Abstract
Nicotine dependence is influenced by chromosome 15q25.1 single nucleotide polymorphisms (SNPs), including the missense SNP rs16969968 that alters function of the α5 nicotinic acetylcholine receptor (CHRNA5) and noncoding SNPs that regulate CHRNA5 mRNA expression. We tested for cis-methylation quantitative trait loci (cis-meQTLs) using SNP genotypes and DNA methylation levels measured across the IREB2-HYKK-PSMA4-CHRNA5-CHRNA3-CHRNB4 genes on chromosome 15q25.1 in the BrainCloud and Brain QTL cohorts [total N = 175 European-Americans and 65 African-Americans (AAs)]. We identified eight SNPs that were significantly associated with CHRNA5 methylation in prefrontal cortex: P ranging from 6.0 × 10−10 to 5.6 × 10−5. These SNP–methylation associations were also significant in frontal cortex, temporal cortex and pons: P ranging from 4.8 × 10−12 to 3.4 × 10−3. Of the eight cis-meQTL SNPs, only the intronic CHRNB4 SNP rs11636753 was associated with CHRNA5 methylation independently of the known SNP effects in prefrontal cortex, and it was the most significantly associated SNP with nicotine dependence across five independent cohorts (total N = 7858 European ancestry and 3238 AA participants): P = 6.7 × 10−4, odds ratio (OR) [95% confidence interval (CI)] = 1.11 (1.05–1.18). The rs11636753 major allele (G) was associated with lower CHRNA5 DNA methylation, lower CHRNA5 mRNA expression and increased nicotine dependence risk. Haplotype analyses showed that rs11636753-G and the functional rs16969968-A alleles together increased risk of nicotine dependence more than each variant alone: P = 3.1 × 10−12, OR (95% CI) = 1.32 (1.22–1.43). Our findings identify a novel regulatory SNP association with nicotine dependence and connect, for the first time, previously observed differences in CHRNA5 mRNA expression and nicotine dependence risk to underlying DNA methylation differences.
Introduction
Cigarette smoking causes cancer, heart disease, stroke, lung disease and many other serious conditions (1). It is the leading cause of preventable death, annually resulting in over 400 000 deaths in the United States and over 5 million deaths worldwide (2). Although smoking rates have greatly declined since the 1960s due to public health campaigns and improved smoking cessation treatment, the rate of decline has slowed in recent years (3). Despite the well-known adverse health effects, 45.3 million adults in the United States are regular smokers (3), even though an estimated 69% report wanting to quit (4). One of the strongest predictors of failing to quit smoking is nicotine dependence (5–7), a heritable trait with an estimated 50% of its variability attributable to genetics (8).
Genome-wide association study (GWAS) analyses of nicotine dependence and other smoking behaviors have unequivocally identified single nucleotide polymorphisms (SNPs) on chromosome 15q25.1, which includes genes encoding three nicotinic acetylcholine receptors (CHRNA5, CHRNA3 and CHRNB4), iron-responsive element binding protein 2 (IREB2), hydroxylysine kinase (HYKK) and proteasome subunit alpha type-4 (PSMA4) (9–15). SNPs in this region have also been associated with lung cancer (11,16,17), airflow obstruction (18), chronic obstructive pulmonary disease (19,20) and behaviors related to other addictive substances such as alcohol (21,22), cocaine (23,24) and opioids (25).
Our prior analyses of the chromosome 15q25.1 region found that nicotine dependence-associated SNPs exert important biological effects: (1) the missense SNP rs16969968 changes CHRNA5 function, and (2) noncoding SNPs located in CHRNA5 (e.g. rs680244) or upstream of its transcription start site (e.g. rs880395) represent expression quantitative trait loci (eQTLs) regulating CHRNA5 mRNA expression levels in human frontal cortex (26–29).
The gene transcription process reflected by mRNA expression is driven, in part, by DNA methylation (biochemical addition of a methyl group to select cytosines in the dinucleotide sequence CpG). SNPs that affect DNA methylation and mRNA expression, methylation QTLs (meQTLs) and eQTLs, respectively, can alter gene transcript levels and thereby influence risk of disease, as evidenced by the enrichment of eQTLs and meQTLs among top GWAS findings for various human phenotypes (30,31). In human brain, eQTLs and meQTLs are abundant and predominantly cis-acting on proximally located genes (31,32).
Prior studies have evaluated methylation across the chromosome 15q25.1 gene region, focusing on lung and other tissues, and associations of their cis-meQTLs with cancer (33–36). However, no studies have reported on brain-specific methylation across this region to help uncover the mechanisms that confer risk for nicotine dependence. In the present study, we used cohorts of European and African ancestries to conduct cis-meQTL mapping in human prefrontal cortex (a highly relevant brain region for studying nicotine dependence) (37,38), extended these cis-meQTL findings to other relevant brain regions and tested the implicated SNPs for association with nicotine dependence. Our findings demonstrate that genetic variation underlying CHRNA5 DNA methylation in disease-relevant brain regions contributes to nicotine dependence and thus expand the known biological basis of this common addiction.
Results
Two cohorts were used to test SNP associations with DNA methylation levels: BrainCloud (39–41) and Brain QTL (32). Five cohorts were used to test SNP associations with nicotine dependence: Collaborative Genetic Study of Nicotine Dependence (COGEND) (9), Study of Addiction: Genetics and Environment [SAGE*; the asterisk (*) denotes SAGE excluding COGEND participants] (42), African American Nicotine Dependence (AAND), Environment and Genetics in Lung Cancer Etiology Study (EAGLE) (43,44) and Atherosclerosis Risk in Communities (ARIC) (45). Participant characteristics are presented in Table 1. The BrainCloud, COGEND, SAGE* and ARIC cohorts were multiancestry, comprised of up to 60% African-Americans (AAs); AAND contained only AAs; and the Brain QTL and EAGLE cohorts represented populations of European ancestry only. The cohorts contained mostly all adults, except for BrainCloud, which had participants spanning the lifespan from the fetal period through elderly ages. All cohorts had a sizable representation of both sexes.
Table 1.
Characteristics of study participants
| Cohorts used to test associations with DNA methylation and mRNA expression |
Cohorts used to test associations with nicotine dependence |
|||||||
|---|---|---|---|---|---|---|---|---|
| BrainCloud | Brain QTL | COGEND | SAGE* | AAND | EAGLE | ARIC | ||
| Brain tissue samples and use | Prefrontal cortex for testing cis-meQTLs (BrainCloudMethyl) | Prefrontal cortex for testing cis-eQTLs (BrainCloud) | Frontal cortex, temporal cortex, pons and cerebellum for testing cis-meQTLs | NA | NA | NA | NA | NA |
| Number of participants | ||||||||
| Europeans, N (%) | 0 | 0 | 0 | 0 | 0 | 0 | 2309 (100) | 0 |
| EAs, N (%) | 43 (40.0)a | 112 (41.6) | 150 (100) | 1933 (73.3) | 710 (57.7) | 0 | 0 | 2906 (79.9) |
| AAs, N (%) | 65 (60.0)a | 147 (54.6) | 0 | 704 (26.7) | 521 (42.3) | 1281 (100) | 0 | 732 (20.1) |
| Hispanics, N (%) | 0 | 6 (2.2) | 0 | 0 | 0 | 0 | 0 | 0 |
| Asians, N (%) | 0 | 4 (1.5) | 0 | 0 | 0 | 0 | 0 | 0 |
| Number of participants, excluding the fetal period | ||||||||
| EAs, N (%) | 40 (51.3) | 110 (49.1) | NA | NA | NA | NA | NA | NA |
| AAs, N (%) | 38 (48.7) | 114 (50.9) | NA | NA | NA | NA | NA | NA |
| Age (years), mean (range) | 36.0 (0–83)b | 32.4 (0–83)b | 45.8 (15–101) | 36.5 (25–46) | 40.0 (18–74) | 35.6 (25–55) | NAc (≤59–79) | 54.1 (44–66) |
| Male, N (%) | 56 (51.9) | 177 (65.8) | 103 (68.7) | 748 (38.7) | 681 (55.3) | 531 (41.5) | 1935 (83.8) | 2008 (55.2) |
AAND, African American Nicotine Dependence; AAs, African-Americans; ARIC, Atherosclerosis Risk in Communities; COGEND, Collaborative Genetic Study of Nicotine Dependence; EAGLE, Environment and Genetics in Lung Cancer Etiology Study; EAs, European-Americans; eQTLs, expression quantitative trait loci; meQTLs, methylation quantitative trait loci; NA, not available; SAGE*, Study of Addiction: Genetics and Environment.
aThe participants with methylation data are a subset of the participants available in the broader BrainCloud cohort, which all have SNP genotype and expression data.
bFor the BrainCloud cohort, mean age was calculated following the exclusion of fetal-aged participants.
cFor EAGLE, age was available as a categorical variable, so average age could not be calculated. The categorical age distributions were as follows: 24.2% aged 59 years or less, 18.6% aged 60–64 years, 22.6% aged 65–69 years, 20.6% aged 70–74 years and 14.0% aged 75–59 years.
SNP associations with DNA methylation and mRNA expression in prefrontal cortex
Eighty SNPs (Supplementary Material, Table S1) were tested in the BrainCloud cohort (N = 108) for association with methylation levels at one or more of the nine CpG sites (Supplementary Material, Table S2) across the IREB2-HYKK-PSMA4-CHRNA5-CHRNA3-CHRNB4 gene region. Eight SNPs were significantly associated with CHRNA5 methylation, and two SNPs were significantly associated with CHRNA3 methylation (P < 1.4 × 10−4 based on Bonferroni correction for 352 tests, Table 2). Of these SNPs, rs2292117 was associated with both CHRNA5 and CHRNA3 methylation. The implicated cis-meQTL SNPs were common (major allele frequencies ranging from 53 to 87%), and almost all showed suggestive (P < 0.05) to statistically significant associations with methylation in both ancestry groups. There was no positional overlap between the implicated cis-meQTL SNPs and the corresponding methylation probes (Table 2). No significant SNP associations were observed for methylation probes in IREB2, PSMA4 or CHRNB4 (Supplementary Material, Table S1).
Table 2.
SNPs significantly associated with methylation level at a nearby CpG site (P < 1.4 × 10−4, corrected for 352 tests) in prefrontal cortex from BrainCloud participants, 43 EAs and 65 AAs, with SNP genotype and methylation data available
| SNP (major allele) | SNP base pair positiona | Allele freq. in EAs | Allele freq. in AAs | Type | SNP location |
Cis-meQTL analysis (N = 108) |
Cis-eQTL analysis (N = 259) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylation probe and base pair positiona | SNP distanceb | P, all | P, EAs only | P, AAs only | Expression probe | P, all | ||||||
| rs12915366 (G) | 76 618 808 | 0.65 | 0.84 | 5′ UTR | PSMA4 | CHRNA5 (cg22563815) located at 76 644 004d | −26 152 | 2.7 × 10−7 | 5.0 × 10−7 | 6.8 × 10−3 | CHRNA5 located at 76 673 092–76 673 161 | 7.2 × 10−6 |
| rs2292117c (G) | 76 621 744 | 0.64 | 0.75 | Intron | PSMA4 | −23 216 | 6.0 × 10−10 | 1.5 × 10−7 | 1.2 × 10−4 | 9.5 × 10−8 | ||
| rs6495306 (A) | 76 652 948 | 0.63 | 0.65 | Intron | CHRNA5 | 0 | 4.2 × 10−8 | 6.3 × 10−8 | 2.4 × 10−3 | 1.3 × 10−8 | ||
| rs680244 (C) | 76 658 343 | 0.62 | 0.53 | Intron | CHRNA5 | 0 | 1.3 × 10−8 | 5.4 × 10−8 | 7.2 × 10−4 | 2.7 × 10−8 | ||
| rs621849 (A) | 76 659 916 | 0.62 | 0.54 | Intron | CHRNA5 | 0 | 6.1 × 10−9 | 5.4 × 10−8 | 4.7 × 10−4 | 2.7 × 10−8 | ||
| rs3743077 (C) | 76 681 951 | 0.62 | 0.83 | Intron | CHRNA3 | 7286 | 1.8 × 10−6 | 2.3 × 10−7 | 0.030 | 2.7 × 10−6 | ||
| rs950776 (T) | 76 713 073 | 0.67 | 0.87 | Intron | CHRNB4 | 38 408 | 5.6 × 10−5 | 1.5 × 10−3 | 5.5 × 10−3 | 7.0 × 10−7 | ||
| rs11636753 (G) | 76 716 001 | 0.64 | 0.73 | Intron | CHRNB4 | 41 336 | 5.8 × 10−5 | 6.4 × 10−6 | 0.10 | 7.7 × 10−3 | ||
| rs2292117c (G) | 76 621 744 | 0.64 | 0.75 | Intron | PSMA4 | CHRNA3 (cg22670733) located at 76 701 339d | −50 704 | 1.3 × 10−4 | 1.3 × 10−3 | 0.019 |
CHRNA3 NA |
NA |
| rs3743075 (C) | 76 696 507 | 0.65 | 0.61 | Coding (synonymous) | CHRNA3 | 0 | 7.7 × 10−6 | 1.1 × 10−3 | 2.1 × 10−3 | NA | ||
SNPs are sorted by chromosomal position for each methylation probe. SNP associations with mRNA expression are also shown in BrainCloud participants with SNP genotype and expression data available (N = 112 EAs and 147 AAs).
AFR, 1000 Genomes populations of African ancestry; eQTL, expression quantitative trait locus; EUR, 1000 Genomes populations of European ancestry; meQTL, methylation quantitative trait locus; NA, not available; SNP, single nucleotide polymorphism; UTR, untranslated region.
aBase pair positions correspond to NCBI build 36. bBase pair distance between the SNP and the start site of the gene whose methylation is significantly associated. A negative value indicates that the SNP is located upstream of the gene, ‘0’ indicates that the SNP is located within the gene and a positive value indicates that the SNP is located downstream of the gene.
cRs2292117 was implicated as a cis-meQTL for both the CHRNA5 and CHRNA3 methylation probes.
dThe CpG islands containing the CHRNA5 and CHRNA3 probes span from 76 643 994 to 76 644 288 and from 76 701 258 to 76 701 471, respectively.
Given the potential for differential methylation patterns between fetal development and postnatal life (39), we tested methylation levels of the implicated probes with age, adjusting for sex, race and all surrogate variables: P = 0.048 for CHRNA3 (CpG probe cg22670733) and P = 0.23 for CHRNA5 (CpG probe cg22563815). We next excluded the fetal-aged samples in the BrainCloud cohort and repeated the SNP association analyses with methylation. As presented in the Supplementary Material, Table S3, all of the implicated SNPs were suggestively (P < 0.05) or significantly (P < 1.4 × 10−4) associated with CHRNA5 or CHRNA3 methylation following the exclusion of fetal-aged samples; the β estimates for seven of the eight CHRNA5-implicated SNPs remained the same or were strengthened despite the reduced sample size, while the β estimates for both CHRNA3-implicated SNPs were weakened. Altogether, these results suggest that the primarily observed SNP associations with CHRNA3 methylation may reflect some differential methylation patterns between fetal and postnatal periods, but the SNP associations with CHRNA5 methylation appear robust to age-related methylation patterns.
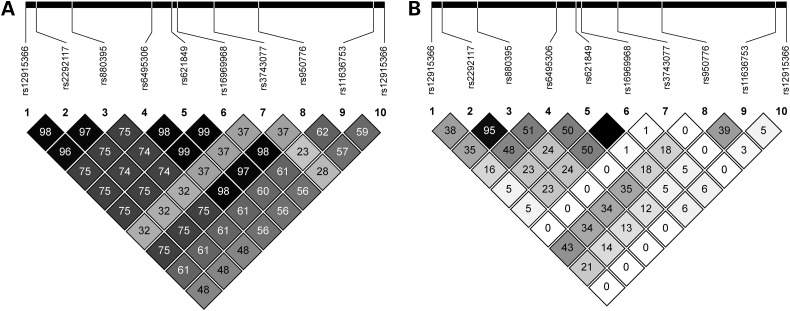
The remaining analyses were focused on the eight CHRNA5-implicated cis-meQTL SNPs. Of these SNPs, four captured the variability at r2 > 0.8 under European ancestry: rs2292117 (tagging rs12915366), rs680244 (tagging rs6495306, rs621849 and rs3743077), rs950776 and rs11636753. In contrast, seven SNPs were needed to capture the variability under African ancestry: rs12915366, rs2292117, rs6495306, rs680244 (tagging rs621849), rs3743077, rs950776 and rs11636753. Among these, rs11636753 (an intronic SNP in CHRNB4) showed the lowest r2 values with all other newly implicated cis-meQTL SNPs (Fig. 1), and it was the only SNP not contained in a linkage disequilibrium (LD) block with a known cis-eQTL or functional missense SNP across both ancestry groups (Supplementary Material, Fig. S1).
Figure 1.
LD structure, based on r2 values, among chromosome 15q25.1 SNPs associated with nicotine dependence: eight SNPs associated with CHRNA5 methylation (rs12915366, rs2292117, rs6495306, rs680244, rs621489, rs3743077, rs950776 and rs11636753) and two other SNPs previously established for their cis-eQTL (rs880395) or functional (rs16969968) effects. Rs680244 and its highly correlated rs621849 represent both newly implicated associations with CHRNA5 methylation and known associations with CHRNA5 expression. Darker gray to black shading indicates higher r2 values in 1000 Genomes reference populations of (A) European ancestry (denoted EUR) and (B) African ancestry (denoted AFR).
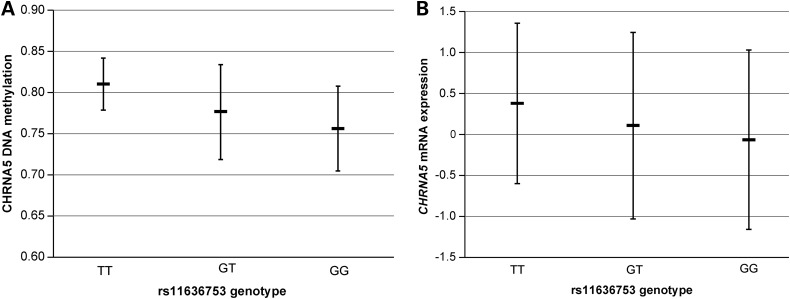
All of the newly implicated SNPs for CHRNA5 methylation were also associated with CHRNA5 mRNA expression in the broader BrainCloud cohort (N = 259, Table 2). The newly implicated cis-meQTL SNPs rs680244 and its highly correlated SNP rs621849 have previously established associations with CHRNA5 mRNA expression (27). Our results show that the SNP major alleles were associated with both lower CHRNA5 methylation and expression in prefrontal cortex, as illustrated in Figure 2 for rs11636753.
Figure 2.
Associations of rs11636753 genotypes with CHRNA5 methylation and expression in prefrontal cortex tissue from the BrainCloud cohort. The rs11636753 genotypes are plotted against (A) CHRNA5 DNA methylation in the 108 BrainCloud participants used for testing cis-meQTLs and (B) CHRNA5 mRNA expression in the 259 BrainCloud participants used for testing cis-eQTLs. The methylation level is presented as a β value: the ratio of signal from a methylated probe relative to the sum of both methylated and unmethylated probes. The expression level is presented as log2 of the ratio of sample signal to the reference signal (pooled RNA from all the participants). Average levels and their standard deviations are shown.
SNP associations with DNA methylation in other brain regions
The SNPs identified as cis-meQTLs in prefrontal cortex from BrainCloud were tested for association with CHRNA5 methylation across four brain regions (frontal cortex, temporal cortex, pons and cerebellum) in the Brain QTL cohort. Methylation levels across the four regions are shown in the Supplementary Material, Figure S2, and the corresponding SNP-methylation results are shown in Table 3. All of the major alleles for CHRNA5-implicated SNPs were significantly (P < 6.3 × 10−3 based on Bonferroni correction for eight SNPs) associated with lower CHRNA5 methylation levels in frontal cortex: P = 3.4 × 10−3 for rs11636753. SNP associations with CHRNA5 methylation were even stronger in temporal cortex and pons: P = 2.2 × 10−4 and 1.8 × 10−6, respectively, for rs11636753. No SNP-methylation associations were found in cerebellum.
Table 3.
Eight cis-meQTL SNPs implicated for CHRNA5 in prefrontal cortex from BrainCloud and their associations with methylation level of the same CHRNA5 CpG probe (cg22563815) across four different brain regions in the Brain QTL cohort
| SNP (major allele) | Frontal cortex (N = 132) |
Temporal cortex (N = 126) |
Cerebellum (N = 120) |
Pons (N = 124) |
||||
|---|---|---|---|---|---|---|---|---|
| β | P | β | P | β | P | β | P | |
| rs12915366 (G) | −0.029 | 5.1 × 10−5 | −0.034 | 2.0 × 10−8 | 0.013 | 0.47 | −0.059 | 4.8 × 10−12 |
| rs2292117 (G) | −0.029 | 5.1 × 10−5 | −0.034 | 2.0 × 10−8 | 0.013 | 0.47 | −0.059 | 4.9 × 10−12 |
| rs6495306 (A) | −0.027 | 1.4 × 10−4 | −0.030 | 8.7 × 10−7 | 0.017 | 0.33 | −0.055 | 9.0 × 10−11 |
| rs680244 (C) | −0.027 | 2.1 × 10−4 | −0.030 | 7.0 × 10−7 | 0.019 | 0.26 | −0.054 | 3.8 × 10−10 |
| rs621849 (A) | −0.027 | 2.1 × 10−4 | −0.030 | 7.0 × 10−7 | 0.019 | 0.26 | −0.054 | 3.8 × 10−10 |
| rs3743077 (C) | −0.025 | 5.4 × 10−4 | −0.029 | 2.1 × 10−6 | 0.018 | 0.30 | −0.056 | 6.4 × 10−11 |
| rs950776 (T) | −0.026 | 6.5 × 10−4 | −0.026 | 9.4 × 10−5 | 0.0088 | 0.61 | −0.047 | 5.0 × 10−7 |
| rs11636753 (G) | −0.022 | 3.4 × 10−3 | −0.025 | 2.2 × 10−4 | 0.012 | 0.47 | −0.046 | 1.8 × 10−6 |
SNPs are sorted by chromosomal position.
P-values shown in bold were statistically significant (P < 6.3 × 10−3, corrected for eight SNP tests).
SNP, single nucleotide polymorphism.
SNP associations with nicotine dependence
The CHRNA5-implicated cis-meQTL SNPs were next tested for association with nicotine dependence in a multiancestral meta-analysis of COGEND, SAGE*, AAND, EAGLE and ARIC (total N = 11 096). As shown in Table 4, the major alleles of all eight SNPs suggested associations with increased risk of nicotine dependence, consistent with the directionality of previous reports of nominal association with Fagerström Test for Nicotine Dependence (FTND)-defined nicotine dependence (rs6495306, rs680244 and rs3743077) (46) or related phenotypes [rs12915366 with cigarette pack-years (47) and rs11636753 with tolerance factor of the Nicotine Dependence Syndrome Scale (48)]. However, only the rs11636753-G allele was significantly associated with increased risk after correcting for multiple testing: P = 6.7 × 10−4, odds ratio (OR) [95% confidence interval (CI)] = 1.11 (1.05–1.18).
Table 4.
Eight cis-meQTL SNPs implicated for CHRNA5 in the prefrontal cortex from BrainCloud and their associations with nicotine dependence
| SNP (major allele) | European ancestry participants only |
AA participants only | Meta-analysis of all participants | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COGEND (N = 1933) | SAGE* (N = 710) | EAGLE (N = 2309) | ARIC (N = 2906) | Meta-analysis P | COGEND (N = 704) | SAGE* (N = 710) | AAND (N = 1281) | ARIC (N = 2906) | Meta-analysis P | P | OR (95% CI) | |||||||||
| P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | |||||
| rs12915366 (G) | 0.39 | 1.06 (0.93–1.21) | 0.54 | 0.90 (0.66–1.25) | 4.3 × 10−3 | 1.21 (1.06–1.37) | 0.88 | 1.01 (0.90–1.14) | 0.047 | 0.57 | 1.09 (0.81–1.46) | 0.54 | 1.15 (0.73–1.82) | 0.086 | 1.25 (0.97–1.62) | 0.014 | 1.52 (1.09–2.08) | 6.5 × 10−3 | 0.014 | 1.08 (1.02–1.16) |
| rs2292117 (G) | 0.33 | 1.07 (0.94–1.22) | 0.68 | 0.93 (0.68–1.28) | 5.4 × 10−3 | 1.20 (1.06–1.37) | 0.89 | 1.01 (0.90–1.14) | 0.039 | 0.20 | 1.18 (0.92–1.53) | 0.44 | 0.84 (0.55–1.30) | 0.063 | 1.25 (0.99–1.58) | 0.19 | 1.20 (0.92–1.59) | 0.027 | 0.011 | 1.09 (1.02–1.16) |
| rs6495306 (A) | 0.17 | 1.10 (0.96–1.25) | 0.87 | 1.03 (0.75–1.40) | 0.055 | 1.14 (1.00–1.30) | 0.78 | 1.02 (0.91–1.14) | 0.046 | 0.046 | 1.27 (1.00–1.62) | 0.74 | 0.94 (0.63–1.39) | 0.18 | 1.16 (0.93–1.44) | 0.17 | 1.19 (0.93–1.52) | 0.013 | 0.010 | 1.08 (1.02–1.15) |
| rs680244 (C) | 0.11 | 1.11 (0.98–1.27) | 0.80 | 1.04 (0.76–1.42) | 0.078 | 1.12 (0.99–1.28) | 0.76 | 1.02 (0.91–1.15) | 0.037 | 0.097 | 1.21 (0.97–1.50) | 0.67 | 0.92 (0.64–1.33) | 0.29 | 1.12 (0.91–1.36) | 0.35 | 1.11 (0.88–1.39) | 0.064 | 0.013 | 1.08 (1.02–1.15) |
| rs621849 (A) | 0.13 | 1.11 (0.97–1.26) | 0.92 | 1.02 (0.75–1.38) | 0.069 | 1.13 (0.99–1.29) | 0.74 | 1.02 (0.91–1.15) | 0.041 | 0.095 | 1.21 (0.97–1.50) | 0.82 | 0.96 (0.67–1.38) | 0.29 | 1.11 (0.91–1.36) | 0.36 | 1.11 (0.88–1.39) | 0.056 | 0.013 | 1.08 (1.02–1.15) |
| rs3743077 (C) | 0.18 | 1.09 (0.96–1.25) | 0.94 | 1.01 (0.74–1.38) | 0.020 | 1.17 (1.03–1.33) | 0.61 | 1.03 (0.92–1.16) | 0.021 | 0.19 | 1.23 (0.91–1.66) | 0.43 | 1.20 (0.76–1.92) | 0.47 | 1.11 (0.83–1.48) | 0.17 | 1.27 (0.91–1.75) | 0.035 | 7.3 × 10−3 | 1.09 (1.02–1.17) |
| rs950776 (T) | 0.83 | 1.02 (0.89–1.16) | 0.67 | 1.07 (0.77–1.50) | 0.028 | 1.17 (1.02–1.34) | 0.51 | 1.04 (0.93–1.18) | 0.072 | 0.54 | 1.11 (0.79–1.56) | 0.58 | 1.17 (0.68–2.02) | 0.49 | 1.12 (0.81–1.55) | 0.13 | 1.32 (0.93–1.89) | 0.088 | 0.044 | 1.07 (1.00–1.15) |
| rs11636753 (G) | 0.26 | 1.08 (0.95–1.23) | 0.35 | 1.16 (0.85–1.60) | 6.9 × 10−3 | 1.20 (1.05–1.37) | 0.22 | 1.08 (0.96–1.22) | 2.5 × 10−3 | 0.33 | 1.13 (0.89–1.43) | 0.61 | 1.10 (0.76–1.59) | 0.14 | 1.18 (0.95–1.46) | 0.98 | 1.00 (0.78–1.28) | 0.12 | 6.7 × 10−4 | 1.11 (1.05–1.18) |
SNP associations were tested across five independent cohorts: COGEND, SAGE*, EAGLE, ARIC and AAND. SNPs are sorted by chromosomal position. P-values of meta-analysis shown in bold were statistically significant (P < 6.3 × 10−3, corrected for eight SNP tests).
CI, confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
SNP associations with DNA methylation, mRNA expression and nicotine dependence conditioned on known SNP effects
We repeated the SNP-methylation association analyses with adjustment for known cis-eQTL SNPs, one tagged by rs680244 within CHRNA5 and the other tagged by rs880395 upstream of CHRNA5 (27,28), in the prefrontal cortex samples from BrainCloud. Although rs11636753 no longer passed the statistical significance threshold used for cis-meQTL mapping (P < 1.4 × 10−4), it remained associated with CHRNA5 methylation at P < 0.05 following adjustment for either cis-eQTL SNP (Supplementary Material, Table S4). There was moderate LD between rs11636753 and the known cis-eQTL SNPs under European ancestry [r2 = 0.48 and 0.56 (Fig. 1A) and D′ = 0.72 and 0.75 (Supplementary Material, Fig. S1A)] but little LD under African ancestry [r2 = 0 and 0.06 (Fig. 1B) and D′ = 0.23 and 0.30 (Supplementary Material, Fig. S1B)]. We also repeated the SNP-methylation association analyses with adjustment for the functional missense SNP rs16969968 (26,27). The SNPs all remained suggestively to significantly associated with CHRNA5 methylation (Supplementary Material, Table S4): P = 1.7 × 10−4 for rs11636753, which had weak to moderate LD with rs16969968 [r2 = 0.48, D′ = 0.86 under European ancestry (Fig. 1A and Supplementary Material, Fig. S1A); r2 = 0, D′ = 0.65 under African ancestry (Fig. 1B and Supplementary Material, Fig. S1B)].
We repeated the SNP-expression association analyses in BrainCloud with adjustment for the cis-eQTL and functional SNPs (Supplementary Material, Table S5). Unlike the independent rs11636753 association with methylation, its association with expression was explained by the known SNP effects.
For the nicotine dependence association testing, we found that the rs11636753-G association with increased risk of nicotine dependence was the only SNP association that persisted at nominal significance following adjustment for the known cis-eQTL SNPs: P = 0.017 following adjustment for rs680244 (Supplementary Material, Table S6) and P = 0.0065 following adjustment for rs880395 (Supplementary Material, Table S7). To evaluate the cis-meQTL SNP associations with nicotine dependence in the context of the functional missense SNP as done previously (27), we constructed haplotypes that aligned the risk-conferring alleles, rs16969968 minor allele [A] and rs11636753 major allele [G]. Rs16969968-A was previously shown to occur primarily on a low mRNA expression background (27), and this alignment enabled us to test the combined genetic associations of decreased CHRNA5 methylation and expression resulting from rs11636753-G and reduced CHRNA5 function resulting from rs16969968-A on risk of nicotine dependence. The haplotype carrying both risk alleles was associated with increased nicotine dependence risk at high statistical significance [P = 3.1 × 10−12, OR (95% CI) = 1.32 (1.22–1.43)], whereas neither of the haplotypes carrying only one risk allele was associated across the ancestry groups (P = 0.32 and 0.18, Supplementary Material, Table S8).
Causal inference
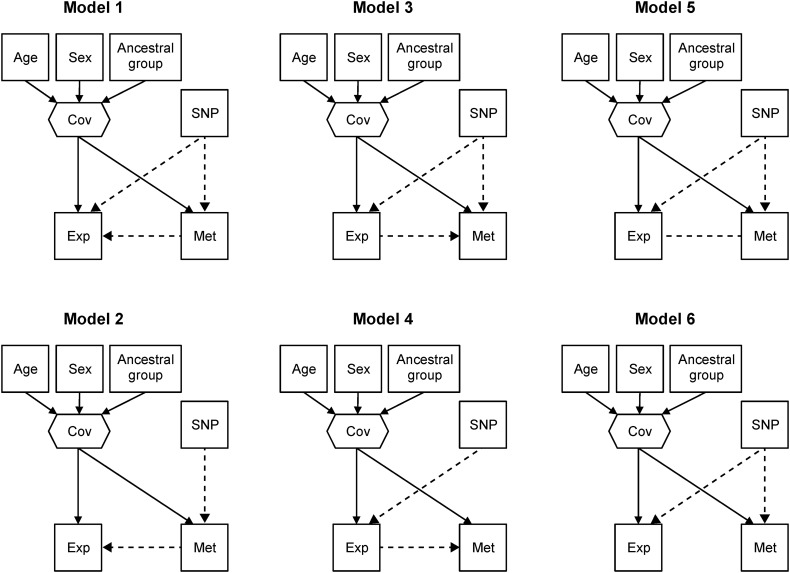
We used structural equation modeling (49) to investigate the potential causal relationship between rs11636753, CHRNA5 methylation and CHRNA5 expression in the BrainCloud cohort. For comparison, the approach was repeated for rs680244, which was both a known cis-eQTL and a newly implicated cis-meQTL for CHRNA5. Alternative models are presented in Figure 3, and the results, as presented in the Supplementary Material, Table S9, showed that the data were more consistent with some possible causal models than others. For both SNPs, the fit indices suggested that Models 4 (the SNP affecting only expression, which in turn affects methylation) and 5 (correlated but noncausal relationship between methylation and expression with the SNP directly affecting each) provided less consistent fits to the data. Models 1, 3 and 6 provided more consistent fits to the data for both rs11636753 and rs680244. Across these models, the SNP directly affected both methylation and expression, but we were unable to distinguish the relationship between methylation and expression. Model 2 provided a more consistent fit to the rs11636753 data (i.e. rs11636753 directly affecting only methylation, which in turn indirectly affects expression) than it provided to the rs680244 data. We cannot rule out the possibility that rs1163673 directly affects both methylation and expression (Models 1, 3 and 6), but the potentially unique result involving Model 2 may explain its association with CHRNA5 expression being captured by the known cis-eQTLs (Supplementary Material, Table S5) but its association with CHRNA5 methylation not being fully captured by the known cis-eQTLs (Supplementary Material, Table S4).
Figure 3.
Six causal models whose fit was assessed using our implicated cis-meQTL SNPs, CHRNA5 expression (Exp) and CHRNA5 methylation (Met). The covariates age, sex and ancestral group were included as a single composite variable. The dotted lines with arrows indicate the direction of effect between the SNP, methylation and/or expression. The dotted line with no arrows (Model 5 only) represents there being correlation between methylation and gene expression due to a hidden confounding effect.
Discussion
The chromosome 15q25.1 gene region has well-established associations with nicotine dependence and smoking-related phenotypes. Our SNP-methylation association findings expand our understanding of known nicotine dependence-associated SNPs by discovering a new biological role for the known variants and highlighting rs11636753 as a novel SNP association that reflects an additional mechanism contributing to nicotine dependence risk. Two mechanisms involving CHRNA5 were previously known: (1) the missense SNP rs16969968 alters the function of CHRNA5, and (2) several upstream and intronic SNPs (rs680244, rs880395 and other correlated SNPs) tag eQTLs regulating CHRNA5 mRNA expression (26–28). Among our identified cis-meQTL SNPs, rs11636753 demonstrated the weakest LD with the known cis-eQTL and functional SNPs, which was most evident under African ancestry, and its association with CHRNA5 methylation was independent of the other SNP effects in the region. The known cis-eQTL SNPs rs680244 and rs880395 and our newly identified cis-meQTL SNP rs11636753 may exert the observed effects themselves or each reside in tight LD with different causal SNPs.
We identified eight SNPs that were significantly associated with CHRNA5 methylation in addiction-relevant human brain regions and were suggestively to significantly associated with risk of nicotine dependence. Bioinformatics analyses provided additional indications that these SNPs have regulatory effects (Supplementary Material, Table S10), including locations within promoters, enhancers and/or DNAse hypersensitivity sites indicative of transcriptional activity and predictions for altering regulatory motifs, with rs11636753 altering the most (24 motifs). Two of the eight CHRNA5-implicated cis-meQTL SNPs (rs680244 and rs621849) have previously been shown to tag a cis-eQTL associated with nicotine dependence (26,27). The six other SNPs (rs12915366, rs2292117, rs6495306, rs3743077, rs950776 and rs11636753) were previously reported for their associations with nicotine dependence, other smoking or alcohol phenotypes or lung cancer (21,27,46–48,50,51), but their biological roles were unknown. There is LD underlying the known cis-eQTL SNPs and several of these newly implicated cis-meQTL SNPs, but our results suggest that DNA methylation helps to further explain known SNP association signals with mRNA expression and risk of nicotine dependence. Lastly, rs11636753 and its associations with CHRNA5 methylation and nicotine dependence were not explained by the known cis-eQTL SNPs, and causal inference modeling suggests that this SNP may uniquely exert an indirect effect on CHRNA5 expression through its direct effect on methylation. Inferences from the structural equation modeling depend on the data meeting several underlying assumptions (e.g. no unmeasured confounders, multivariate normality and linearity and correct model specification). Models with less consistent fits to our data may have correctly inferred nonexistent effects, or they may have effect sizes too weak to detect with our sample size. Repeating this modeling using a larger sample size with the SNP genotype, methylation and expression data in addiction-relevant brain regions, once available, is needed to strengthen the evidence for causal inference. Nonetheless, our results suggest that rs11636753 represents a novel signal that underlies CHRNA5 methylation and contributes to nicotine dependence risk.
Studies of nicotine dependence and related phenotypes have demonstrated SNP associations spanning IREB2-HYKK-PSMA4-CHRNA5-CHRNA3-CHRNB4, so our cis-meQTL mapping was conducted across the entire region. Similar to our prior cis-eQTL studies in human brain, we found significant SNP-methylation associations for CHRNA5 and CHRNA3. The identified SNPs had consistent directions of association with CHRNA5 and/or CHRNA3 methylation in postmortem prefrontal cortex samples from both ancestry groups. CHRNA5 associations were stronger than CHRNA3 associations, and their differences increased after excluding the fetal samples. No significant associations were observed for IREB2, PSMA4 or CHRNB4.
The major alleles of the CHRNA5-implicated SNPs were associated with lower methylation levels and lower expression levels. This pattern highlights two important points. First, our findings provide an uncommon example of SNPs being associated with both DNA methylation and mRNA expression. The Brain QTL cohort investigators (32) and others (31) reported that SNPs implicated as eQTLs and SNPs implicated as meQTLs in human brain most often do not overlap with one another, with each type of QTL representing a distinct regulatory feature. SNPs that are associated with both DNA methylation and mRNA expression tend to associate with methylation probes that are located close to transcription start sites: mean distance of 27.5 kb (32). Indeed, all of our tested methylation probes were located within 1.5 kb of transcription start sites for genes in the chromosome 15q25.1 region. Second, the pattern of SNPs being positively correlated with methylation and expression in the same direction might appear to contrast the traditional view of an inverse correlation, whereby lower methylation levels lead to higher expression levels for nearby transcripts. However, when considering only the subset of SNPs that act as both eQTLs and meQTLs, the Brain QTL study investigators found that almost half show patterns similar to ours, where the direction of correlation is the same for methylation and expression (32).
Besides being associated with lower CHRNA5 methylation and expression levels, the rs11636753 major allele (G) was associated with an altered risk of nicotine dependence. Rs11636753-G was associated with an increased risk of nicotine dependence when not accounting for rs16969968, and it is consistent with prior studies (27,52). This overall pattern is consistent with rodent models, which demonstrate that eliminating CHRNA5 expression leads to markedly increased nicotine intake by reducing inhibitory motivational signals in the brain's reward pathways at high nicotine doses (53,54). Importantly, however, the rs16969968 risk allele (A) falls almost exclusively on a low-expression background in European and African ancestries, whereas the wild-type rs16969968-G occurs on both high- and low-expression backgrounds. Our prior studies showed that low-expression alleles associate with lower risks for both nicotine dependence and lung cancer when occurring on the wild-type rs16969968-G background (27,55,56). In the current study, we similarly found that the haplotype carrying both rs16969968-G and the low-methylation/low-expression allele (rs11636753-G) was associated with decreased nicotine dependence risk in European-Americans (EAs) specifically (Supplementary Material, Table S8).
Our cis-meQTL mapping was focused on prefrontal cortex, a highly relevant brain region for studying nicotine dependence (37,38) given its role in self-control/response inhibition, emotional regulation, flexibility/control of attention, and planning and goal formation. Dysregulation of this brain region can increase impulsivity, risk taking and stress reactivity and bias attention and reward anticipation toward immediate rather than delayed gratification: all characteristics generally associated with greater risk of addiction (57). Because physiological response to nicotine is a complex process involving multiple brain regions (58), we evaluated the cis-meQTL patterns in other brain regions and found that our findings extended into the frontal cortex along with the temporal cortex and pons. The frontal cortex encompasses the prefrontal cortex and other components. The temporal cortex is involved in visual memories and sensory processing among other functions, and it has been directly linked to processing and reacting to smoking-related cues (59–61). The pons relays signals that require passage from the left side of the brain to the right, and activity in this region has been correlated with craving following nicotine abstinence (62). Furthermore, the mesopontine tegmentum tissue that lies at the junction of the mesencephalon and the pons has been described as a ‘master regulator of nicotinic dopaminergic signaling in the brain’ (63). Our cis-meQTL SNP findings were not present in cerebellum, where CHRNA5 methylation levels were higher and more variable than the levels measured in the other brain regions (Supplementary Material, Fig. S2); this observation is consistent with the original Brain QTL cohort investigators’ report that the most distinct patterns across all methylation probes were observed in the cerebellum (32). The distinct CHRNA5 methylation patterns warrant future study, as the cerebellum has some indications in addiction (e.g. drug cue reactivity) (64,65). Altogether, our study provided evidence for several SNPs that tag a cis-regulatory effect on CHRNA5 methylation across biologically relevant brain regions for addiction. Our findings merit extension into other relevant brain regions, such as the nucleus accumbens and ventral tegmental area (58).
A limitation of our study was the lack of nicotine dependence data in the publically available cohorts used for cis-meQTL mapping, so linking DNA methylation, mRNA expression and nicotine dependence in the same participants was not feasible. Other limitations included different sample sizes being available to compare SNP associations with methylation and expression and diverse ancestries not being universally represented across the cohorts. We also recognize that the methylation and expression array data reflect important features of gene regulation but not the full scope of this complex process. Nonetheless, DNA methylation is viewed as an important and relatively stable epigenetic change that can drive long-lasting changes in gene expression and consequently influence susceptibility to developing addiction (66).
A major strength of our study was the availability of multifaceted biological data across several disease-relevant brain regions. A prior study evaluating SNPs implicated in a variety of phenotypes from the GWAS catalog, and their eQTL patterns across the brain and blood tissues showed clear examples of both shared and distinct eQTL effects between these two tissue types (67). For example, the major allele of rs880395 is reproducibly associated with lower CHRNA5 expression levels in the brain; however, its association in the blood has been inconsistent (28,29). Such tissue specificity has been observed for both eQTLs (67) and meQTLs (31), thus emphasizing the importance of using expression and methylation data from disease-relevant tissues for QTL mapping. Moreover, the different patterns observed in the cerebellum compared with the prefrontal cortex, frontal cortex, temporal cortex and pons in our study demonstrate that regional specificity is highly important when studying QTLs in the brain.
Prior studies identified biologically important SNP associations with nicotine dependence by integrating mRNA expression data. We found several SNPs associated with both CHRNA5 methylation and expression in addiction-relevant brain regions—prefrontal cortex, frontal cortex, temporal cortex and pons—and with risk of nicotine dependence across diverse ancestry groups. Among these SNPs, we identified rs11636753 as exerting an effect on CHRNA5 methylation that may operate independently of the known cis-eQTL and functional SNP effects in the region. Although future studies are needed to firmly establish the causal relationship between chromosome 15q25.1 SNPs, CHRNA5 methylation and expression in the implicated brain regions, and risk of nicotine dependence using a sample with all of these facets of data available, our findings show, for the first time, that genetic variation underlying CHRNA5 methylation in human brain contributes to risk of nicotine dependence.
Materials and Methods
Cohorts for cis-meQTL mapping
We used the BrainCloud cohort to find SNPs associated with methylation across chromosome 15q25.1, using SNP genotypes (Illumina Human1M-Duo and HumanHap650Y arrays) and DNA methylation levels (Illumina HumanMethylation27 array) available from postmortem prefrontal cortex of human participants who had no neuropathological or neuropsychiatric diagnoses, no reported alcohol or drug abuse and no positive toxicology result (39–41). We obtained these data via the database of Genotypes and Phenotypes (dbGaP; accession number phs000417.v2.p1) and the BrainCloud project website (http://braincloud.jhmi.edu/). SNP genotypes and mRNA expression data were available (Illumina Human 49 K Oligo array) on all participants (147 AAs, 112 EAs, 6 Hispanics and 4 Asians). DNA methylation data were available on a subset of 65 AAs and 43 EAs. Further details on the BrainCloud cohort and quality control (QC) procedures have been described elsewhere (39,40).
SNPs implicated in BrainCloud were tested with the same DNA methylation probes across four brain regions (frontal cortex, temporal cortex, cerebellum and pons) using up to 132 unrelated EA samples from the Brain QTL cohort (32). None of these participants had a clinical history of neurological or cerebrovascular disease or a diagnosis of cognitive impairment. A few (N = 12) had a drug-related cause of death, but smoking history was unknown. We obtained their SNP genotypes (Illumina HumanHap550v3 array) via dbGaP accession number phs000249.v1.p1 and their DNA methylation levels (Illumina HumanMethylation27 array) via the Gene Expression Omnibus series number GSE15745. Details on this cohort and their QC procedures have been described elsewhere (32).
Cis-meQTL mapping
We used the BrainCloudMethyl software application to test additively coded SNP genotypes for association with methylation levels using the best fit procedure under a general linear model, as described at http://braincloud.jhmi.edu/. The methylation level of each CpG dinucleotide probe corresponded to the ratio of methylated probe signal to the sum of both methylated and unmethylated probe signals (39). The resulting β values ranged from 0 (completely unmethylated) to 1 (fully methylated). The range of covariates included sex, ancestry group, age, life stage [fetal, childhood (≤10 years old), and adolescence/adulthood (>10 years old)], age-by-stage interaction and up to 18 surrogate variables to account for batch effects and other sources of measurement heterogeneity (39,68).
Eighty SNPs were tested for association with methylation levels reported for one or more probes over the 375 kb region of interest (Supplementary Material, Table S1). There were nine DNA methylation probes available for testing: two IREB2, two PSMA4, one CHRNA5, two CHRNA3 and two CHRNB4 probes (Supplementary Material, Table S2). There was no probe available for the sixth gene in this region, HYKK located between IREB2 and PSMA4. A total of 352 association tests were conducted using a sliding window-based approach, whereby SNPs located from 100 kb upstream to 100 kb downstream of each gene were tested. SNP-methylation associations with P < 1.4 × 10−4, based on Bonferroni correction for 352 tests, were declared statistically significant.
We used the BrainCloud software, analogous to BrainCloudMethyl, to test SNP associations with mRNA expression. The best fit procedure was used under a general linear model to test the log2 of the normalized ratio of sample-to-reference mRNA expression levels for association with additive SNP genotypes (40). The range of covariates included sex, ancestry group, age, life stage and age-by-stage interaction (40). Secondary analyses of the BrainCloud SNP genotype, methylation and expression data were run using SAS software (SAS, Cary, NC).
We used the Brain QTL cohort to validate the SNP-methylation associations and compare their patterns across four different brain regions. We tested the SNP associations with methylation level in PLINK (69), using linear regression under an additive genetic model with covariates of age, sex, postmortem interval, tissue source and hybridization batch.
Cohorts for nicotine dependence association testing
SNPs were tested for association with nicotine dependence across five independent cohorts comprised of European ancestry and/or AA participants. Study protocols were approved by the Institutional Review Boards at the participating sites.
COGEND
Using a community-based case–control study design, COGEND contrasts nicotine-dependent smokers with smokers who never developed symptoms of nicotine dependence, as previously described (9). Beginning in 2001, study participants were recruited from St Louis and Detroit through telephone screening to identify current smokers aged 25–44 years. Study eligibility was determined by administering the FTND (70). Current smokers with FTND ≥ 4 were recruited as nicotine-dependent cases, and smokers (>100 cigarettes during their lifetime) with FTND≤1 were recruited as nicotine-dependent controls.
A subset of 1406 COGEND participants were genotyped on the Illumina Human1M-Duo BeadChip array as part of SAGE (42) and had call rates >97%. Another 1480 COGEND participants were genotyped on the Illumina HumanOmni2.5 array as part of the Gene Environment Association Studies Initiative (GENEVA) (71) and had call rates >95%. In each subset, genotyped SNPs with call rate >98% and Hardy Weinberg equilibrium P ≥ 1 × 10−4 were retained. The subsets were combined, and after removing duplicates and first-degree relatives, there remained 2752 participants with genome-wide SNP genotype data available: 1961 self-identified EAs, 712 self-identified AAs and 79 participants from other ancestry groups. All SNP-level and participant-level QC steps were conducted in PLINK (69).
To capture all of the cis-meQTL SNPs, genotype imputation was needed. We previously found that combined imputation of subjects genotyped on different arrays may lead to biased association test results, and we circumvented this potential bias by using only the genotyped SNPs at the intersection of the different arrays as the basis for imputation (72). After taking the intersection of SNPs across the genome, we additionally excluded 318 SNPs with call rate <97% in either ancestry group (EAs or AAs) and 767 SNPs with P < 1 × 10−4 in any of six Hardy Weinberg equilibrium analyses (all participants, controls only or cases only in either ancestry group). For the participant-level QC, we excluded 12 genotyped participants with call rate <97% and another 24 participants with incomplete FTND data, and we confirmed that none of the remaining participants were duplicates, first-degree relatives, sex discordant or had excessive homozygosity. The final COGEND analysis dataset included 1933 EAs (1000 cases and 933 controls) and 704 AAs (459 cases and 245 controls).
SAGE*
Our use of SAGE (dbGaP accession number phs000092.v1.p1) included participants from the Collaborative Study on the Genetics of Alcoholism (COGA) (73) and the Family Study of Cocaine Dependence (FSCD) (74). COGEND participants were excluded from the overall SAGE dataset to avoid redundancy. This cohort of COGA and FSCD participants is thus referred to as SAGE*. The SAGE* participants were all ascertained as part of case–control studies of addictive disorders, and we analyzed them together, as done in previous GWAS analyses (42), using 710 EAs (589 cases with FTND ≥ 4 and 121 controls with FTND ≤ 1) and 521 AAs (422 cases and 99 controls) who were genotyped on the Illumina Human1M-Duo array, passed QC mimicking the criteria used for COGEND and had complete FTND and covariate data.
AAND
The AAND cohort was designed to mimic COGEND, whereby nicotine-dependent cases were compared with non-dependent smoking controls. AAND participants were recruited from the Chicago area (2011–2013) and administered the FTND to determine their study eligibility. Genotyping was conducted using the Illumina Omni Express array. Following the same QC procedures used in COGEND, the final analysis dataset included 1281 AAs (1038 cases with FTND ≥ 4 and 243 controls with FTND ≤ 1).
EAGLE
The EAGLE cohort, a population-based study of newly diagnosed lung cancer cases and matched controls from the Lombardy region of Italy (43,44), was genotyped using the Illumina HumanHap550v3 array as part of GENEVA. We obtained the SNP genotypes and the FTND and other phenotype data for the EAGLE cohort via dbGaP accession number phs000093.v2.p2. We began with the genotyped SNPs and participants that passed all QC criteria recommended by the original study investigators and then removed additional participants with chromosomal anomalies or with incomplete FTND data. Unlike COGEND participants who were ascertained specifically for nicotine dependence, EAGLE participants were ascertained for lung cancer. Cigarette smoking behavior is influenced by the onset and progression of lung cancer. However, because FTND data in EAGLE were collected based on lifetime smoking habits among current and former smokers, both lung cancer cases and controls were included in the SNP association analyses with nicotine dependence. The final analysis dataset included 2309 Italians who were former or current smokers (1596 cases with FTND ≥ 4 and 713 controls with FTND ≤ 1).
ARIC
The ARIC study (45) (dbGaP accession number phs000090.v1.p1) is a community-based longitudinal study of 12 771 participants, aged 45–64 years at the baseline examination, who were genotyped using the Affymetrix 6.0 array. We created a nested case–control study by taking EAs and AAs in ARIC who passed QC and reported ever smoking (as defined during ascertainment by smoking >400 cigarettes during their lifetime). FTND questionnaire data were not collected in ARIC, so we used cigarettes per day (CPD) as a proxy measure and defined 2145 heavy smoking cases (1907 EAs and 238 AAs) based on ≥21 CPD and 1493 controls (999 EAs and 494 AAs) based on ≤10 CPD.
Genotype imputation
Genotype imputation was conducted, separately by study cohort and ancestry group, with reference to the 1000 Genomes ALL Phase I integrated variant set release (v3) haplotype panel (April 19, 2012 release available at http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html). Genotype imputation was preceded by pre-phasing of the study genotypes using SHAPEIT2 (75); default settings were used except for specifying 500 conditioning states and effective population sizes of 11 418 for EAs/Europeans and 15 000 for AAs. Imputation based on the resulting haplotype estimates was performed using IMPUTE2 (76) on 5 MB chromosomal chunks with 1 MB flanking buffers. We chose to impute SNP genotypes using IMPUTE2 because our previous evaluation of imputation in AAs showed that IMPUTE2 resulted in higher imputation quality when compared with three other programs (77). The default settings in IMPUTE2 were used, except the expected numbers of most useful reference haplotypes were specified as 468 for AAs [number of haplotypes from AA (denoted ASW), EA (denoted CEU) and West African (denoted YRI) reference populations], 170 for EAs (number of CEU haplotypes) and 758 for Europeans (number of haplotypes from 5 European reference populations, collectively denoted as EUR). Following imputation, all 80 SNPs tested in BrainCloud were available for association testing with nicotine dependence across COGEND, SAGE*, EAGLE, ARIC, and AAND. The SNP genotype probabilities were converted to dosages and used in the regression model for association testing with nicotine dependence to account for any imputation uncertainty (78).
Association analyses with nicotine dependence
Nicotine-dependent cases and controls were defined using the FTND, a six-item questionnaire with scores ranging from 0 to 10 to indicate the level of physiological dependence (70,79), in COGEND, SAGE*, AAND and EAGLE. Among individuals who smoked >100 cigarettes during their lifetime, we defined dependent cases based on FTND ≥ 4 and non-dependent controls based on FTND ≤ 1. FTND data were not collected in ARIC, so we used CPD as a proxy measure for nicotine dependence, defining cases (≥21 CPD) and controls (≤10 CPD) among participants who reported smoking. Prior studies have shown high agreement between heavy smoking defined by CPD and nicotine dependence defined by FTND (80). The final analyses included 7858 European ancestry participants (5549 EAs and 2309 Italians) and 3238 AAs (Table 1).
We used logistic regression models in ProbABEL (81) to test SNPs or haplotypes for association with nicotine dependence, adjusting for age, sex and up to three principal component eigenvectors to limit any bias due to population stratification. SNPs were either genotyped or 1000 Genomes-imputed with high quality (info > 0.9) across the cohorts. The principal component eigenvectors were generated using genotyped SNPs in linkage in EIGENSTRAT (82). In SAGE*, additional adjustment was needed for DSM-IV-defined cocaine dependence and alcohol dependence given its ascertainment for these other addiction disorders. Models were run separately by cohort and by ancestry group, and results were combined using fixed-effects, inverse, variance-weighted meta-analysis.
Causal inference
To infer the causal relationships between our implicated SNPs, CHRNA5 methylation and CHRNA5 expression, we used structural equation modeling in the R package lavaan (83), following the approach taken by Liu et al. (84). SNP genotype and covariates of age, sex and ancestral group were included as predictors across all six alternative models (Supplementary Material, Fig. S2), and expression and methylation were varied as the dependent outcome or a mediating variable in the BrainCloud cohort with the fetal samples excluded. The covariates were bundled into a single composite variable, which enabled us to incorporate the multifaceted influences of age, sex and ancestral group and focus our interpretation on the pathways describing the relationship between SNP genotype, methylation and expression; the valid use of a composite variable relies on it influencing at least two distinct outcomes, which is a reasonable assumption in our models with the composite variable influencing both methylation and expression (85). The fit for each of six alternative causal models (Fig. 3) was assessed using standard structural equation modeling fit indices (86).
The tested SNPs were not associated with ancestry group (P > 0.05), so models were run with the ancestry groups combined. All methylation and expression levels were observed within 3.1 standard deviations from the mean levels. Besides no population stratification and no outliers, the inferences drawn from the causal models rely on other underlying assumptions, including sufficient sample size, no unmeasured confounders, multivariate normality and linearity, and correct model specification. Missing data are also assumed to be completely random; 76 of the 78 participants input into our analyses had no missing data.
Bioinformatics analyses
LD patterns and the number of tag SNPs were discerned using Haploview (87) and its embedded Tagger program (88), respectively, with reference to the 1000 Genomes populations of European (denoted EUR) or African (denoted AFR) ancestry. SNPs were also annotated for their regulatory potential using the HaploReg v2 database (89), which provides information on chromatin states, conservation and regulatory motif alterations from the Encyclopedia of DNA Elements and elsewhere.
Supplementary material
Funding
This work was supported by the National Institutes of Health (NIH)/National Institute on Drug Abuse (NIDA) (grant numbers R01 DA035825, R01 DA025888 and R01 DA036583). Funding acknowledgements for each of the cohorts used in this study are provided below.
Collection of the BrainCloud cohort data was conducted through a collaborative study sponsored by the NIH/National Institute of Mental Health (NIMH) Intramural Research Program. Submission of the data to dbGaP was provided by Drs Barbara Lipska and Joel Kleinman.
Funding support for the Brain QTL study was provided through the Division of Aging Biology and the Division of Geriatrics and Clinical Gerontology, NIH/National Institute on Aging (NIA). The Brain QTL study includes a GWAS funded as part of the Intramural Research Program, NIA.
Funding support for collection of the COGEND dataset and its analyses was provided by the NIH/National Cancer Institute (NCI) (grant number P01 CA089392). Funding support for genotyping, which was performed at the Center for Inherited Disease Research (CIDR), was provided by grant number X01 HG005274 and by the NIH Genes, Environment and Health Initiative (GEI) (grant number U01 HG004422) of the National Human Genome Research Institute (NHGRI). CIDR is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract number HHSN268200782096C). Assistance with genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (grant number U01 HG004446).
Funding support for SAGE was provided through the NIH GEI (grant number U01 HG004422). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (grant number U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information (NCBI). Support for collection of datasets and samples used for this manuscript was provided through COGA (grant number U10 AA008401) and FSCD (grant number R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University CIDR, was provided by the NIH GEI (grant number U01 HG004438), the National Institute on Alcohol Abuse and Alcoholism (NIAAA), NIDA and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (contract number HHSN268200782096C).
Funding support for the EAGLE participants derived from the GWAS of Lung Cancer and Smoking as provided through the NIH GEI (contract number Z01 CP 010200). This study is supported by intramural resources of the NCI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (grant number U01 HG004446). Assistance with data cleaning was provided by NCBI. Funding support for genotyping, which was performed at the Johns Hopkins University CIDR, was provided by the NIH GEI (grant number U01 HG004438).
The ARIC cohort was carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (NHLBI) (contract numbers HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C). Genotyping was conducted by GENEVA. Funding for GENEVA was provided by the NHGRI (grant number U01HG004402).
Supplementary Material
Acknowledgements
The authors thank the BrainCloud investigators, staff and participants for their contributions that made this study possible, including the sharing of surrogate variables to conduct the follow-up SNP-methylation association analyses. The authors also thank the investigators, staff and participants of the Brain QTL, COGEND, SAGE, EAGLE, AAND and ARIC study cohorts for their important contributions that are represented in this study.
The data used for the cis-meQTL testing were obtained via dbGaP for the BrainCloud (accession number phs000417.v1.p1) and Brain QTL (accession number phs000249.v1.p1) cohorts. The BrainCloud and BrainCloudMethyl applications were downloaded from http://braincloud.jhmi.edu/.
Data used for nicotine dependence association testing have also been made available to the scientific community via dbGaP: COGEND (accession number phs000404.v1.p1), SAGE (accession number phs000092.v1.p1), EAGLE (accession number phs000093.v2.p2) and ARIC (accession number phs000090.v1.p1).
Conflict of Interest statement. J.-C.W., A.G. and L.J.B. are listed as inventors on an issued patent ‘Markers for Addiction’ (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. N.L.S. is the spouse of another inventor listed on this same patent.
References
- 1.U.S. Department of Health and Human Services. (2014) The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. Atlanta, GA. [Google Scholar]
- 2.World Health Organization. (2008) WHO Report on the Global Tobacco Epidemic, 2009. Geneva, Switzerland. [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC). (2011) Vital signs: current cigarette smoking among adults aged >/=18 years—United States, 2005–2010. MMWR Morb. Mortal. Wkly. Rep., 60, 1207–1212. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). (2011) Quitting smoking among adults—United States, 2001–2010. MMWR Morb. Mortal. Wkly. Rep., 60, 1513–1519. [PubMed] [Google Scholar]
- 5.Breslau N., Johnson E.O. (2000) Predicting smoking cessation and major depression in nicotine-dependent smokers. Am. J. Public Health, 90, 1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker T.B., Piper M.E., McCarthy D.E., Bolt D.M., Smith S.S., Kim S.Y., Colby S., Conti D., Giovino G.A., Hatsukami D. et al. (2007) Time to first cigarette in the morning as an index of ability to quit smoking: implications for nicotine dependence. Nicotine Tob. Res., 9 (Suppl. 4), S555–S570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlowski L.T., Porter C.Q., Orleans C.T., Pope M.A., Heatherton T. (1994) Predicting smoking cessation with self-reported measures of nicotine dependence: FTQ, FTND, and HSI. Drug Alcohol Depend., 34, 211–216. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan P.F., Kendler K.S. (1999) The genetic epidemiology of smoking. Nicotine Tob. Res., 1 (Suppl. 2), S51–S57; discussion S69–S70. [DOI] [PubMed] [Google Scholar]
- 9.Bierut L.J., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O.F., Swan G.E., Rutter J., Bertelsen S., Fox L. et al. (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet., 16, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccone S.F., Hinrichs A.L., Saccone N.L., Chase G.A., Konvicka K., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O. et al. (2007) Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet., 16, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorgeirsson T.E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K.P., Manolescu A., Thorleifsson G., Stefansson H., Ingason A. et al. (2008) A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature, 452, 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S. et al. (2010) Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature Genet., 42, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobacco and Genetics Consortium. (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genet., 42, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J.Z., Tozzi F., Waterworth D.M., Pillai S.G., Muglia P., Middleton L., Berrettini W., Knouff C.W., Yuan X., Waeber G. et al. (2010) Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature Genet., 42, 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David S.P., Hamidovic A., Chen G.K., Bergen A.W., Wessel J., Kasberger J.L., Brown W.M., Petruzella S., Thacker E.L., Kim Y. et al. (2012) Genome-wide meta-analyses of smoking behaviors in African Americans. Transl. Psychiatry, 2, e119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amos C.I., Wu X., Broderick P., Gorlov I.P., Gu J., Eisen T., Dong Q., Zhang Q., Gu X., Vijayakrishnan J. et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P. et al. (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 18.Wilk J.B., Shrine N.R., Loehr L.R., Zhao J.H., Manichaikul A., Lopez L.M., Smith A.V., Heckbert S.R., Smolonska J., Tang W. et al. (2012) Genome wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am. J. Respir. Crit. Care Med., 186, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A. et al. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet., 5, e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho M.H., Castaldi P.J., Wan E.S., Siedlinski M., Hersh C.P., Demeo D.L., Himes B.E., Sylvia J.S., Klanderman B.J., Ziniti J.P. et al. (2012) A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet., 21, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joslyn G., Brush G., Robertson M., Smith T.L., Kalmijn J., Schuckit M., White R.L. (2008) Chromosome 15q25.1 genetic markers associated with level of response to alcohol in humans. Proc. Natl. Acad. Sci. USA, 105, 20368–20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlaepfer I.R., Hoft N.R., Collins A.C., Corley R.P., Hewitt J.K., Hopfer C.J., Lessem J.M., McQueen M.B., Rhee S.H., Ehringer M.A. (2008) The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol. Psychiatry, 63, 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grucza R.A., Wang J.C., Stitzel J.A., Hinrichs A.L., Saccone S.F., Saccone N.L., Bucholz K.K., Cloninger C.R., Neuman R.J., Budde J.P. et al. (2008) A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol. Psychiatry, 64, 922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherva R., Kranzler H.R., Yu Y., Logue M.W., Poling J., Arias A.J., Anton R.F., Oslin D., Farrer L.A., Gelernter J. (2010) Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology, 35, 1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erlich P.M., Hoffman S.N., Rukstalis M., Han J.J., Chu X., Linda Kao W.H., Gerhard G.S., Stewart W.F., Boscarino J.A. (2010) Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum. Genet., 128, 491–499. [DOI] [PubMed] [Google Scholar]
- 26.Bierut L.J., Stitzel J.A., Wang J.C., Hinrichs A.L., Grucza R.A., Xuei X., Saccone N.L., Saccone S.F., Bertelsen S., Fox L. et al. (2008) Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry, 165, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J.C., Cruchaga C., Saccone N.L., Bertelsen S., Liu P., Budde J.P., Duan W., Fox L., Grucza R.A., Kern J. et al. (2009) Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum. Mol. Genet., 18, 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J.C., Spiegel N., Bertelsen S., Le N., McKenna N., Budde J.P., Harari O., Kapoor M., Brooks A., Hancock D. et al. (2013) Cis-regulatory variants affect CHRNA5 mRNA expression in populations of African and European ancestry. PLoS One, 8, e80204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith R.M., Alachkar H., Papp A.C., Wang D., Mash D.C., Wang J.C., Bierut L.J., Sadee W. (2011) Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5′ upstream polymorphisms. Eur. J. Hum. Genet., 19, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet., 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamazon E.R., Badner J.A., Cheng L., Zhang C., Zhang D., Cox N.J., Gershon E.S., Kelsoe J.R., Greenwood T.A., Nievergelt C.M. et al. (2013) Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol. Psychiatry, 18, 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J. et al. (2010) Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet., 6, e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherf D.B., Sarkisyan N., Jacobsson H., Claus R., Bermejo J.L., Peil B., Gu L., Muley T., Meister M., Dienemann H. et al. (2013) Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene, 32, 3329–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paliwal A., Vaissiere T., Krais A., Cuenin C., Cros M.P., Zaridze D., Moukeria A., Boffetta P., Hainaut P., Brennan P. et al. (2010) Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res., 70, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balassiano K., Lima S., Jenab M., Overvad K., Tjonneland A., Boutron-Ruault M.C., Clavel-Chapelon F., Canzian F., Kaaks R., Boeing H. et al. (2011) Aberrant DNA methylation of cancer-associated genes in gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Cancer Lett., 311, 85–95. [DOI] [PubMed] [Google Scholar]
- 36.Lambert M.P., Paliwal A., Vaissiere T., Chemin I., Zoulim F., Tommasino M., Hainaut P., Sylla B., Scoazec J.Y., Tost J. et al. (2011) Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J. Hepatol., 54, 705–715. [DOI] [PubMed] [Google Scholar]
- 37.Yalachkov Y., Kaiser J., Naumer M.J. (2009) Brain regions related to tool use and action knowledge reflect nicotine dependence. J. Neurosci., 29, 4922–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson S.J., Sayette M.A., Fiez J.A. (2004) Prefrontal responses to drug cues: a neurocognitive analysis. Nature Neurosci., 7, 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Numata S., Ye T., Hyde T.M., Guitart-Navarro X., Tao R., Wininger M., Colantuoni C., Weinberger D.R., Kleinman J.E., Lipska B.K. (2012) DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet., 90, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colantuoni C., Lipska B.K., Ye T., Hyde T.M., Tao R., Leek J.T., Colantuoni E.A., Elkahloun A.G., Herman M.M., Weinberger D.R. et al. (2011) Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature, 478, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipska B.K., Deep-Soboslay A., Weickert C.S., Hyde T.M., Martin C.E., Herman M.M., Kleinman J.E. (2006) Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry, 60, 650–658. [DOI] [PubMed] [Google Scholar]
- 42.Rice J.P., Hartz S.M., Agrawal A., Almasy L., Bennett S., Breslau N., Bucholz K.K., Doheny K.F., Edenberg H.J., Goate A.M. et al. (2012) CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction, 107, 2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landi M.T., Chatterjee N., Yu K., Goldin L.R., Goldstein A.M., Rotunno M., Mirabello L., Jacobs K., Wheeler W., Yeager M. et al. (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landi M.T., Consonni D., Rotunno M., Bergen A.W., Goldstein A.M., Lubin J.H., Goldin L., Alavanja M., Morgan G., Subar A.F. et al. (2008) Environment and genetics in lung cancer etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health, 8, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The ARIC investigators. (1989) The atherosclerosis risk in communities (ARIC) study: design and objectives. Am. J. Epidemiol., 129, 687–702. [PubMed] [Google Scholar]
- 46.Saccone N.L., Wang J.C., Breslau N., Johnson E.O., Hatsukami D., Saccone S.F., Grucza R.A., Sun L., Duan W., Budde J. et al. (2009) The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res., 69, 6848–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamidovic A., Kasberger J.L., Young T.R., Goodloe R.J., Redline S., Buxbaum S.G., Benowitz N.L., Bergen A.W., Butler K.R., Franceschini N. et al. (2011) Genetic variability of smoking persistence in African Americans. Cancer Prev. Res. (Phila.), 4, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broms U., Wedenoja J., Largeau M.R., Korhonen T., Pitkaniemi J., Keskitalo-Vuokko K., Happola A., Heikkila K.H., Heikkila K., Ripatti S. et al. (2012) Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits. Nicotine Tob. Res., 14, 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith G.D., Ebrahim S. (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol., 32, 1–22. [DOI] [PubMed] [Google Scholar]
- 50.Pedneault M., Labbe A., Roy-Gagnon M.H., Low N.C., Dugas E., Engert J.C., O'Loughlin J. (2014) The association between CHRN genetic variants and dizziness at first inhalation of cigarette smoke. Addict. Behav., 39, 316–320. [DOI] [PubMed] [Google Scholar]
- 51.Wang J.C., Grucza R., Cruchaga C., Hinrichs A.L., Bertelsen S., Budde J.P., Fox L., Goldstein E., Reyes O., Saccone N. et al. (2009) Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry, 14, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falvella F.S., Galvan A., Colombo F., Frullanti E., Pastorino U., Dragani T.A. (2010) Promoter polymorphisms and transcript levels of nicotinic receptor CHRNA5. J. Natl. Cancer Inst., 102, 1366–1370. [DOI] [PubMed] [Google Scholar]
- 53.Fowler C.D., Lu Q., Johnson P.M., Marks M.J., Kenny P.J. (2011) Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature, 471, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fowler C.D., Tuesta L., Kenny P.J. (2013) Role of alpha5* nicotinic acetylcholine receptors in the effects of acute and chronic nicotine treatment on brain reward function in mice. Psychopharmacology, 229, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J.C., Bierut L.J., Goate A.M. (2009) Variants weakly correlated with CHRNA5 D398N polymorphism should be considered in transcriptional deregulation at the 15q25 locus associated with lung cancer risk. Clin. Cancer Res., 15, 5599; author reply 5599. [DOI] [PubMed] [Google Scholar]
- 56.Saccone N.L., Culverhouse R.C., Schwantes-An T.H., Cannon D.S., Chen X., Cichon S., Giegling I., Han S., Han Y., Keskitalo-Vuokko K. et al. (2010) Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet ., 6, e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldstein R.Z., Volkow N.D. (2011) Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat. Rev. Neurosci., 12, 652–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wise R.A. (1996) Neurobiology of addiction. Curr. Opin. Neurobiol., 6, 243–251. [DOI] [PubMed] [Google Scholar]
- 59.Yalachkov Y., Kaiser J., Gorres A., Seehaus A., Naumer M.J. (2013) Sensory modality of smoking cues modulates neural cue reactivity. Psychopharmacology, 225, 461–471. [DOI] [PubMed] [Google Scholar]
- 60.Goudriaan A.E., de Ruiter M.B., van den Brink W., Oosterlaan J., Veltman D.J. (2010) Brain activation patterns associated with cue reactivity and craving in abstinent problem gamblers, heavy smokers and healthy controls: an fMRI study. Addict. Biol., 15, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang O.S., Chang D.S., Jahng G.H., Kim S.Y., Kim H., Kim J.W., Chung S.Y., Yang S.I., Park H.J., Lee H. et al. (2012) Individual differences in smoking-related cue reactivity in smokers: an eye-tracking and fMRI study. Prog. Neuropsychopharmacol. Biol. Psychiatry, 38, 285–293. [DOI] [PubMed] [Google Scholar]
- 62.Rose J.E., Behm F.M., Salley A.N., Bates J.E., Coleman R.E., Hawk T.C., Turkington T.G. (2007) Regional brain activity correlates of nicotine dependence. Neuropsychopharmacology, 32, 2441–2452. [DOI] [PubMed] [Google Scholar]
- 63.Maskos U. (2008) The cholinergic mesopontine tegmentum is a relatively neglected nicotinic master modulator of the dopaminergic system: relevance to drugs of abuse and pathology. Br. J. Pharmacol., 153 (Suppl. 1), S438–S445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miquel M., Toledo R., Garcia L.I., Coria-Avila G.A., Manzo J. (2009) Why should we keep the cerebellum in mind when thinking about addiction? Curr. Drug Abuse Rev., 2, 26–40. [DOI] [PubMed] [Google Scholar]
- 65.Jasinska A.J., Stein E.A., Kaiser J., Naumer M.J., Yalachkov Y. (2014) Factors modulating neural reactivity to drug cues in addiction: a survey of human neuroimaging studies. Neurosci. Biobehav. Rev., 38, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng J., Nestler E.J. (2013) Epigenetic mechanisms of drug addiction. Curr. Opin. Neurobiol., 23, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernandez D.G., Nalls M.A., Moore M., Chong S., Dillman A., Trabzuni D., Gibbs J.R., Ryten M., Arepalli S., Weale M.E. et al. (2012) Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol. Dis., 47, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leek J.T., Storey J.D. (2007) Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet., 3, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heatherton T.F., Kozlowski L.T., Frecker R.C., Fagerstrom K.O. (1991) The Fagerstrom test for nicotine dependence: a revision of the Fagerstrom tolerance questionnaire. Br. J. Addict., 86, 1119–1127. [DOI] [PubMed] [Google Scholar]
- 71.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J. et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol., 34, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson E.O., Hancock D.B., Levy J.L., Gaddis N.C., Saccone N.L., Bierut L.J., Page G.P. (2013) Imputation across genotyping arrays for genome-wide association studies: assessment of bias and a correction strategy. Hum. Genet., 132, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edenberg H.J. (2002) The collaborative study on the genetics of alcoholism: an update. Alcohol Res. Health., 26, 214–218. [PMC free article] [PubMed] [Google Scholar]
- 74.Bierut L.J., Strickland J.R., Thompson J.R., Afful S.E., Cottler L.B. (2008) Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend., 95, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delaneau O., Zagury J., Marchini J. (2013) Improved whole chromosome phasing for disease and population genetic studies. Nat. Methods, 10, 5–6. [DOI] [PubMed] [Google Scholar]
- 76.Howie B., Marchini J., Stephens M. (2011) Genotype imputation with thousands of genomes. G3 (Bethesda), 1, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hancock D.B., Levy J.L., Gaddis N.C., Bierut L.J., Saccone N.L., Page G.P., Johnson E.O. (2012) Assessment of genotype imputation performance using 1000 Genomes in African American studies. PLoS One, 7, e50610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchini J., Howie B. (2010) Genotype imputation for genome-wide association studies. Nat Rev. Genet., 11, 499–511. [DOI] [PubMed] [Google Scholar]
- 79.Piper M.E., McCarthy D.E., Baker T.B. (2006) Assessing tobacco dependence: a guide to measure evaluation and selection. Nicotine Tob. Res., 8, 339–351. [DOI] [PubMed] [Google Scholar]
- 80.Stevens V.L., Bierut L.J., Talbot J.T., Wang J.C., Sun J., Hinrichs A.L., Thun M.J., Goate A., Calle E.E. (2008) Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol. Biomarkers Prev., 17, 3517–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinf., 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 83.Rosseel Y. (2012) Lavaan: an R package for structural equation modeling. J. Stat. Softw., 48, 1–36. [Google Scholar]
- 84.Liu Y., Ding J., Reynolds L.M., Lohman K., Register T.C., De La Fuente A., Howard T.D., Hawkins G.A., Cui W., Morris J. et al. (2013) Methylomics of gene expression in human monocytes. Hum. Mol. Genet., 22, 5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grace J.B., Bollen K.A. (2006) The interface between theory and data in structural equation models: U.S. Geological Survey Open-File Report 2006–1363, p. 33. [Google Scholar]
- 86.Hooper D., Coughlan J., Mullen M.R. (2008) Structural equation modeling: guidelines for determining model fit. Electron. J. Bus. Res. Methods, 6, 53–60. [Google Scholar]
- 87.Barrett J.C. (2009) Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc., 2009, pdb ip71. [DOI] [PubMed] [Google Scholar]
- 88.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. (2005) Efficiency and power in genetic association studies. Nature Genet., 37, 1217–1223. [DOI] [PubMed] [Google Scholar]
- 89.Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.