Abstract
Epidemiological studies have demonstrated associations between endometriosis and certain histotypes of ovarian cancer, including clear cell, low-grade serous and endometrioid carcinomas. We aimed to determine whether the observed associations might be due to shared genetic aetiology. To address this, we used two endometriosis datasets genotyped on common arrays with full-genome coverage (3194 cases and 7060 controls) and a large ovarian cancer dataset genotyped on the customized Illumina Infinium iSelect (iCOGS) arrays (10 065 cases and 21 663 controls). Previous work has suggested that a large number of genetic variants contribute to endometriosis and ovarian cancer (all histotypes combined) susceptibility. Here, using the iCOGS data, we confirmed polygenic architecture for most histotypes of ovarian cancer. This led us to evaluate if the polygenic effects are shared across diseases. We found evidence for shared genetic risks between endometriosis and all histotypes of ovarian cancer, except for the intestinal mucinous type. Clear cell carcinoma showed the strongest genetic correlation with endometriosis (0.51, 95% CI = 0.18–0.84). Endometrioid and low-grade serous carcinomas had similar correlation coefficients (0.48, 95% CI = 0.07–0.89 and 0.40, 95% CI = 0.05–0.75, respectively). High-grade serous carcinoma, which often arises from the fallopian tubes, showed a weaker genetic correlation with endometriosis (0.25, 95% CI = 0.11–0.39), despite the absence of a known epidemiological association. These results suggest that the epidemiological association between endometriosis and ovarian adenocarcinoma may be attributable to shared genetic susceptibility loci.
Introduction
Ovarian cancer is the most fatal gynaecological cancer. It is not a single disease but comprises a number of histotypes (1). The most common subtype is serous carcinoma, accounting for over 50% of all invasive epithelial ovarian cancers (EOC). Recent morphological and molecular genetic studies have confirmed that invasive serous EOC should be further sub-categorized into high-grade and low-grade types (2,3). The clear cell, endometrioid and mucinous EOCs are three most common non-serous tumours. The heterogeneity among EOC subtypes is manifested in differences in risk factors, germline and somatic mutations, gene expression and chemotherapy responsiveness (4).
Endometriosis is a common gynaecological disorder associated with pelvic pain and sub-fertility, affecting 10–15% of women of reproductive age (5). The disease is defined as the presence of endometrial-like tissue outside the uterine cavity, primarily on pelvic organs. The disease is typically staged using the revised American Fertility Society (rAFS) classification (6) based on the differential location of the lesions (ovarian, peritoneal, recto-vaginal), extent of disease and adhesion formation.
The epidemiological link between endometriosis and ovarian cancer was first identified in 1925 (7) and has been largely replicated since (8). Some studies have suggested that the link might differ between ovarian cancer histotypes (9–11). Recently, a large collaborative effort by the Ovarian Cancer Association Consortium (OCAC) reported that endometriosis increases the risk of clear cell ovarian cancer by 3-fold, and the risk of low-grade serous and endometrioid subtypes by ∼2-fold, but that endometriosis was not associated with other histotypes (12).
Despite the convincing epidemiological link, the mechanisms underlying the co-occurrence of endometriosis and ovarian cancer are unknown. Several studies have attempted to demonstrate a causal relationship between the two conditions via the identification of somatic mutations that may represent early events in the transformation of benign endometriotic lesions in the ovary (endometrioma) to malignancy. For example, one study identified truncating mutations in ARID1A that cause loss of expression of BAF250 protein in both clear cell and endometrioid tumours, and in two cases the same mutations were found in endometriotic lesions adjacent to the tumours (13).
An alternative explanation is that the observed associations may arise from shared germline genetic risk factors, but this has not been well explored (5,14). Traditionally, studying genetic correlation requires large family or twin studies with phenotypic data for both diseases. However, because of the rare co-occurrence of EOC (particularly of the rare histotypes) and endometriosis in families, ascertaining sufficient cases for family studies is difficult. Recently, germline genetic variants predisposing women to the two diseases have been identified from genome-wide association studies (GWAS) (15–21). With the exception of the locus at 1p36 (nearest gene: WNT4) (20,22), there is limited overlap in the susceptibility loci for the two diseases, which seems to lend little support to the hypothesis of shared genetic factors underlying the two diseases. However, we and others have previously shown that germline genetic contributions to ovarian cancer and to endometriosis are not simply limited to the genome-wide significant variants from GWAS; instead, many more genetic variants that do not pass the genome-wide significant threshold contribute to the disease heritability (23,24). Therefore, to study shared genetic risks between the two diseases, we need to extend beyond genome-wide significant variants.
In the current study, we applied two complementary statistical genetic methods, genomic-relatedness-matrix restricted maximum likelihood (GREML) (25,26) and genetic risk prediction (GRP) (27), both of which evaluate the joint contribution of all germline genetic variants captured on the genotyping arrays. The first method predicts the phenotypic similarities from the genomic relatedness between individuals with distant relationships, thus estimates array heritability (that is, the heritability attributable to the variants on the genotyping array) in the univariate case or genetic correlation/co-heritability in the bivariate case. The second method selects genetic variants from a discovery set according to their associations with the trait assessed and constructs polygenic risk scores in an independent replication set using the selected variants and weights from the discovery. An association of risk scores with the trait in the replication set indicates genetic risks overlap between the two traits assessed. Both methods allow the use of independent datasets for each phenotype of interest, thus circumventing the ascertainment issue in traditional quantitative genetic designs. In addition to investigating the shared genetic risks between endometriosis and ovarian cancer, we also estimated the array heritability specific for EOC histotypes.
Results
Array heritabilities of EOC histotypes
In previous work, we applied the GREML method implemented in the software Genome-wide Complex Trait Analysis (GCTA) to ovarian cancer (all histotypes combined) and estimated that the array heritability was 30% (24). That is, 30% of variance in risk of ovarian cancer was attributable to the common variants on commercial genotyping arrays. However, the sample size in the previous study was insufficient for the estimation of array heritability for the various histotypes. Here, we used a larger collection of ovarian cancer case–control data from the OCAC genotyped using the customized Illumina Infinium iSelect arrays (iCOGS; sample sizes by histotypes and study sites in Supplementary Material, Table S1). We found significant, albeit small, array heritabilities for all major invasive EOC histotypes except for mucinous and low-grade serous diseases (Table 1). The most common EOC subtype, high-grade serous, had the highest array heritability, 8.8% (95% CI = 6.8–10.8%), followed by clear cell, 6.7% (95% CI = 0.3–13.1%), and endometrioid, 3.2% (95% CI = 0.06–6.3%). The mucinous and low-grade serous EOCs were estimated to have null array heritability. Overall, 5.6% (95% CI = 4.4–6.7%) of variance on the liability scale can be explained by the SNPs on the iCOGS array for all invasive EOC, regardless of histotypes. We also carried out a control–control contrast study, dividing 21 663 controls into four pseudo case–control sets (see the Materials and Methods section). We estimated the array heritability in each pseudo case–control set, none of which were significantly different from zero (maximum P = 0.37; Supplementary Material, Table S2).
Table 1.
Array heritability estimated from iCOGS array for invasive EOC, stratified by histotypes
| EOC histotype | N case/control (relatedness <0.1) | K | (95% CI) |
|---|---|---|---|
| High-grade serous | 4121/21 663 (4098/21 242) |
0.0055 | 8.8 (6.8–10.8) |
| Low-grade serous | 363/21 663 (362/21 242) |
0.0005 | 0.0 (0–9.3) |
| Endometrioid | 1350/21 663 (1342/21 242) |
0.001 | 3.2 (0.06–6.3) |
| Mucinous | 662/21 663 (658/21 242) |
0.0005 | 0.0 (0–5.4) |
| Clear cell | 621/21 663 (620/21 242) |
0.0005 | 6.7 (0.3–13.1) |
| All invasive EOC | 10 065/21 663 (10 014/21 242) |
0.009 | 5.6 (4.4–6.7) |
N case/control (relatedness <0.1): case/control sample size, and case/control sample size restricting pairwise genetic relatedness between individuals <0.1 (see the Supplementary Material, Table S3, for comparison of results using the cut-off of 0.05); K: disease prevalence of EOC histotype, defined as the lifetime risk of invasive ovarian cancer (∼0.9%, see the Materials and Methods section) times the fraction of the corresponding EOC histotype; (95% CI): array heritability and its 95% confidence intervals that were estimated using ovarian cancer iCOGS data, while adjusting for 10 PCs and study sites.
Shared genetics between EOC subtypes and endometriosis
When assessing shared genetics between ovarian cancer subtypes and endometriosis, the number of SNPs common to the arrays used in these two datasets was reduced to ∼84 K. Nonetheless, we found strong genetic correlations between endometriosis and clear cell EOC (rg = 0.51, 95% CI = 0.18–0.84) and endometriosis and endometrioid EOC (rg = 0.48, 95% CI = 0.07–0.89), and a moderate genetic correlation between endometriosis and serous EOC (rg = 0.29, 95% CI = 0.16–0.42). Interestingly, despite a null array heritability for low-grade serous EOC, its genetic correlation with endometriosis was significant (rg = 0.40, 95% CI = 0.05–0.75). The correlation coefficient was lower for high-grade serous EOC (rg = 0.25, 95% CI = 0.11–0.39) compared with low-grade serous disease. In contrast, no evidence was apparent for genetic correlation between endometriosis and the mucinous histotype (rg = −0.01, 95% CI = −0.44–0.42). Overall, the genetic correlation between all invasive EOC and endometriosis was estimated to be 0.40 (95% CI = 0.26–0.54).
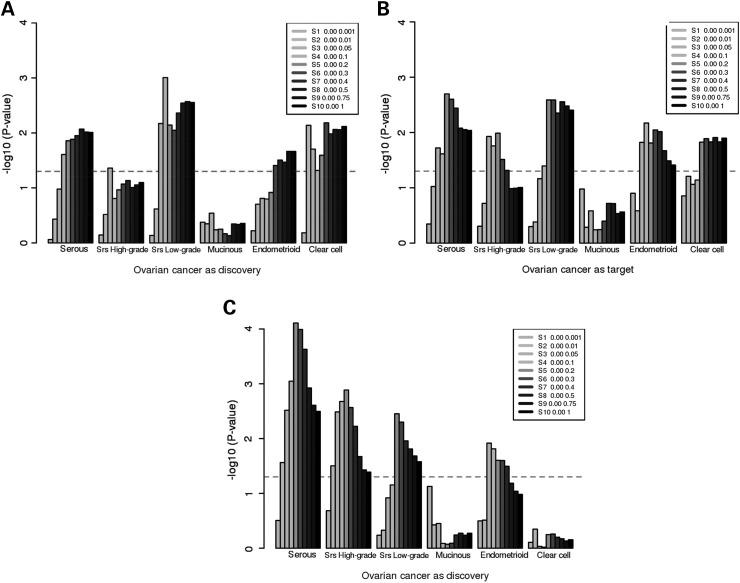
We then performed the GRP analyses in two ways, firstly assessing whether the risk scores calculated from summary results of EOC subtypes were associated with endometriosis, and secondly, whether the risk scores calculated from endometriosis were associated with EOC histotypes. The results from the risk score analyses were highly consistent with the estimated genetic correlations (Fig. 1A and B). The risk scores from low-grade serous, endometrioid and clear cell EOC histotypes predicted endometriosis risks, and the risk scores of endometriosis predicted all EOC subtypes but mucinous. Also, the risk scores of endometriosis were more strongly associated with low-grade serous EOC than with high-grade serous disease. The results remained almost unchanged in sensitivity analyses where we restricted the GRP analysis to the less-related samples from endometriosis and OCAC datasets (genetic relatedness <0.1; Supplementary Material, Fig. S1) or to the set of SNPs clumped to reduce LD (∼44 K SNPs with r2 < 0.2; Supplementary Material, Fig. S2), except that the association between endometriosis and endometrioid EOC was weaker in the latter case.
Figure 1.
Genetic risk prediction between endometriosis and EOC histotypes [serous, high-grade and low-grade serous (denoted as “Srs High-grade” and “Srs Low-grade” respectively), mucinous, endometrioid and clear cell]. (A) EOC histotypes as discovery and endometriosis as target; (B) endometriosis as discovery and EOC histotypes as target; and (C) endometriosis as discovery and EOC histotypes as target, after the exclusion of women with endometriosis in the ovarian cancer case–control set. The figures show the association (y-axis: −log10P) between genetic risk scores calculated from the discovery set and the target disease. The shading denotes the P-value bins used to select SNPs from the discovery set. The dashed horizontal line marks the significance threshold (P = 0.05). The genetic risk scores were calculated from all platform-overlapping SNPs without LD clumping.
The epidemiological association between EOC and endometriosis means that women with ovarian cancer are more likely to have a history of endometriosis compared with the general female population. Hence, we investigated whether the observed genetic overlaps between EOC and endometriosis arose from women with both diseases. We used self-reported data on endometriosis from the OCAC (12) to exclude women with histories of endometriosis or with missing data. A limitation here was that a few OCAC studies did not ascertain endometriosis status; thus, the sample size for this sub-analysis was substantially reduced (nearly 45% reduction). Nonetheless, we found that the risk scores calculated from the discovery set of endometriosis were still significantly associated with high-grade serous, low-grade serous and endometrioid EOC (Fig. 1C). The significance of the genetic overlap between endometriosis and clear cell EOC was, however, considerably attenuated.
Psychiatric Genomics Consortium (PGC) data as negative controls
We used the PGC data for negative control experiments because psychiatric disorders are not likely to share genetic risk factors with the two gynaecological diseases studied here. We examined the genetic overlaps of endometriosis and ovarian cancer with schizophrenia and bipolar disorder in the GRP analyses (28,29). Based on ∼130–170 K overlapping SNPs, we found no associations of risk scores from schizophrenia or bipolar disorder with all invasive EOC (P > 0.1 for all P-values; Supplementary Material, Fig. S3). Since the control subjects from the Wellcome Trust Case Control Consortium (WTCCC) study were included in the PGC data, and also used as the controls for the UK endometriosis study, we only assessed the polygenic risk scores from the two psychiatric disorders in the Australian endometriosis data. Based on analysis of 440–470 K overlapping SNPs, we found no evidence for genetic overlap between endometriosis and schizophrenia or bipolar disorder (P > 0.1 for all P-values; Supplementary Material, Fig. S3).
Discussion
While various studies have demonstrated a consistent epidemiological association between endometriosis and risk of ovarian cancer, the underlying mechanism is not clear. Here, we examined whether shared genetic risk factors, beyond the limited number of genome-wide significant variants, might underlie the observed association.
On the premise of polygenic architecture of both diseases (23,24), we derived the shared genetics from the aggregate effect of germline genetic variants captured on genotyping arrays, using two complementary statistical genetic methods, GREML and GRP. Both methods have been widely applied to investigate shared genetics between traits and diseases (25,27,30–32). The bivariate GREML method provides unbiased estimates of genetic correlation. However, it requires genotype data that may not be readily available in many cases, whereas the GRP method only requires summary results from the discovery set (although care needs to be taken to ensure that no subject overlaps between the discovery and target sets). The results from our control–control contrast and negative control experiments showed that the two methods are reliable and that the results we presented here are unlikely to be artefacts.
Further to our previous work that estimated array heritability for ovarian cancer (all histotypes combined), we reported the array heritabilities from the customized iCOGS genotyping array, for individual histotypes of ovarian cancer. Among five major EOC histotypes, we found significant array heritability for high-grade serous (8.8%, 95% CI = 6.8–10.8%), clear cell (6.7%, 0.3–13.1%) and endometrioid EOC (3.2%, 0.06–6.3%), but not in mucinous or low-grade serous disease. To our knowledge, because of the low disease prevalence, no family or twin studies have examined EOC histotypes. Through the large OCAC dataset, we were able to estimate, for the first time, the array heritabilities specific for the histotypes. However, compared with our previous estimate of 30% heritability for all invasive EOC using Illumina Human610-Quad and Human1M-Duo arrays (24), the estimates presented here are small due to the limited genome coverage on the iCOGS array. The iCOGS array includes 211 K SNPs [195 K post quality control (QC) in our analyses], although many of these are concentrated on particular regions of the genome, with little or no coverage of some genomic regions. In contrast, the 471 K SNPs common to Illumina Human610-Quad and Human1M-Duo arrays provide an even coverage of most of the genome. For the histotypes that did not show significant array heritabilities (mucinous and low-grade serous EOC), the true heritabilities may be too low to be detected with the current case numbers, or the underlying SNPs may not be well represented on the iCOGS array. We acknowledge that the estimates would be more informative if such large dataset were genotyped on the arrays with full-genome coverage.
We found widespread shared genetics between endometriosis and most EOC histotypes. The strong genetic correlations of endometriosis with clear cell EOC (rg = 0.51, 95% CI = 0.18–0.84), with endometrioid EOC (rg = 0.48, 95% CI = 0.07–0.89) and with low-grade serous EOC (rg = 0.40, 95% CI = 0.05–0.75) were consistent with epidemiological links between endometriosis and these EOC histotypes (12). Our results, therefore, suggest that shared genetics partly explains the observed link between endometriosis and EOC for these histotypes. Interestingly, we observed a weaker but significant genetic correlation between endometriosis and high-grade serous EOC (rg = 0.25, 95% CI = 0.11–0.39), despite the absence of a known epidemiological association (12). The weaker correlation may reflect that high-grade serous carcinoma often arises from fallopian tubes (33). The specific lack of association with mucinous EOC dovetails with the lack of epidemiological risk factors for that tumour type (11). Overall, the genetic correlation between all invasive EOC and endometriosis was 0.40 (95% CI = 0.26–0.54). In addition, the shared genetics between endometriosis and EOC histotypes, except for clear cell, remained after the exclusion of women with histories of endometriosis, suggesting that our results were unlikely to be solely attributable to women with both diseases. Compared with the other EOC histotypes, the attenuated results in clear cell EOC were likely due to the largest reduction in the case numbers, which was expected given that the epidemiological association is the most pronounced between endometriosis and this histotype. The genetic overlap between endometriosis and most EOC histotypes suggests that women with endometriosis may be at elevated risk of later EOC. Bearing in mind that the germline genetic contribution to absolute risk is relatively small, it will be of interest in future studies to quantify better how genetic predisposition to endometriosis confers risk of EOC.
In light of substantial heterogeneity among ovarian cancer histotypes (4), here we investigated the shared genetics between endometriosis and ovarian cancer, stratified by ovarian cancer histotypes. Further investigation of the links stratified by different stages of endometriosis may be worthwhile, especially in the localized form of ovarian endometriosis (also known as endometriomas). By rAFS classification, endometriomas fall in Stage III/IV (moderate to severe endometriosis) (6). Interestingly, a previous study showed increased genetic loading in women with moderate to severe endometriosis compared with those with minimal disease (19).
It is worth noting that our results were derived either from SNPs on the iCOGS array or from the overlap of SNPs on the iCOGS and commercial GWAS arrays. Hence, the numbers of SNPs included in the analyses were appreciably smaller than the numbers in a typical GWAS. Analyses can be performed on imputed data; however, since the SNPs on the iCOGS array were not designed to tag the whole genome, imputation would still be limited to certain genomic regions that are represented on the array. Further analyses will be warranted when the more comprehensive genotyping array, Infinium OncoArray-500K BeadChip, which integrates a genome-wide backbone of 250 K tag SNPs, becomes available.
In summary, using the large OCAC dataset genotyped on the iCOGS array, we established that the majority of ovarian cancer histotypes have a polygenic architecture. More importantly, we found that genetic risks overlap between endometriosis and all histotypes of ovarian cancer, except for mucinous. These results suggest that the epidemiological association between endometriosis and ovarian cancer is, at least partly, attributable to shared genetics. Therefore, future studies should focus on identifying common molecular pathways underlying both diseases.
Materials and Methods
Data
We used two GWAS datasets of surgically confirmed endometriosis cases from International Endogene Consortium (IEC) in this study, one from Australia and the other from the UK (19,23). In the Australian study, 2270 women with endometriosis were recruited from QIMR Berghofer Medical Research Institute and genotyped using Illumina Human670Quad BeadArrays. The controls were 1870 unrelated Australians from an adolescent twin study recruited in the same institute, genotyped using Illumina Human610-Quad arrays. The UK cases (n = 924) were recruited through the University of Oxford and also genotyped on Illumina Human670Quad BeadArray. We used 5190 individuals from the WTCCC who were genotyped on the Illumina Human1M-Duo array as the UK controls. We applied standard QC to the Australian set, that is, subjects with >5% missing genotypes were excluded, as were SNPs with minor allele frequencies <0.01, call rates <0.99 or P-values from testing Hardy–Weinberg equilibrium <0.0001 (24). As noted before (23,24), the UK dataset using WTCCC controls required more stringent QC, so additional QC criteria including differential missingness between cases and controls (P < 0.001) and two-locus QC (34) (P < 0.02) were applied. We merged the two datasets for analyses (SNPs with strand-ambiguous alleles were excluded), yielding genotype data on a common set of 483 940 SNPs for 3194 cases and 7060 controls, all of European ancestry (19).
For ovarian cancer, we used data from OCAC study that comprises 47 630 cases and controls from 43 studies genotyped using the iCOGS arrays. This array was designed to accommodate 211 155 SNPs that were selected as either GWAS replication, fine-mapping or candidate SNPs from breast, prostate and ovarian cancer consortia. The details of array design and QC have been described elsewhere (15). We applied similar QC, except that we selected only subjects of European ancestry and with invasive EOC tumours of clearly identified histotypes. In total, data from 10 065 women with invasive EOC (4121, 1350, 662, 621 and 363 for the high-grade serous, endometrioid, mucinous, clear cell and low-grade serous histotypes, respectively) and 21 663 controls, typed for 195 183 SNPs, were available for analyses (see the Supplementary Material, Table S1, for details of individual OCAC studies).
Analysis
We applied two statistical genetic approaches in this study. The first approach was GREML, which involved the estimation of genetic relatedness between subjects using genotyping array data and links the resultant relationship matrix to univariate or bivariate phenotype(s). We used GCTA (35) to construct the relationship matrix using all available SNPs post QC. In the univariate analysis of EOC histotypes, the genetic relatedness of subjects with the corresponding EOC histotype was estimated using variants on the iCOGS arrays and compared with the relatedness among OCAC controls in order to estimate array heritabilities Intuitively, the array heritability is high when case–case and control–control pairs are more genetically similar than case–control pairs (that is, individuals with higher genetic relatedness share more similar phenotype). In the bivariate analyses of endometriosis and EOC histotypes, the genetic relatedness of subjects in one case–control set was compared with relatedness in the other set, thus to estimate genetic correlation (rg) between the two diseases. The genetic correlation is zero when genetic relatedness among cross-trait case–case pairs is the same as cross-trait case–control pairs; it is positive when relatedness among cross-trait case–case pairs is higher than among cross-trait case–control pairs, and negative when relatedness among cross-trait case–case pairs is lower than cross-trait case–control pairs (32).
In the GREML analyses, we excluded closely related individuals to avoid confounding from shared environmental factors. For endometriosis datasets, a very stringent relatedness threshold was applied (genetic relatedness between samples <0.025, approximately equivalent to the relatedness between 3rd and 4th cousins); while for EOC data on iCOGS arrays, we applied a less stringent threshold in order to retain sufficient sample size for rare EOC histotypes (relatedness <0.1, less than first cousin; we also applied a threshold of 0.05 as a sensitivity check, see the Supplementary Material, Table S3). All analyses were adjusted for 10 principal components (PCs) and for study site. We also transformed the array heritability on the observed binary scale to an underlying quantitative liability scale taking into account the disease prevalence. According to the Surveillance, Epidemiology, and End Results Program DevCan database (SEER 18 Incidence and Mortality, 2014 submission; http://surveillance.cancer.gov/devcan/canques.html), the lifetime risk of ovarian cancer by the age of 80 is 1.07% (1.37% by age 95+) in 2010–2012 for non-Hispanic whites (the race of subjects in the present study; 1.00% for all races). Since 80–90% of all ovarian malignancies are invasive disease (36), we used 0.9% as the prevalence of invasive EOC in this study (also used 1% as a sensitivity check; Supplementary Material, Table S3). The prevalence of individual EOC histotype was then calculated as the prevalence of all invasive EOC times the fraction of the corresponding histotype (37) (values of prevalence are listed in Table 1). rg estimates are approximately the same on the observed and the liability scales, thus not dependent on disease prevalence (25).
Our second approach was GRP based on the aggregate effects of many genetic variants; one dataset serves as the discovery set, with associations examined in a second replication set (27). The genetic risk scores of individuals in the replication set were calculated as the sum of their risk alleles weighted by the allelic effects that were estimated from the discovery set. These risk scores were then examined for associations with disease status in the replication set, while adjusting for PCs and study site. This method requires duplicate samples in the discovery and replication sets to be removed (e.g. the same individual present in the endometriosis data and OCAC studies). We used genetic relationship calculated from GCTA to identify duplicate samples (with relatedness >0.85) and excluded one in each pair from the analyses. To assess the impact of relatedness, we also applied the relatedness threshold of 0.1 as a sensitivity check (Supplementary Material, Fig. S1). When calculating risk scores, we used SNPs without clumping for high linkage disequilibrium (LD; we also applied LD clumping, i.e. dropping the SNPs in high LD with index SNPs, as a sensitivity check, Supplementary Material, Fig. S2). The risk scores were calculated in PLINK (38), and logistic regression was performed in R.
Control–control contrast studies and negative control experiments
To evaluate the reliability of the two approaches, we conducted control–control contrast studies using OCAC controls and negative control experiments using data from the PGC. For control–control contrast studies, we split 21 663 OCAC controls into 8 sets (7 sets had 2694 subjects and 1 set had 2805 subjects), which were then randomly assigned as 4 pseudo case–control sets. We performed the univariate GREML analyses on these sets. Since we did not have access to the full genotype data from the PGC, we only performed the GRP analyses in the negative control experiments. The publically available GWAS summary results for the two major psychiatric conditions, schizophrenia and bipolar disorder (both from studies with ∼20 000 case and control subjects) (28,29) were used to calculate the genetic risk scores in our endometriosis and OCAC datasets.
Supplementary Material
Funding
This study was supported by the QIMR Berghofer-Weekend to End Women's Cancers Research Grant (WEWC140014). The endometriosis GWAS was supported by a grant from the Wellcome Trust (WT084766/Z/08/Z) and makes use of WTCCC2 control data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of these data is available from http://www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under awards 076113 and 085475. The QIMR study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (241944, 339462, 389927, 389875, 389891, 389892, 389938, 443036, 442915, 442981, 496610, 496739, 552485 and 552498), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne) and donations from N. Hawkins and S. Hawkins. The COGS project is funded through a European Commission's Seventh Framework Programme Grant (agreement number 223175 HEALTH F2 2009-223175); the Genetic Associations and Mechanisms in Oncology (GAME-ON): A NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). Funding of the constituent studies was provided by the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes of Health Research (MOP-86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA80668, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC67001, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA072720, P30-CA15083, P30-CA008748, P50-CA159981, P50-CA105009, P50-CA136393, R01-CA149429, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063678, R01-CA063682, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA080978, R01-CA083918, R01-CA087538, R01-CA092044, R01-CA095023, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA136924, R03-CA113148, R03-CA115195, U01-CA069417, U01-CA071966, UM1-CA186107, UM1-CA176726 and Intramural research funds); the NIH/National Center for Research Resources/General Clinical Research Center (MO1-RR000056); the US Army Medical Research and Material Command (DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449, W81XWH-10-1-02802); the US Public Health Service (PSA-042205); the National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden-Wurttemberg through Medical Faculty of the University of Ulm (P.685); the German Cancer Research Center; the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; Eve Appeal; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital ‘Womens Health Theme’ and the Royal Marsden Hospital; and WorkSafeBC 14. Investigator-specific funding: Y.L. was supported by the NHMRC Early Career Fellowship. D.R.N. was supported by the NHMRC Fellowship (339462 and 613674) and the Australian Research Council (ARC) Future Fellowship (FT0991022) schemes. A.P.M. was supported by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science (WT098017). G.W.M. was supported by the NHMRC Fellowships Scheme (339446, 619667). K.T.Z. was supported by a Wellcome Trust Research Career Development Fellowship (WT085235/Z/08/Z). G.C.T. is supported by the National Health and Medical Research Council. S.M. was supported by an ARC Future Fellowship.
Supplementary Material
Acknowledgements
We acknowledge with appreciation all the individuals who participated in the QIMR and Oxford endometriosis studies and in the OCAC studies, and the many hospital directors and staff, gynaecologists, general practitioners and pathology services who provided assistance with confirmation of diagnoses, and the many research assistants and interviewers for assistance with the studies. For OCAC studies, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer-Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WYL (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila and Y. Bensman (NJO); M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini, P. Harrington the SEARCH team and ECRIC (SEA); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan, C. Karpinskyj and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, J. Dennis, E. Dicks, K. Michilaidou and K. Kuchenbaker (COGS).
Conflict of Interest Statement. K.T.Z. has been a member of scientific advisory boards for AbbVie, Inc., Bayer Pharma AG and Roche Diagnostics.
Appendix The International Endogene Consortium
Carl A. Anderson1,2, Scott D. Gordon3, Qun Guo4, Anjali K. Henders3, Ann Lambert5, Sang Hong Lee6, Peter Kraft7, Stephen H. Kennedy5, Stuart MacGregor3, Nicholas G. Martin3, Stacey A. Missmer4, Grant W. Montgomery3, Andrew P. Morris1, Dale R. Nyholt3, Jodie N. Painter3, Fenella Roseman5, Susan A. Treloar8, Peter M. Visscher9, Leanne Wallace3 and Krina T. Zondervan1,5.
1Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK; 2Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK; 3Queensland Institute of Medical Research, Herston, QLD, Australia; 4Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA; 5Nuffield Department of Obstetrics and Gynaecology, University of Oxford, John Radcliffe Hospital, Oxford, UK; 6Queensland Brain Institute, The University of Queensland, Brisbane, QLD 4072, Australia; 7Harvard School of Public Health, Boston, MA, USA; 8Centre for Military and Veterans' Health, The University of Queensland, Mayne Medical School, QLD, Australia; 9The University of Queensland Diamantina Institute, Princess Alexandra Hospital, Brisbane, QLD 4102, Australia.
Contributor Information
Collaborators: The International Endogene Consortium (IEC), Carl A. Anderson, Scott D. Gordon, Qun Guo, Anjali K. Henders, Ann Lambert, Sang Hong Lee, Peter Kraft, Stephen H. Kennedy, Stuart Macgregor, Nicholas G. Martin, Stacey A. Missmer, Grant W. Montgomery, Andrew P. Morris, Dale R. Nyholt, Jodie N. Painter, Fenella Roseman, Susan A. Treloar, Peter M. Visscher, Leanne Wallace, and Krina T. Zondervan
References
- 1.Gilks C.B., Prat J. (2009) Ovarian carcinoma pathology and genetics: recent advances. Hum. Pathol., 40, 1213–1223. [DOI] [PubMed] [Google Scholar]
- 2.Kurman R.J., Shih Ie M. (2008) Pathogenesis of ovarian cancer: lessons from morphology and molecular biology and their clinical implications. Int. J. Gynecol. Pathol., 27, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shih Ie M., Kurman R.J. (2004) Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am. J. Pathol., 164, 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bast R.C. Jr., Hennessy B., Mills G.B. (2009) The biology of ovarian cancer: new opportunities for translation. Nat. Rev. Cancer, 9, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayasneh A., Tsivos D., Crawford R. (2011) Endometriosis and ovarian cancer: a systematic review. ISRN Obstet. Gynecol., 2011, 140310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American Society for Reproductive Medicine. (1997) Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril., 67, 817–821. [DOI] [PubMed] [Google Scholar]
- 7.Sampson J.A. (1925) Endometrial carcinoma of the ovary, arising in endometrial tissue in that organ. Arch. Surg., 10, 1–72. [Google Scholar]
- 8.Somigliana E., Vigano P., Parazzini F., Stoppelli S., Giambattista E., Vercellini P. (2006) Association between endometriosis and cancer: a comprehensive review and a critical analysis of clinical and epidemiological evidence. Gynecol. Oncol., 101, 331–341. [DOI] [PubMed] [Google Scholar]
- 9.Gates M.A., Rosner B.A., Hecht J.L., Tworoger S.S. (2010) Risk factors for epithelial ovarian cancer by histologic subtype. Am. J. Epidemiol., 171, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurian A.W., Balise R.R., McGuire V., Whittemore A.S. (2005) Histologic types of epithelial ovarian cancer: have they different risk factors? Gynecol. Oncol., 96, 520–530. [DOI] [PubMed] [Google Scholar]
- 11.Risch H.A., Marrett L.D., Jain M., Howe G.R. (1996) Differences in risk factors for epithelial ovarian cancer by histologic type. Results of a case-control study. Am. J. Epidemiol., 144, 363–372. [DOI] [PubMed] [Google Scholar]
- 12.Pearce C.L., Templeman C., Rossing M.A., Lee A., Near A.M., Webb P.M., Nagle C.M., Doherty J.A., Cushing-Haugen K.L., Wicklund K.G. et al. (2012) Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol., 13, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiegand K.C., Shah S.P., Al-Agha O.M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M.K., Anglesio M.S., Kalloger S.E. et al. (2010) ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med., 363, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigano P., Somigliana E., Chiodo I., Abbiati A., Vercellini P. (2006) Molecular mechanisms and biological plausibility underlying the malignant transformation of endometriosis: a critical analysis. Hum. Reprod. Update, 12, 77–89. [DOI] [PubMed] [Google Scholar]
- 15.Pharoah P.D., Tsai Y.Y., Ramus S.J., Phelan C.M., Goode E.L., Lawrenson K., Buckley M., Fridley B.L., Tyrer J.P., Shen H. et al. (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet., 45, 362–370, 370e361-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode E.L., Chenevix-Trench G., Song H., Ramus S.J., Notaridou M., Lawrenson K., Widschwendter M., Vierkant R.A., Larson M.C., Kjaer S.K. et al. (2010) A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet., 42, 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uno S., Zembutsu H., Hirasawa A., Takahashi A., Kubo M., Akahane T., Aoki D., Kamatani N., Hirata K., Nakamura Y. (2010) A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat. Genet., 42, 707–710. [DOI] [PubMed] [Google Scholar]
- 18.Song H., Ramus S.J., Tyrer J., Bolton K.L., Gentry-Maharaj A., Wozniak E., Anton-Culver H., Chang-Claude J., Cramer D.W., DiCioccio R. et al. (2009) A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet., 41, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Painter J.N., Anderson C.A., Nyholt D.R., MacGregor S., Lin J., Lee S.H., Lambert A., Zhao Z.Z., Roseman F., Guo Q. et al. (2011) Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet., 43, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyholt D.R., Low S.K., Anderson C.A., Painter J.N., Uno S., Morris A.P., MacGregor S., Gordon S.D., Henders A.K., Martin N.G. et al. (2012) Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat. Genet., 44, 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolton K.L., Tyrer J., Song H., Ramus S.J., Notaridou M., Jones C., Sher T., Gentry-Maharaj A., Wozniak E., Tsai Y.Y. et al. (2010) Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet., 42, 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchenbaecker K.B., Ramus S.J., Tyrer J., Lee A., Shen H.C., Beesley J., Lawrenson K., McGuffog L., Healey S., Lee J.M. et al. (2015) Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet., 47, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S.H., Harold D., Nyholt D.R.. ANZGene Consortium, International Endogene Consortium, Genetic and Environmental Risk for Alzheimer's disease Consortium Goddard M.E., Zondervan K.T., Williams J., Montgomery G.W., Wray N.R., Visscher P.M. (2013) Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum. Mol. Genet., 22, 832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y., Ek W.E., Whiteman D., Vaughan T.L., Spurdle A.B., Easton D.F., Pharoah P.D., Thompson D.J., Dunning A.M., Hayward N.K. et al. (2014) Most common ‘sporadic’ cancers have a significant germline genetic component. Hum. Mol. Genet., 23, 6112–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S.H., Yang J., Goddard M.E., Visscher P.M., Wray N.R. (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics, 28, 2540–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. (2011) Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet., 88, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International Schizophrenia Consortium Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Psychiatric GWAS Consortium Bipolar Disorder Working Group. (2011) Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet., 43, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. (2011) Genome-wide association study identifies five new schizophrenia loci. Nat. Genet., 43, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witte J.S., Hoffmann T.J. (2011) Polygenic modeling of genome-wide association studies: an application to prostate and breast cancer. OMICS, 15, 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deary I.J., Yang J., Davies G., Harris S.E., Tenesa A., Liewald D., Luciano M., Lopez L.M., Gow A.J., Corley J. et al. (2012) Genetic contributions to stability and change in intelligence from childhood to old age. Nature, 482, 212–215. [DOI] [PubMed] [Google Scholar]
- 32.Cross-Disorder Group of the Psychiatric Genomics Consortium Lee S.H., Ripke S., Neale B.M., Faraone S.V., Purcell S.M., Perlis R.H., Mowry B.J., Thapar A., Goddard M.E. et al. (2013) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet., 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., Coffey D.M., Creighton C.J., Yu Z., Hawkins S.M., Matzuk M.M. (2012) High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc. Natl. Acad. Sci. U. S. A., 109, 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.H., Nyholt D.R., Macgregor S., Henders A.K., Zondervan K.T., Montgomery G.W., Visscher P.M. (2010) A simple and fast two-locus quality control test to detect false positives due to batch effects in genome-wide association studies. Genet. Epidemiol., 34, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granstrom C., Sundquist J., Hemminki K. (2008) Population attributable fractions for ovarian cancer in Swedish women by morphological type. Br. J. Cancer, 98, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soslow R.A. (2008) Histologic subtypes of ovarian carcinoma: an overview. Int. J. Gynecol. Pathol., 27, 161–174. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.