Moshkina N.P., Peralta Z., Noel J., Melegari C., Maestre A.M. et al. (2015) Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat. Immunol., 16, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. et al. (2008) Genetics of gene expression and its effect on disease. Nature, 452, 423–428. [DOI] [PubMed] [Google Scholar]
- 25.MacLennan N.K., Dong J., Aten J.E., Horvath S., Rahib L., Ornelas L., Dipple K.M., McCabe E.R. (2009) Weighted gene co-expression network analysis identifies biomarkers in glycerol kinase deficient mice. Mol. Genet. Metab., 98, 203–214. [DOI] [PubMed] [Google Scholar]
- 26.Saris C.G., Horvath S., van Vught P.W., van Es M.A., Blauw H.M., Fuller T.F., Langfelder P., DeYoung J., Wokke J.H., Veldink J.H. et al. (2009) Weighted gene co-expression network analysis of the peripheral blood from amyotrophic lateral sclerosis patients. BMC Genomics, 10, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winden K.D., Karsten S.L., Bragin A., Kudo L.C., Gehman L., Ruidera J., Geschwind D.H., Engel J. Jr. (2011) A systems level, functional genomics analysis of chronic epilepsy. PloS ONE, 6, e20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J.A., Woltjer R.L., Goodenbour J.M., Horvath S., Geschwind D.H. (2013) Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med., 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y., Voineagu I., Pasca S.P., Won H., Chandran V., Horvath S., Dolmetsch R.E., Geschwind D.H. (2014) Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med., 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin M.F., Yeo A.J., Becherel O.J. (2013) Senataxin protects the genome: implications for neurodegeneration and other abnormalities. Rare Dis., 1, e25230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee D.Y., Clayton D.A. (1998) Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem., 273, 30614–30621. [DOI] [PubMed] [Google Scholar]
- 32.Lin M.T., Beal M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443, 787–795. [DOI] [PubMed] [Google Scholar]
- 33.Lobsiger C.S., Cleveland D.W. (2007) Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat. Neurosci., 10, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kailasan Vanaja S., Rathinam V.A., Atianand M.K., Kalantari P., Skehan B., Fitzgerald K.A., Leong J.M. (2014) Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proc. Natl Acad. Sci. USA, 111, 7765–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mankan A.K., Schmidt T., Chauhan D., Goldeck M., Honing K., Gaidt M., Kubarenko A.V., Andreeva L., Hopfner K.P., Hornung V. (2014) Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J., 33, 2937–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigby R.E., Webb L.M., Mackenzie K.J., Li Y., Leitch A., Reijns M.A., Lundie R.J., Revuelta A., Davidson D.J., Diebold S. et al. (2014) RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J., 33, 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason S., Piper M., Gronostajski R.M., Richards L.J. (2009) Nuclear factor one transcription factors in CNS development. Mol. Neurobiol., 39, 10–23. [DOI] [PubMed] [Google Scholar]
- 38.Uittenbogaard M., Baxter K.K., Chiaramello A. (2010) The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN Neuro, 2, e00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greer E.L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet., 13, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X.M., Yu C., Wang Z.W., Zhang Y.L., Liu X.M., Zhou D., Sun Q.Y., Fan H.Y. (2013) DNA topoisomerase II is dispensable for oocyte meiotic resumption but is essential for meiotic chromosome condensation and separation in mice. Biol. Reprod., 89, 118. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L., Mandler M.D., Yi H., Feng Y. (2010) Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl Acad. Sci. USA, 107, 19061–19066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martins-Taylor K., Schroeder D.I., LaSalle J.M., Lalande M., Xu R.H. (2012) Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics, 7, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashimoto H., Horton J.R., Zhang X., Cheng X. (2009) UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics, 4, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeo A.J., Becherel O.J., Luff J.E., Graham M.E., Richard D., Lavin M.F. (2015) Senataxin controls meiotic silencing through ATR activation and chromatin remodelling. Cell Discovery, [in press]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardy R.J. (1998) QKI expression is regulated during neuron-glial cell fate decisions. J. Neurosci. Res., 54, 46–57. [DOI] [PubMed] [Google Scholar]
- 46.Kitaoka Y., Munemasa Y., Kojima K., Hirano A., Ueno S., Takagi H. (2013) Axonal protection by Nmnat3 overexpression with involvement of autophagy in optic nerve degeneration. Cell Death Dis., 4, e860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer-Lindenberg A., Straub R.E., Lipska B.K., Verchinski B.A., Goldberg T., Callicott J.H., Egan M.F., Huffaker S.S., Mattay V.S., Kolachana B. et al. (2007) Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J. Clin. Invest., 117, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oda K., Shiratsuchi T., Nishimori H., Inazawa J., Yoshikawa H., Taketani Y., Nakamura Y., Tokino T. (1999) Identification of BAIAP2 (BAI-associated protein 2), a novel human homologue of hamster IRSp53, whose SH3 domain interacts with the cytoplasmic domain of BAI1. Cytogenet. Cell Genet., 84, 75–82. [DOI] [PubMed] [Google Scholar]
- 49.Shi G.X., Cai W., Andres D.A. (2013) Rit subfamily small GTPases: regulators in neuronal differentiation and survival. Cell Signal., 25, 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimura-Akiyoshi S., Niimi K., Nakashiba T., Itohara S. (2007) Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc. Natl Acad. Sci. USA, 104, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pappas D.J., Coppola G., Gabatto P.A., Gao F., Geschwind D.H., Oksenberg J.R., Baranzini S.E. (2009) Longitudinal system-based analysis of transcriptional responses to type I interferons. Physiol. Genomics, 38, 362–371. [DOI] [PubMed] [Google Scholar]
- 52.Yu Q., Katlinskaya Y.V., Carbone C.J., Zhao B., Katlinski K.V., Zheng H., Guha M., Li N., Chen Q., Yang T. et al. (2015) DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep., 11, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavin M.F. (2013) The appropriateness of the mouse model for ataxia-telangiectasia: neurological defects but no neurodegeneration. DNA Repair (Amst.), 12, 612–619. [DOI] [PubMed] [Google Scholar]
- 54.Rulten S.L., Caldecott K.W. (2013) DNA strand break repair and neurodegeneration. DNA Repair (Amst.), 12, 558–567. [DOI] [PubMed] [Google Scholar]
- 55.Muguruma K., Nishiyama A., Ono Y., Miyawaki H., Mizuhara E., Hori S., Kakizuka A., Obata K., Yanagawa Y., Hirano T. et al. (2010) Ontogeny-recapitulating generation and tissue integration of ES cell-derived Purkinje cells. Nat. Neurosci., 13, 1171–1180. [DOI] [PubMed] [Google Scholar]
- 56.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin II., Thomson J.A. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science, 324, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu R.H., Peck R.M., Li D.S., Feng X., Ludwig T., Thomson J.A. (2005) Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Meth., 2, 185–190. [DOI] [PubMed] [Google Scholar]
- 58.Wettenhall J.M., Smyth G.K. (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics, 20, 3705–3706. [DOI] [PubMed] [Google Scholar]
- 59.Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L. et al. (2015) BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell, 57, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vantaggiato C., Cantoni O., Guidarelli A., Romaniello R., Citterio A., Arrigoni F., Doneda C., Castelli M., Airoldi G., Bresolin N. et al. (2004) Novel SETX variants in a patient with ataxia, neuropathy, and oculomotor apraxia are associated with normal sensitivity to oxidative DNA damaging agents. Brain Dev., 36, 682–689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.