Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is caused by chromatin relaxation that results in aberrant expression of the transcription factor Double Homeobox 4 (DUX4). DUX4 protein is present in a small subset of FSHD muscle cells, making its detection and analysis of its effects historically difficult. Using a DUX4-activated reporter, we demonstrate the burst expression pattern of endogenous DUX4, its method of signal amplification in the unique shared cytoplasm of the myotube, and FSHD cell death that depends on its activation. Transcriptome analysis of DUX4-expressing cells revealed that DUX4 activation disrupts RNA metabolism including RNA splicing, surveillance and transport pathways. Cell signaling, polarity and migration pathways were also disrupted. Thus, DUX4 expression is sufficient for myocyte death, and these findings suggest mechanistic links between DUX4 expression and cell migration, supporting recent descriptions of phenotypic similarities between FSHD and an FSHD-like condition caused by FAT1 mutations.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most prevalent neuromuscular disorders (1) and is characterized by progressive asymmetric skeletal muscle weakness that begins in the face, shoulder girdle, and upper arms (2). FSHD-affected individuals also commonly exhibit tortuosity of retinal vessels and sensorineural hearing loss (3,4). The causative genetic defect is the loss of transcriptional repression of the Double Homeobox Protein 4 (DUX4) gene present in each repeat of the macrosatellite array (D4Z4) at chromosome 4q35 (5–7). Chromatin is relaxed as a result of array contraction to <11 repeats (FSHD1) (8) or mutation of epigenetic modifiers of the region (FSHD2) (9) and results in aberrant expression of DUX4 in muscle cells (10–12).
Controlling the pathogenesis of FSHD by halting or reversing its progression will require a detailed understanding of the events that occur downstream of DUX4 activation. The forced expression of DUX4 using strong viral promoters in cultured cells leads to aberrant activation of a cascade of diverse targets and produces transcripts from normally dormant transposable elements, transcripts characteristic of an innate immune response and germline-specific transcripts normally silenced in muscle cells (13). Exogenous expression of DUX4 is also toxic to cultured mouse myoblasts (14), disrupts Xenopus development (15) and results in p53-dependent muscle damage in adult mice and zebrafish (16). Germline expression in mice results in embryonic lethality and/or runting and produces a unique skin phenotype due to failure of basal keratinocyte migration. These mice also exhibit retinal vessel tortuosity reminiscent of that found in FSHD (17).
While much has been learned from analyzing cells where DUX4 has been forcibly expressed, the pattern and level of endogenous DUX4 expression in FSHD myoblasts are substantially more subtle. DUX4 transcripts are found in a small percentage of cultured mononuclear FSHD myoblasts that appear to grow and divide without an obvious phenotype (18). Recently, we demonstrated that sporadic DUX4 expression occurs almost exclusively in differentiated FSHD myotubes. When myoblast fusion is extensive, DUX4 protein can be detected in many myonuclei and cultures of FSHD myotubes demonstrate DUX4-mediated cytotoxicity, even when only a fraction of nuclei are actively transcribing DUX4 (19). Importantly, we fused human FSHD myoblasts with mouse C2C12 cells to demonstrate that DUX4 expression from a single nucleus can result in diffusion of DUX4 protein to adjacent nuclei within the same myotube.
The spatial and temporal relationship between DUX4 expression and the induction of transcription from DUX4 target genes is a less studied but important feature of DUX4-mediated cytotoxicity. Several groups have correlated marks of apoptosis with DUX4 expression, including events resulting from forced expression in adult mouse muscle (16), human cell lines (15) or from presumably endogenous DUX4 expression in FSHD muscle tissue (20). TUNEL-positive foci exist in human FSHD myotube cultures but do not co-localize with DUX4 immunofluorescence, suggesting that apoptosis may only occur when DUX4 is expressed at very high supraphysiologic levels, or that apoptosis is temporally disconnected from DUX4 protein in FSHD myotubes. Supporting the latter, we found that the treatment with anti-apoptotic chemicals could prevent death seen in FSHD myotube cultures (21). This finding led us to hypothesize that the expression of DUX4 is momentary, though impactful enough to leave a lasting and detrimental transcriptional signature that results in muscle death.
Forced overexpression of DUX4 could cause molecular signatures that may be unrelated to FSHD. Given the possibly unique transcription effects of endogenous levels of DUX4 expressed in a unique temporal and spatial pattern, we chose to analyze its effects when expressed from its endogenous arrayed subtelomeric location in muscle cells from people affected by FSHD. A DUX4 target gene reporter was developed that responds to the presence of DUX4 and reflects the transcriptional activation of DUX4 target genes and retrotransposons. We show that DUX4 activation in differentiated FSHD myoblasts is asynchronous and transient. Co-localization of DUX4 and DUX4 targets in myotube nuclei reveals a temporal progression of DUX4 protein spreading from a sentinel nucleus and results in DUX4 target gene activation in nearby nuclei. Live cell imaging of differentiating FSHD myoblasts demonstrates that all cells exposed to DUX4 undergo death within a constricted timeframe, suggesting that the immune system is not essential for disease pathogenesis and that therapeutic interventions should be targeted to events early in the DUX4 activation cascade. Reporter fluorescence allowed us to isolate DUX4-expressing and DUX4-non-expressing cells for transcriptome analysis. Here, we report the transcriptional cascade that results from endogenous DUX4 burst expression and the major molecular pathways disrupted as a result. These findings shed new light on the pathogenic transcriptome induced by DUX4 and suggest mechanisms for the distribution of muscle weakness found in FSHD.
Results
DUX4 expression is necessary and sufficient to activate a DUX4-responsive fluorescent reporter
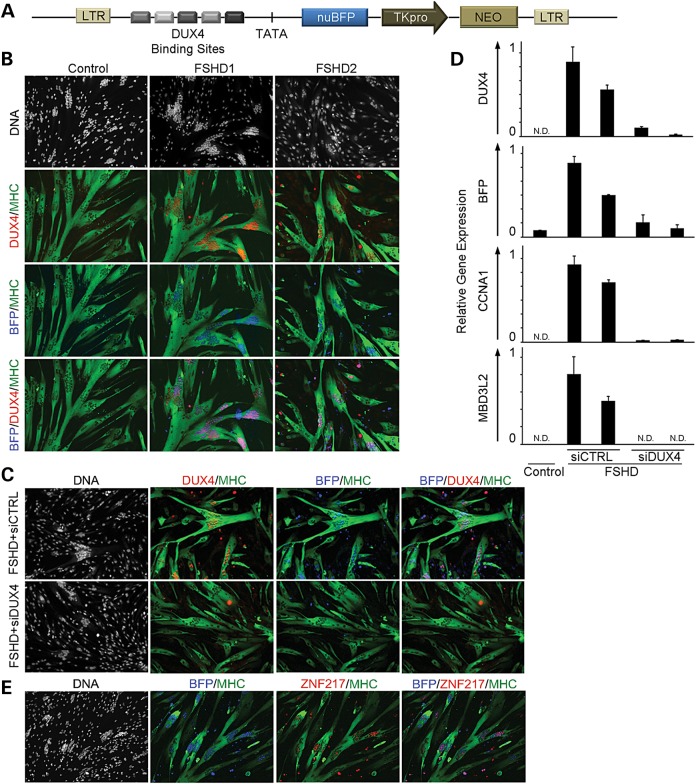
A DUX4 target gene reporter was constructed by placing an array of five DUX4 binding sites upstream of a TATA box and nuclear-localized Blue or Green Fluorescence Protein (BFP or GFP) gene. The expression cassette was delivered to FSHD myoblasts with a lentivirus vector, and growth of transduced cells was selected using G418 resistance provided by a thymidine kinase promoter (pTK) driving neomycin phosphotransferase (Fig. 1A). BFP fluorescence co-localized with DUX4 in FSHD myonuclei after differentiation of myoblasts derived from either FSHD1 or FSHD2 muscle biopsies (Fig. 1B). Vector-transduced myoblasts derived from a control muscle biopsy did not have detectable DUX4 protein (Fig. 1B) or mRNA (Fig. 1D) and did not activate the fluorescent reporter (Fig. 1B). FSHD cells treated with a small interfering RNA (siRNA) designed to target the DUX4 transcript (siDUX4) expressed very low levels of DUX4 transcript (Fig. 1D) and protein (Fig. 1C) and had significantly reduced reporter fluorescence and transcript levels compared to a non-targeting siRNA (siCTRL) (Fig. 1C and D). Endogenous DUX4 activated transcripts CCNA1 and MBD3L2 were also significantly reduced (Fig. 1D) after treatment of cells with siDUX4, demonstrating that BFP fluorescence mimics expression of endogenous DUX4 targets. Additionally, immunofluorescent staining of endogenous DUX4 target gene ZNF217 during FSHD myoblast differentiation co-localized with reporter fluorescence (Fig. 1E). Taken together, these findings demonstrate that the vector is a sensitive and specific reporter for DUX4 expression and mimics the expression profile of endogenous targets.
Figure 1.
Characterization of a DUX4-activated fluorescent reporter. (A) A schematic diagram of the lentiviral vector encoding a DUX4 reporter with five unique DUX4 binding site sequences, identified from individual DUX4 genomic targets and a TATA box located upstream of the sequence for nuclear-localized Blue Fluorescent Protein (nuBFP). A pTK and neomycin phosphotransferase gene (NEO) were included to allow for selection of transduced cells. (B) BFP is specifically present in MHC-positive myotubes (green) derived from individuals with FSHD and co-localizes with DUX4 protein (red) by immunofluorescence. (C) Delivery of an siRNA targeting the DUX4 transcript (siDUX4) during FSHD myoblast differentiation prohibits BFP reporter activation by eliminating DUX4 protein expression. A universal non-targeting control siRNA (siCTRL) transfected in parallel results in BFP and DUX4 expression typical of FSHD myotubes. (D) qRT-PCR of DUX4 and DUX4 targets BFP, CCNA1 and MBD3L2 shows reduced mRNA levels after siDUX4 delivery during FSHD myoblast differentiation. DUX4 and DUX4 target mRNA levels are typical of FSHD cells when siDUX4 is substituted with siCTRL. (N.D., not detected). (E) Immunostaining of endogenous DUX4 target ZNF217 (red) co-localizes with BFP fluorescence (blue) during FSHD myoblast differentiation.
Asynchronous burst expression of DUX4 protein in sentinel nuclei results in DUX4 target expression in neighboring nuclei within multinucleated myotubes
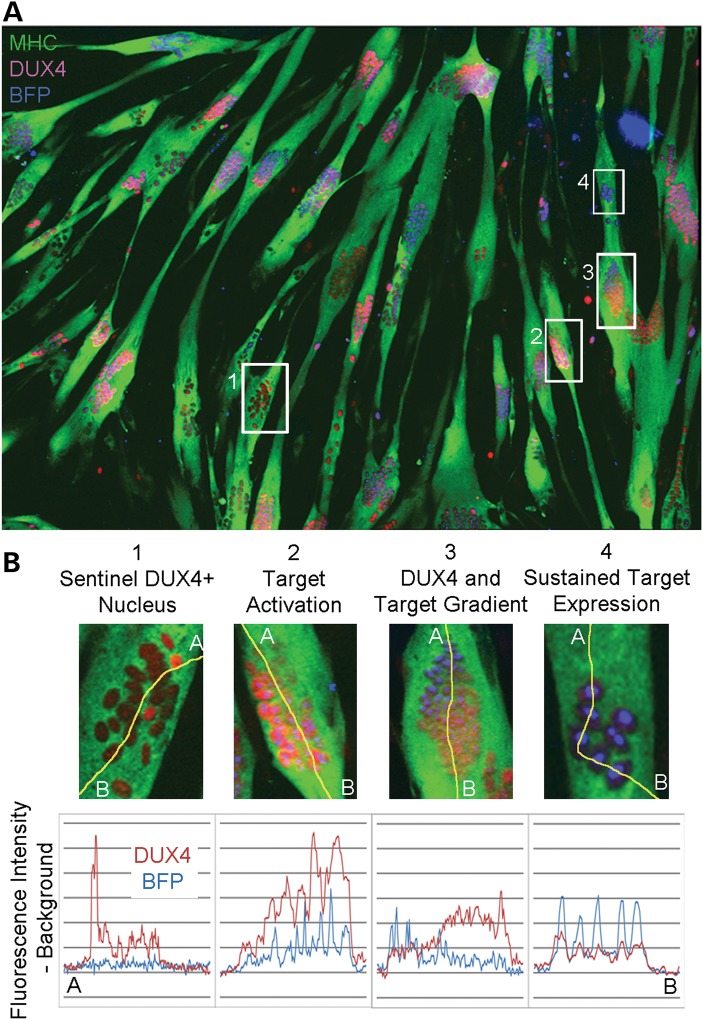
We previously demonstrated that DUX4 protein will diffuse from an activated nucleus through the shared cytoplasm of the myotube to neighboring nuclei (21). We determined the temporal sequence of DUX4 expression and subsequent DUX4 target gene activation during FSHD myoblast differentiation by comparing DUX4 immunostaining with BFP fluorescence in the same cells. The dependence of target gene activation on DUX4 protein allowed us to construct the history of DUX4 expression in each cluster of myonuclei by comparing DUX4 protein immunofluorescence intensity and BFP reporter fluorescence intensity. All DUX4+ nuclear clusters could be categorized into four distinct patterns of DUX4 and BFP localization, providing a temporal model of DUX4 and DUX4 target activation (Fig. 2A and B). The earliest pattern consists of nuclear clusters that contain sentinel bright-staining DUX4+ nuclei associated with other non-staining nuclei or adjacent nuclei containing lower levels of DUX4. BFP fluorescence could not be identified above background levels in these clusters (Fig. 2B, Panel 1). Nuclear clusters that showed both DUX4 staining and DUX4 target activation represent the next step in the temporal progression (Fig. 2B, Panel 2) and precede nuclear clusters containing gradient patterns, where BFP expression was inversely correlated with DUX4 immunofluorescence (Fig. 2B, Panel 3). This latter pattern demonstrates that the half-life of DUX4 protein is shorter than that of BFP and that the effects of DUX4 expression persist even after expression has halted. This pattern is caused by DUX4's diffusion across the nuclear cluster originating from the sentinel nucleus marked by the brightest BFP fluorescence (Fig. 2B, Panel 3, Point A) and continuing to nuclei at the opposite end (Fig. 2B, Panel 3, Point B). Because DUX4 signal was close to background levels in these bright BFP+ nuclei, we conclude that DUX4 activation occurs as a temporary burst of transcription. This finding is supported by identification of a fourth pattern, where DUX4 signal is relatively low throughout the clustered nuclei but BFP signal is consistently bright (Fig. 2B, Panel 4). Here, a burst of DUX4 protein has diffused and activated its targets in all clustered nuclei and dissipated while target expression persists in those nuclei most recently exposed to DUX4 protein. We conclude that bursts of DUX4 expression are asynchronously activated in sentinel nuclei during FSHD myoblast differentiation and activate DUX4 targets in all neighboring nuclei.
Figure 2.
DUX4 and target expression reveal temporal stages of the transcriptional cascade resulting from DUX4 activation. (A) Four distinct patterns identified by line scan fluorescence intensity analysis suggest that DUX4-activated target expression follows transient asynchronous pulses of DUX4 expression and can persist even after DUX4 is no longer detectable. (B) DUX4 protein originates from sentinel DUX4+ nuclei that do not express DUX4 targets (1), then diffuses to nearby nuclei in the shared cytoplasm of the myotube and activates target genes (2). This momentary expression of DUX4 followed by diffusion results in a gradient of the protein and its targets (3), followed by sustained target expression in nuclei previously exposed to DUX4 (4).
Endogenous DUX4 activation during differentiation of FSHD muscle cultures results in cell death
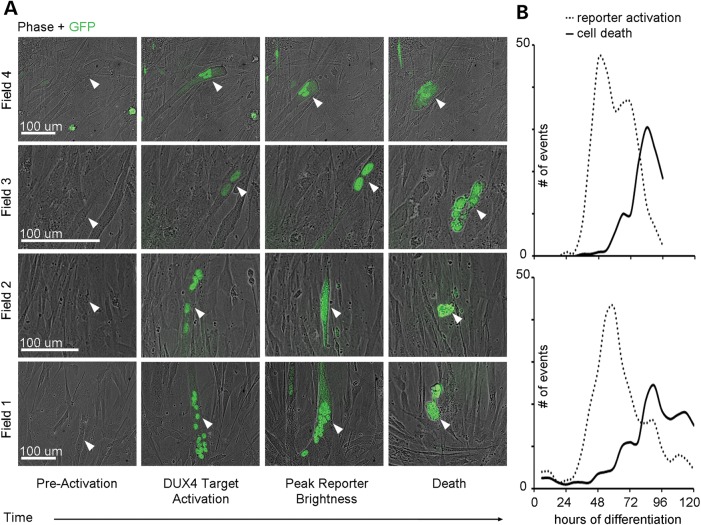
We photographed differentiating FSHD myoblasts containing a DUX4-activated GFP reporter every 15 min for 120 h in order to identify phenotypic changes that result from DUX4 expression. We observed that DUX4-expressing cells universally died (Fig. 3A and B, Supplementary Material, Videos S1 and S2) ∼20.2 h after fluorescence detection (±8.7 h). FSHD cells activated GFP and succumbed to DUX4-mediated death continuously throughout the 120 h of observed differentiation (Fig. 3B). The number of DUX4-expressing cells reached a maximum at 48–54 h into differentiation and was followed by a peak of death events at 70–84 h (Fig. 3B). Regular periods of increased cell death that occurred on average every 12 h were also observed and decreased in amplitude over time, likely due to the eventual depletion of myoblasts in the differentiating cultures (Fig. 3B). These findings demonstrate the first evidence of endogenous DUX4-induced cell death in live FSHD myoblasts. Additionally, the observation of rapid death after fluorescence detection suggests that FSHD treatments aimed downstream of DUX4 activation should target very early events of the DUX4-induced cascade in order to prevent cell death in FSHD muscle.
Figure 3.
Imaging and quantification of DUX4 target activation and cell death events during FSHD myoblast differentiation. (A) Brightfield and GFP live cell imaging of differentiating FSHD myoblasts associates DUX4-activated reporter expression with cell death (white arrows) occurring ∼20.2 h after GFP is detectable (data not shown). (B) Quantification of DUX4-activated reporter detection in sentinel events and associated cell death events during 120 h of FSHD myoblast differentiation, where each graph describes an independent differentiation experiment and reports total events observed over 11.7 mm2.
DUX4-expressing cells can be isolated from FSHD cultures and carry the expected RNA signature
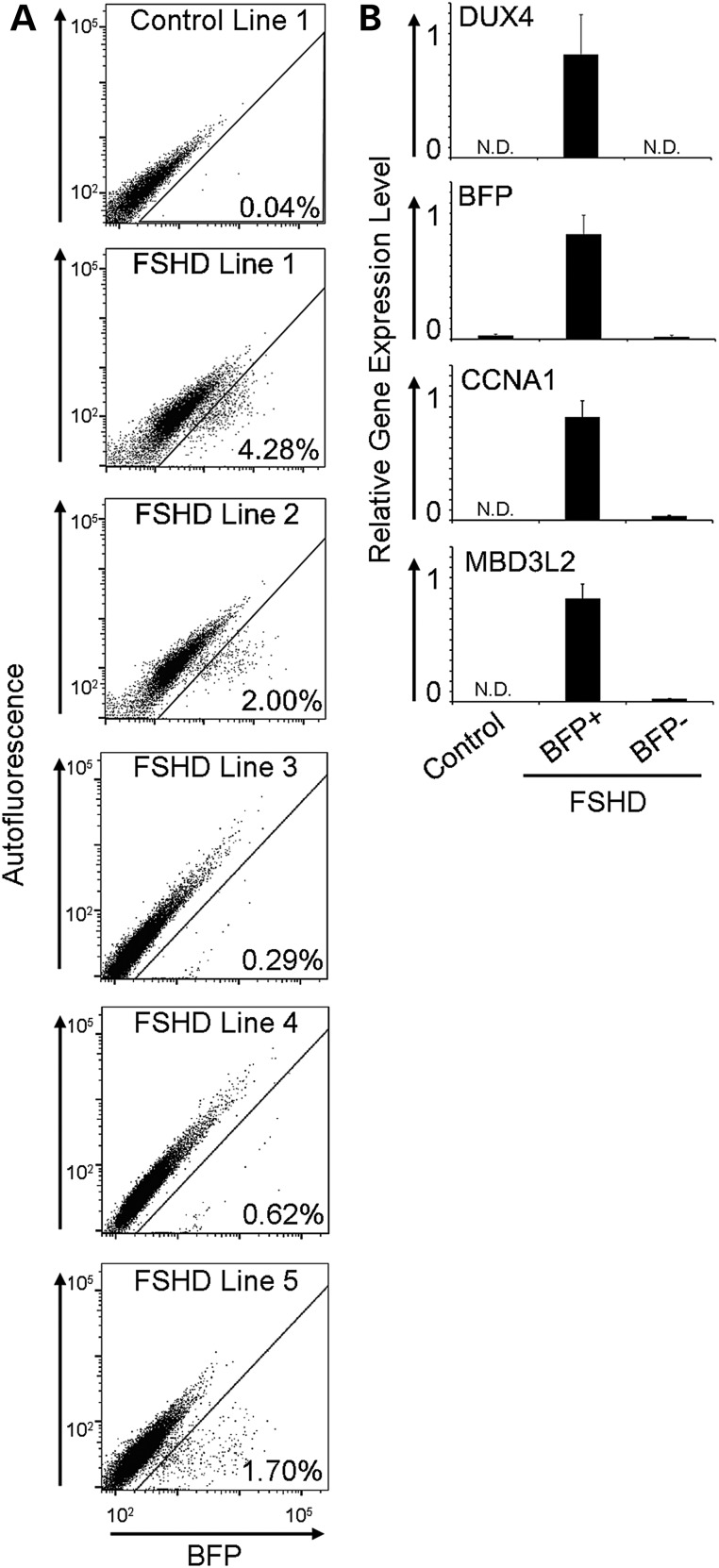
Calcium chelation with ethylene glycol tetraacetic acid (EGTA) has been shown to prevent myoblast fusion during terminal differentiation (22), and inhibiting cell fusion would allow us to quantify DUX4 activation events and examine the consequences of DUX4 expression in differentiated mononuclear myocytes using fluorescence-activated cell sorting. We used the DUX4-BFP fluorescence reporter to screen myoblast samples from five different FSHD-affected individuals to identify myoblast cultures where DUX4 activation was most frequent during differentiation. DUX4 was expressed in 0.29–4.28% of mononuclear FSHD cells after 72 h of differentiation (Fig. 4A), demonstrating the inter-individual variability that has been previously described (21,23). Two different FSHD samples that contained the most DUX4-activated cells (Fig. 4A, FSHD Lines 1 and 2) were selected for subsequent experiments.
Figure 4.
Quantification of DUX4 activation events and DUX4 and DUX4 target mRNA expression analysis in sorted myoblasts. (A) Representative flow cytometry plots of BFP fluorescence intensity (X-axis) versus autofluorescence (Y-axis) in 72 h differentiated control and FSHD myoblasts harboring the DUX4-activated BFP reporter with % reporter + cells displayed for each line. (B) Populations of BFP+ and BFP– myoblasts were collected by flow sorting and mRNA levels of DUX4, and DUX4 targets BFP, CCNA1 and MBD3L2 were measured by qRT-PCR. (N.D., not detected).
The percentage of cells expressing DUX4 in proliferating myoblast cultures was determined to establish a baseline for experiments to be conducted in differentiated cells. DUX4 expression was present in 0.62% of FSHD Line 1 cells and 0.18% of FSHD Line 2 cells and increased by 3.5- to 5-fold after a 48 h induction of differentiation in the absence of EGTA (Supplementary Material, Fig. S1A). We tested the effects of EGTA treatment on DUX4 activation by counting DUX4-positive cells differentiated in the presence of EGTA at the same time points and found that approximately the same number of cells activate DUX4 expression whether or not EGTA is present in the differentiation culture medium (Supplementary Material, Fig. S1B). In addition, we found no significant difference in DUX4 activation events whether cells were differentiated in horse serum or knockout serum replacer (KOSR) (Supplementary Material, Fig. S2A).
The fluorescence signal from the DUX4 reporter also allowed for the comparison of transcriptomes of DUX4-expressing and DUX4-non-expressing cells from the same FSHD myoblast cultures. FSHD Lines 1 and 2 were differentiated for 72 h in the presence of EGTA to prevent fusion, sorted for BFP+ and BFP– cell populations, and RNA was isolated from the collected cells. To control for culture conditions and biological variation, we analyzed RNA from three biological replicates of each cell line prepared on different days. The percent of fluorescing cells in each replicate was similar and averaged 4.56% for Line 1 and 3.42% for Line 2 (Fig. 4A shows the fluorescence distribution of a representative sort for each line). DUX4 transcripts were undetectable by quantitative real-time polymerase chain reaction (qRT-PCR) in BFP– cells and readily detectable in sorted populations of BFP+ cells. Additionally, transcripts from DUX4 target genes CCNA1 and MBD3L2 were enriched by 25- and 40-fold, respectively, in BFP+ cell populations (Fig. 4B).
DUX4-induced transcripts are mostly up-regulated and come from genes with associated DUX4 binding sites and genes transcriptionally activated by secondary events
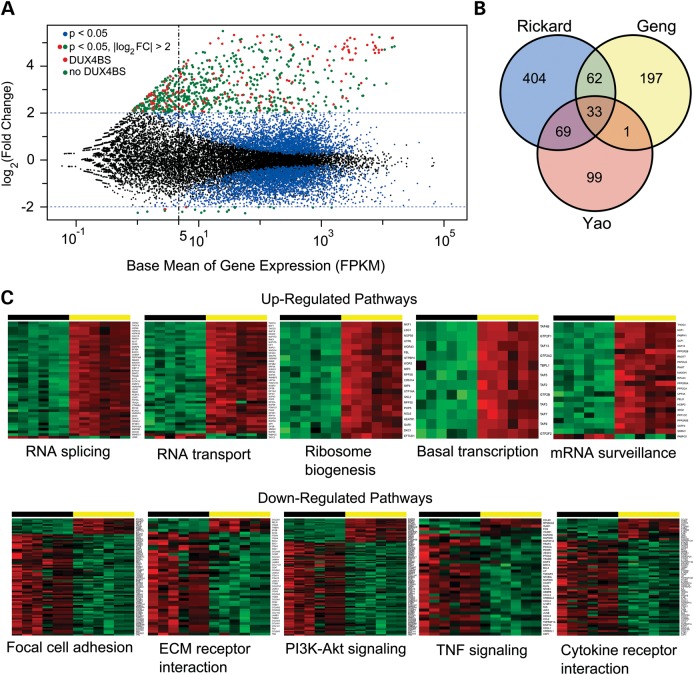
DUX4-expressing cells produced by forcing high levels of DUX4 expression from a retroviral vector and comparing the resulting transcriptome to the same cells transduced with a GFP-expressing vector were previously analyzed (13). While these studies have shed light on the transcriptional effects of DUX4 in myogenic cells, the high level of DUX4 expression, enhanced expression of genes at retroviral insertion sites and cellular responses to retroviral transduction may obscure important transcriptional effects of DUX4 expression. We prepared sequencing libraries from RNA isolated from sorted cell populations where the DUX4 target reporter identified cells were expressing DUX4 from pathogenic D4Z4 arrays at their subtelomeric location on chromosome 4 and compared them with isogenic reporter-negative cells sorted from the same population. We found that physiologic DUX4 expression activates a unique group of genes in addition to a subset of those previously identified (Fig. 5A and B, Supplementary Material, Table S1). Five hundred and fifty-two genes had transcript levels that increased by at least 4-fold [Log2 fold change (log2FC) >2, P-value <0.005] when comparing DUX4-expressing and DUX4-non-expressing cells from the same culture. Eighteen genes had expression levels that were reduced by 4-fold in DUX4-expressing cells (Fig. 5A).
Figure 5.
RNA-seq analysis of differentially expressed genes in DUX4-expressing cells. (A) Scatter plot of the log2FC in gene expression verses the mean expression level for each gene. All points from three biological replicates of two different FSHD myoblast cell lines are shown (6 samples sorted for DUX4-expressing and DUX4-non-expressing cells, 12 samples total). Gene expression levels with significant (P < 0.05) log2FC values are shown as colored dots (blue, red and green). Data points for genes with log2FC change >2 (linear fold change >4) are colored in red if they contain a DUX4 binding site within 5 kb of the transcription start site and green if they do not. DUX4 binding sites were determined by analysis of publically available ChIP-seq data (13). (B) Venn diagram comparing DUX4 mis-expressed genes found in previous RNA-seq studies (Geng and Yao) to those identified here. (C) Examples of several significantly dysregulated gene sets listed in Table 1. Differentially expressed genes identified using the DESeq2 R package and plotted in (A) were used for Generally Applicable Gene-set/Pathway Analysis (GAGE) (25). The genes from a particular pathway that showed significant up- or down-regulation are displayed as heat maps (green low, red high) for all 12 data points. As in (A), three biological replicates of two FSHD cell lines are shown, and data points for DUX4 reporter-negative cells (black horizontal bar) and DUX4 reporter-positive cells (yellow horizontal bar) are shown as different color shades based on the regularized log transformation (log2(counts)) of normalized counts. Gene symbols are listed vertically along the right border of each heat map.
DUX4-activated genes include both direct targets of the DUX4 transcription factor and those resulting from a transcriptional cascade initiated by DUX4. We utilized previously published chromatin immunoprecipitation with paralleled sequencing (ChIP-seq) data (13) to determine the genomic location of DUX4 binding sites and then identified DUX4-activated genes that contained DUX4 binding site sequences within 5 kb upstream or downstream of a known transcriptional start site. Of the 570 DUX4-altered transcripts, 122 contained a DUX4 binding site by ChIP-seq analysis (Fig. 5A points in red; Supplementary Material, Table S2). The other 448 either had a DUX4 binding site beyond our window or were activated by the secondary effects of an initial cascade of DUX4 target gene activation. Of the 18 genes that were down-regulated >4-fold in DUX4-expressing cells, one contained a DUX4 binding site within the specified window. These findings have important implications for the design of therapies for FSHD because the majority of DUX4-mediated dysregulation of transcription appears to be secondary.
Endogenous DUX4-expressing differentiated myoblasts have a unique transcriptome
We compared the list of genes identified here with those reported to be up-regulated as a result of forced DUX4 expression (13), and with reprocessed analysis of data from Yao et al. (24) that compared the transcriptomes of FSHD and control myotubes, in order to determine which genes are differentially identified depending on the approach. Of the 293 genes with log2FC >2 identified by Geng, 95 genes were in common, including most (13/17) of the cancer testis antigens previously described. Four hundred and seventy-three novel transcripts were identified here, and 197 genes were unique to the list identified by Geng et al. Because the subset of DUX4-expressing cells was not isolated prior to RNA-seq in the study performed by Yao et al., the fold change values identified there are expected to be lower, so we compared the list of Yao genes with P < 0.05 (202 total) to the list of genes identified here with log2FC > 2 (P < 0.05). Of the 568 DUX4-activated genes identified using our approach, 102 genes were found to be in common between both lists, leaving 466 unique genes identified here (Fig. 5B). The difference between these lists suggests that endogenous DUX4 expression levels, isogenic controls and identical culture conditions are important variables for determining the consequences of DUX4 expression in diseased cells. As a result, the transcriptional consequences of DUX4 expression identified here are critically distinct from those previously reported.
DUX4 induces a specific gene expression signature that includes disruption of pathways for RNA splicing, RNA surveillance and nuclear to cytoplasmic RNA transport
The large number of differentially expressed transcripts in DUX4-expressing cells requires higher-order grouping to determine the cellular consequence of DUX4 expression. Gene set enrichment analysis is a strategy of grouping gene dysregulation based on knowledge of molecular pathways and offers increased sensitivity and biological relevance (25). Using this approach, we found a specific DUX4 transcriptional signature that involves disruption of RNA metabolism including splicing, surveillance and transport (Fig. 5C, Table 1). The spliceosome pathway consisting of 130 genes was the most differentially regulated group in DUX4-expressing cells, with consistent up-regulation of a significant number of members (43/130 genes; Fig. 5C, Table 1). Nuclear–cytoplasmic RNA transport was also significantly disrupted (43/163 genes). Finally, members of the RNA-surveillance pathway were significantly up-regulated (22/90 genes; Fig. 5C, Table 1), consistent with a previous report that ectopic DUX4 causes UPF1 protein degradation and disrupts processing of transcripts containing premature termination codons in myoblasts (26). These results suggest that the molecular pathophysiology of FSHD may be similar to that of myotonic dystrophy, where splicing, RNA transport and RNA surveillance are also among the major pathways disrupted by gain-of-function RNAs in muscle cells (27–29).
Table 1.
GAGE analysis of DUX4 differentially expressed genes identified by RNA-seq
| KEGG gene set | KEGG Ida | Meanb | P-value | # Genesc |
|---|---|---|---|---|
| Up-regulated pathways | ||||
| Spliceosome | hsa03040 | 6.7 | 2.63E − 55 | 130 |
| RNA transport | hsa03013 | 4.7 | 2.36E − 30 | 163 |
| Ribosome biogenesis in eukaryotes | hsa03008 | 4.7 | 2.50E − 28 | 75 |
| Basal transcription factors | hsa03022 | 3.2 | 2.27E − 14 | 44 |
| mRNA-surveillance pathway | hsa03015 | 2.9 | 5.61E − 13 | 90 |
| RNA polymerase | hsa03020 | 2.1 | 4.11E − 07 | 31 |
| Down-regulated pathways | ||||
| Focal adhesion | hsa04510 | −5.9 | 2.79E − 46 | 207 |
| ECM-receptor interaction | hsa04512 | −4.9 | 1.16E − 31 | 86 |
| PI3K-Akt signaling pathway | hsa04151 | −4.5 | 4.76E − 28 | 346 |
| TNF signaling pathway | hsa04668 | −3.6 | 4.21E − 18 | 110 |
| Cytokine–cytokine receptor interaction | hsa04060 | −3.3 | 2.93E − 16 | 264 |
| Apoptosis | hsa04210 | −3.2 | 5.29E − 15 | 86 |
| Regulation of actin cytoskeleton | hsa04810 | −2.8 | 7.89E − 12 | 214 |
| Lysosome | hsa04142 | −2.7 | 3.06E − 11 | 122 |
| Jak-STAT signaling pathway | hsa04630 | −2.6 | 1.99E − 10 | 156 |
| Toll-like receptor signaling pathway | hsa04620 | −2.5 | 4.46E − 10 | 105 |
| NF-kappa B signaling pathway | hsa04064 | −2.5 | 7.10E − 10 | 90 |
| Glutathione metabolism | hsa00480 | −2.4 | 2.24E − 09 | 50 |
| Gap junction | hsa04540 | −2.4 | 5.12E − 09 | 89 |
| Drug metabolism—cytochrome P450 | hsa00982 | −2.4 | 5.27E − 09 | 67 |
| Vascular smooth muscle contraction | hsa04270 | −2.4 | 5.53E − 09 | 121 |
| Other glycan degradation | hsa00511 | −2.5 | 8.17E − 09 | 18 |
| Notch signaling pathway | hsa04330 | −2.3 | 1.26E − 08 | 48 |
| Protein digestion and absorption | hsa04974 | −2.3 | 1.33E − 08 | 88 |
| ABC transporters | hsa02010 | −2.3 | 1.67E − 08 | 44 |
| Other types of O-glycan biosynthesis | hsa00514 | −2.3 | 1.87E − 08 | 31 |
| Glycosaminoglycan biosynthesis | hsa00532 | −2.4 | 1.87E − 08 | 20 |
| Osteoclast differentiation | hsa04380 | −2.2 | 3.94E − 08 | 131 |
| Axon guidance | hsa04360 | −2.1 | 1.09E − 07 | 127 |
| p53 signaling pathway | hsa04115 | −2.1 | 1.50E − 07 | 68 |
| Melanogenesis | hsa04916 | −2.1 | 2.99E − 07 | 101 |
| Adipocytokine signaling pathway | hsa04920 | −2.0 | 3.82E − 07 | 70 |
| Hippo signaling pathway | hsa04390 | −2.0 | 4.20E − 07 | 153 |
| Wnt signaling pathway | hsa04310 | −2.0 | 5.77E − 07 | 139 |
| GnRH signaling pathway | hsa04912 | −2.0 | 7.56E − 07 | 92 |
| Val, leu and ile degradation | hsa00280 | −2.0 | 1.10E − 06 | 47 |
| Bidirectionally disrupted pathways | ||||
| Endocytosis | hsa04144 | 3.1 | 5.29E − 15 | 86 |
| MAPK signaling pathway | hsa04010 | 2.7 | 4.46E − 10 | 105 |
| TGF-beta signaling pathway | hsa04350 | 2.4 | 2.24E − 09 | 50 |
| Adherens junction | hsa04520 | 2.1 | 5.27E − 09 | 67 |
aKyoto Encyclopedia of Genes and Genomes identification number.
bStatistical mean.
cThe number of genes in the KEGG pathway gene set.
Gene pathways involved in cell adhesion, extracellular matrix signaling, cell proliferation and cell orientation are down-regulated in DUX4-expressing cells
Many of the affected muscle groups in FSHD have unique developmental origins, suggesting that differential expression of DUX4 during development of specific muscles could explain the unique pattern of muscle weakness. Although direct targets of DUX4 were generally up-regulated, DUX4 expression had a repressive effect on a group of related pathways involved in cell adhesion (81/207 genes), extracellular matrix (ECM) signaling (44/86 genes), cell proliferation (PI3K-Akt, TNF, cytokine, apoptosis and Hippo), and cell orientation and migration (actin cytoskeleton, Gap junction, adherens junction, Notch and Wnt signaling) (Table 1, Fig. 5C). Cell migration defects due to FAT1 mutations in humans (30) and mice (31) have recently been shown to affect muscle groups similar to those most severely affected in FSHD, suggesting that the DUX4-induced disruption of cell signaling described here may partly explain the propensity for weakness of certain muscle groups in FSHD.
DUX4-induced splicing disruption affects genes involved in muscle structure and function
The observation of DUX4-mediated global disruption of RNA processing led us to examine the RNA-seq data for alternatively spliced transcripts. Significant alterations in all major classes of splicing aberrations, including alternative 5′ and 3′ splice sites (A5SS, A3SS), retained introns (RI), skipped exons (SE) and shifts in the selection of mutually exclusive exons (MXE), were observed (Table 2). To determine the significance and reproducibility of these aberrations, we calculated and averaged the percent spliced in (Ψ) values for each event when comparing DUX4-expressing cells with DUX4-non-expressing cells from the same culture. Splicing events were ordered by the number of comparisons that showed the aberration (out of six total) and the Bayes score (corresponding to the odds ratio of differential expression calculated with MISO (mixture of isoforms)) (32) (Table 2). The list contains splicing events in genes involved in splicing and RNA processing (LUC7L3, CSE1L, DDX39B, EIF4A2, SF1, HNRNPAB, STAU1, TRA2B, AGFG1, ZFAND5), cell polarity and signaling (ENAH, CERS5, ACHE, FEZ2, DNM2), mitochondrial energy production (SLC25A3, PTRH2, FIS1), muscle structure (MYL6, TPM2, PALLD) and ubiquitination (RPN2, URI1). Aberrant forms of the serine/arginine-rich splicing factor 2 (SRSF2) were among the transcripts with significant differences between DUX4-expressing and DUX4-non-expressing cells. This observation is consistent with previous reports where alterations in transcript stability were proposed to result from DUX4-mediated inhibition of the nonsense-mediated decay pathway (26). Aberrant forms of the SRSF3 gene (also described by Feng et al.) were noted in some of our samples, but significant changes were not present in at least four biological replicates and therefore did not meet our cut-off for inclusion in Table II. In addition to the disruption of RNA surveillance, another important consequence of aberrant DUX4 expression is a significant change in splice site choice within a large number of genes. Here, we describe new candidates resulting from DUX4-mediated splice site disruption that may be important targets for therapeutic intervention.
Table 2.
Splicing events in genes subject to DUX4-induced alternative splicing
| Symbol | Descriptive name | Ψconta (%) | ΨDUXb (%) | ΔΨc (%) | Bayesd | Typee | #f |
|---|---|---|---|---|---|---|---|
| SRSF2 | Serine/arginine-rich splicing factor 2 | 10 | 32 | −23 | 8.52E11 | SE | 6 |
| SLC25A3 | Solute carrier family 5 member 3 | 48 | 74 | −26 | 8.33E11 | MXE | 6 |
| SLC25A3 | Solute carrier family 5 member 3 | 28 | 49 | −21 | 6.67E11 | MXE | 6 |
| PRRC2A | Proline-rich coiled-coil 2A | 6 | 37 | −31 | 5.00E11 | RI | 6 |
| MYL6 | Myosin light polypeptide 6 | 82 | 64 | 18 | 3.33E11 | SE | 6 |
| EIF4B | Eukaryotic initiation factor 4B | 79 | 63 | 16 | 1.67E11 | MXE | 6 |
| ARGLU1 | Arginine and glutamate rich 1 | 6 | 27 | −22 | 1.83E10 | SE | 6 |
| MEG3 | Maternally expressed 3 | 34 | 14 | 20 | 1.00E12 | RI | 5 |
| MEG3 | Maternally expressed 3 | 39 | 16 | 23 | 1.00E12 | RI | 5 |
| LUC7L3 | LUC7-like 3 (S. cerevisiae) | 70 | 47 | 23 | 1.00E12 | RI | 5 |
| MEG3 | Maternally expressed 3 | 41 | 19 | 22 | 1.00E12 | A5SS | 5 |
| MEG3 | Maternally expressed 3 | 42 | 20 | 23 | 1.00E12 | A5SS | 5 |
| BRD2 | Bromodomain containing 2 | 4 | 17 | −12 | 8.65E11 | A3SS | 5 |
| BRD2 | Bromodomain containing 2 | 8 | 30 | −22 | 8.41E11 | A3SS | 5 |
| TPM2 | Tropomyosin 2 | 12 | 26 | −14 | 6.00E11 | A3SS | 5 |
| FERMT2 | Fermitin family member 2 | 22 | 59 | −37 | 6.00E11 | SE | 5 |
| PTRH2 | Peptidyl-tRNA hydrolase 2 | 9 | 23 | −14 | 4.00E11 | SE | 5 |
| CSE1L | CSE1 chromosome segregation 1-like | 5 | 23 | −18 | 4.00E11 | A3SS | 5 |
| ENAH | Enabled homolog | 15 | 31 | −16 | 4.00E11 | A3SS | 5 |
| YPEL5 | Yippee-like 5 | 9 | 25 | −16 | 4.00E11 | SE | 5 |
| RPN2 | Ribophorin II | 14 | 30 | −16 | 2.00E11 | SE | 5 |
| FIS1 | Fission 1 homolog | 81 | 41 | 40 | 2.00E11 | SE | 5 |
| DDX39B | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B | 31 | 50 | −19 | 28722 | RI | 5 |
| WDR46 | WD repeat domain 46 | 6 | 23 | −17 | 2514 | RI | 5 |
| EIF4A2 | Eukaryotic initiation factor 4A-II | 9 | 27 | −17 | 1.00E12 | SE | 4 |
| MYL6 | Myosin light polypeptide 6 | 11 | 15 | −4 | 7.50E11 | SE | 4 |
| SF1 | Homo sapiens splicing factor 1 | 41 | 26 | 15 | 5.00E11 | A3SS | 4 |
| ENAH | Enabled homolog | 28 | 64 | −36 | 5.00E11 | SE | 4 |
| PPP4C | Protein phosphatase 4, catalytic subunit | 11 | 45 | −34 | 5.00E11 | RI | 4 |
| PPP4C | Protein phosphatase 4, catalytic subunit | 4 | 17 | −14 | 5.00E11 | RI | 4 |
| PALLD | Palladin | 36 | 58 | −23 | 5.00E11 | SE | 4 |
| APLP2 | Amyloid beta (A4) precursor-like protein 2 | 32 | 53 | −21 | 2.50E11 | SE | 4 |
| HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | 7 | 33 | −26 | 2.50E11 | RI | 4 |
| ATXN2L | Ataxin 2-like | 46 | 66 | −20 | 2.40E11 | RI | 4 |
| STAU1 | Staufen double-stranded RNA binding protein 1 | 39 | 73 | −35 | 2.50E11 | SE | 4 |
| ATXN2L | Ataxin 2-like | 56 | 79 | −22 | 2.50E11 | RI | 4 |
| ATXN2L | Ataxin 2-like | 57 | 77 | −20 | 2.50E11 | RI | 4 |
| CERS5 | Ceramide synthase 5 | 22 | 50 | −28 | 2.27E11 | SE | 4 |
| FEZ2 | Fasciculation and elongation protein zeta 2 | 6 | 29 | −23 | 3.42E09 | SE | 4 |
| ACHE | Acetylcholinesterase | 10 | 26 | −16 | 2.07E07 | A3SS | 4 |
| URI1 | Prefoldin-like chaperone (URI1) transcript 1 | 94 | 81 | 13 | 1.28E07 | A3SS | 4 |
| CSNK1D | Casein kinase 1, delta | 83 | 59 | 24 | 1539819 | SE | 4 |
| DNM2 | Dynamin 2 | 71 | 45 | 26 | 371049 | MXE | 4 |
| TRA2B | Transformer 2 beta homolog | 20 | 34 | −14 | 8660 | SE | 4 |
| SF1 | Homo sapiens splicing factor 1 | 24 | 12 | 13 | 5658 | SE | 4 |
| AGFG1 | ArfGAP with FG repeats 1 | 22 | 49 | −27 | 333 | SE | 4 |
| BUB3 | BUB3 Mitotic checkpoint protein | 50 | 33 | 17 | 88 | RI | 4 |
| ZFAND5 | Zinc finger, AN1-type domain 5 | 62 | 41 | 21 | 83 | SE | 4 |
| TMEM185A | Transmembrane protein 185A | 48 | 76 | −28 | 30 | MXE | 4 |
| MED15 | Mediator complex subunit 15 | 7 | 24 | −17 | 26 | RI | 4 |
aΨcont ‘% spliced in’, The average percentage of transcripts that are aberrantly spliced in control samples.
bΨDUX ‘% spliced in’, The average percentage of transcripts that are aberrantly spliced in DUX4+ samples.
cΔΨ (Ψcont–ΨDUX).
dBayes factor for the comparison, corresponding to the odds of differential expression (where the change in Ψ is non-zero) over no differential expression (where the change is zero).
eType of splicing aberration (SE, skipped exon; RI, retained intron; MXE, mutually exclusive exons; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site).
fThe number of comparisons (out of six total) showing this particular splicing aberration.
Discussion
We previously fused mouse and human myoblasts to show that DUX4 produced in one nucleus could diffuse to a nucleus incapable of producing DUX4 transcripts (21). These studies suggested a mechanism of signal amplification that could result in transcriptional disruption of all nuclei in a myotube, despite the possibility that only a single nucleus may be transcribing DUX4. Here, we demonstrate the temporal sequence of DUX4 and DUX4 target gene activation both statically and in real time. We show that DUX4 transcription is pulsatile and originates from single myonuclei and that activation of downstream target genes is lasting and inevitably results in death of the myotube. The time course of this process is relatively quick in cell culture, where densely clustered myonuclei quickly take up DUX4 protein. The density and distribution of myonuclei in vivo are different from cultured cells and may provide for conditions where transient DUX4 expression is not fatal to the fiber or takes considerably longer to disrupt fibers. Our data also suggest that regions of muscle tissue and muscle types containing densely packed nuclei may be more sensitive to DUX4 expression than fibers containing widely spaced myonuclei. The observation that DUX4 expression is pulsatile suggests the possibility that DUX4-primed muscle tissue may not be effectively treated by decreasing DUX4 transcripts.
Our findings have important implications for therapy development. Because no animal model produced to date reliably recapitulates disease (17,33,34), the discovery of a cell culture phenotype that can be used as a platform for therapeutic screening is important for the development of treatments for FSHD. We previously demonstrated that myotube cultures from FSHD biopsies die in a DUX4-dependent manner (21) and others have described an atrophic phenotype of FSHD myotubes (35). Here, we show specific myotubes identified by a fluorescent DUX4 target gene reporter that disintegrate in culture, providing a target phenotype for intervention that is preceded by visible reporter activation. This system allows for high-throughput screening of repression of reporter activity and serves as a platform to identify and test therapeutics. Importantly, DUX4 expression in these cells is under normal epigenetic and genetic control, so interventions that suppress transcription at the earliest stage of activation could be identified using FSHD myoblasts containing this reporter as a screening platform.
These findings also have implications for our understanding of the FSHD disease mechanism. One proposed theory is that expression of antigenic proteins (cancer testes antigens) induced by DUX4 transcriptional activity may induce an immune response against DUX4-expressing muscle fibers and suggests that immune-mediated muscle destruction is a primary mechanism of FSHD (13). While several groups have demonstrated muscle and myoblast death after forced overexpression of DUX4, here we show that endogenous levels of DUX4 are sufficient to kill myocytes and demonstrate that the immune system is not essential for DUX4-mediated cell death and therefore could play a secondary role in FSHD pathogenesis, perhaps more similar to its role in Duchenne muscular dystrophy (DMD).
The transcriptomes generated for analysis in this study are also unique. Here, we separated DUX4-expressing and DUX4-non-expressing cells from the same culture and compared transcriptional differences between the isogenic cell populations. This comparison is fundamentally different from comparing diseased and control samples from different people and different biopsies. As a consequence, we obtained unique results that generate new insights about the effects of DUX4 transcription on muscle cells. While we observed some altered expression of germline genes, we also saw a more specific DUX4 signature that suggests DUX4-mediated toxicity is a consequence of disruption of specific set of molecular pathways rather than mis-expression of tissue-specific and developmentally inappropriate genes that leads to cell ‘confusion’ and triggers apoptosis. Our findings suggest that a disruption in RNA metabolism is central to DUX4-mediated toxicity and are intriguing given the similarities to myotonic dystrophy.
An interesting observation resulting from this study is that DUX4 disrupts a number of pathways involved in cell migration. The unique pattern of weakness in facial, scapular, biceps and lower leg musculature seen in FSHD-affected individuals has intrigued researchers and suggests a developmental mechanism for creating muscle groups that are spared or vulnerable to the disease process. FAT1 mutations in animals (31) and humans (30) phenocopy FSHD with respect to the distribution of affected muscles and retinal changes. Altered levels of FAT1 have been reported in muscle groups first affected in FSHD (36), suggesting that developmental disruption of planar cell polarity pathways may be a component of FSHD. Here, we show that DUX4-induced mis-splicing of 67% of the Enabled Homolog gene transcripts (ENAH) leads to the addition of 230 amino acids or the deletion of 36 amino acids from the wild-type protein (Supplemental Material, Fig. S3). Because ENAH is a direct binding partner of FAT1, we speculate that these findings may connect DUX4 expression, and resultant mis-splicing of transcripts like ENAH, with FAT1 disruption causing a sequence of events that could pattern muscles differentially during development and create both spared and vulnerable muscle groups.
Materials and Methods
Cell culture
FSHD and control primary human myoblasts were provided by Dr. Rabi Tawil and members of the Fields Center at the University of Rochester, whose patients gave written informed consent for sample use (http://www.urmc.rochester.edu/fields-center/protocols/myoblast-cell-cultures.cfm). Proliferating myoblasts were cultured in F10 medium (Life Technologies, Carlsbad, CA) supplemented with 20% fetal bovine serum (Thermo Scientific, Waltham, MA), 10 ng bFGF (Life Technologies), 1 µM dexamethasone (Sigma) and 50 U/50 µg penicillin/streptomyocin (Life Technologies). Differentiation was induced by plating cells at 70 000 cells per cm2 then changing media after 24 h to DMEM:F12 (1:1, Life Technologies) supplemented with 20% KOSR (Life Technologies) or 2% horse serum (Life Technologies) and 50 U/50 µg penicillin/streptomycin (Life Technologies). Cells were allowed to differentiate in these conditions for 48–120 h.
Lentivirus reporter cloning, production and delivery
Six individual sequence motifs of the previously identified DUX4 DNA binding site ((13) sequence: AGATAATTGAATCATGGGGTAATCCAATCATGGAGTAATTTAATCAGCCGTTAATTGAATCATGGGGTAATCCAATCATGGAGTAATTTAATCAGCCG) followed by a minimal TATA box promoter upstream of the nuclear BFP (Addgene #14893, Cambridge, MA) or nuclear turbo GFP (Open Biosystems #140544, Thermo Scientific) were amplified by PCR. The reporter cassette was cloned into the pRRLsincPPT-wpre third-generation lentivirus backbone (37). The pTK promoter driving the neomycin resistance gene was then cloned downstream of BFP to confer G418 selection capability independent of reporter activation by the DUX4 protein. The lentiviral vector was packaged, by polyethylenimine-mediated (PEI) co-transfection with plasmids containing viral genome components, in HEK-293T cells. Vector preparations were concentrated 100 fold. CDK4-expressing FSHD and control myoblasts were plated at a density of 26 000 cells per cm2 and, after 24 h of proliferation, were transduced with 50 µl of concentrated virus in 1 ml of myoblast proliferation medium containing 8 µg/ml polybrene (Sigma-Aldrich, St. Louis, MO) for 2 h. Cells were allowed to recover and proliferate overnight, then growth of transduced cells was selected in myoblast proliferation medium containing 300 µg/ml active G418 (Life Technologies).
siRNA delivery
For imaging experiments, siRNAs were delivered to myoblasts after 24 h of high-density growth in 384-well tissue culture dishes and subsequent media change into 25 µl of KOSR differentiation medium. One hundred microliters of room temperature, pH-equilibrated OptiMEM I Reduced Serum Media (Life Technologies) was mixed with 1.8 µl of DharmaFECT 1 Transfection Reagent (Dharmacon, GE Healthcare #T-2001-01, Lafayette, CO) and incubated at room temperature for 5 min. Nineteen microliters of the transfection mix was mixed with 1 µl of 10 µM Universal Non-Targeting siRNA (Dharmacon siGenome #D001210-01-05) or DUX4-targeting siRNA (Dharmacon siGenome, custom design: CCGAGCCUUUGAGAAGAUCGCUUU), and incubated at 37°C for 1 h. Ten microliters of the siRNA mix was then combined with 200 µl of differentiation medium, and 25 µl of this transfection mixture was added to the 25 µl of differentiation medium in each well. As a positive control for siRNA transfection efficiency, BLOCK-iT Alexa Fluor Red Fluorescent siRNA Control (Life Technologies) was transfected in parallel wells, and fluorescence was analyzed (data not shown). For RNA preparation, cells were plated at high density in 35 mm tissue culture dishes and changed to KOSR differentiation medium 24 h after plating. Five microliters of DharmaFECT 1 was mixed with 125 µl of equilibrated OptiMEM. Separately, 100 picomols of non-targeting or DUX4-targeting siRNA was mixed with 125 µl of OptiMEM. The transfection reagent and siRNA dilutions were then combined and incubated for 30 min at room temperature, and then added drop wise to the cells. In all experiments, cells were differentiated in the siRNA-containing KOSR differentiation medium for 48–72 h.
Immunofluorescent staining, microscopy and line scan analysis
Immunofluorescent staining was performed by first fixing adherent cells in 4% paraformaldehyde (Thermo Scientific) for 10 min at room temperature, then washing with phosphate-buffered saline (PBS, Life Technologies) containing 0.005% Triton X-100 (Sigma-Aldrich), permeablizing with 0.5% Triton X-100 in PBS at room temperature for 10 min, and washing again. Cells were treated with Image-iT FX signal enhancer (Life Technologies) for 30 min at room temperature in a humidified chamber. Immunostaining was performed overnight at 4°C with a 1:5000 dilution of DUX4 E5-5 rabbit monoclonal antibody (Abcam, Cambridge, UK) in PBS, a 1:5000 dilution of MF20 mouse monoclonal antibody against myosin heavy chain (MHC) (R&D Systems, Minneapolis, MN) or a 1:1000 dilution of ZNF217 rabbit polyclonal antibody (Sigma-Aldrich). Cells were then washed and labeled with fluorescent-conjugated Alexa 488 or 594 goat anti-rabbit or goat anti-mouse secondary antibodies (Life Technologies) for 2 h at room temperature, then washed again. Counterstaining with 200 nM 4′,6-diamidino-2-phnylindole dihydrochloride (DAPI, Sigma-Aldrich) or 1 µM 1,5-bis([2-(di-methylamino)ethyl]amino)-4, 8-dihydroxyanthracene-9,10-dione (Draq5, Cell Signaling Technologies, Danvers, MA) for 2 h at room temperature was completed to allow visualization of nuclei by staining DNA. Images of immunofluorescent staining were obtained with a High-Resolution-Optimized Nikon TiE Inverted Widefield Microscope. Line scan analyses were performed using Nikon's NIS-Elements Image Analysis software. For live cell experiments, myoblasts were differentiated (see ‘Cell Culture’ section) in an environmentally controlled microscope enclosure and bright field, and green fluorescence images were captured every 15 min for 120 h using a Nikon TiE Inverted Widefield Fluorescence Microscope. All imaging experiments were performed at the University of Washington's Lynn and Mike Garvey Cell Imaging Lab.
Flow cytometry
CDK4-expressing FSHD and control myoblasts, previously transduced with the reporter vector and selected with G418, were plated at high density, and differentiation was induced (see ‘Cell Culture’ section) in the presence of 1 mm EGTA (Sigma-Aldrich) to inhibit cell fusion. After 48–72 h of differentiation, cells were treated with 0.05% trypsin–EDTA (Life Technologies), collected, pelleted by centrifugation, and resuspended in PBS containing 1% fetal bovine serum (Thermo Scientific). Cells strained through a Cell-Strainer Cap (Falcon by Corning, Corning, NY), were analyzed and sorted in the University of Washington Cell Analysis Facility using a four-laser Aria II cell sorter. Gates were set by comparison of BFP fluorescence against a control laser to identify autofluorescence, and both BFP+ and BFP– populations were collected from FSHD cells.
RNA preparation and qRT-PCR analysis
RNA isolation was performed by exposing sorted or adherent cells to 500 μl or 1 ml of TRIzol Cell Lysis Reagent (Life Technologies) at room temperature for 10 min. One-fifth volume of chloroform was added, and samples were mixed vigorously and centrifuged to separate phases. The aqueous phase was extracted with chloroform, and RNA was precipitated with ½ volume of 2-propanol. RNA pellets were washed with 75% EtOH, allowed to dry briefly and resuspended in RNAse-free water. Ten micrograms of RNA was incubated with 5 U DNAseI (NEB, Ipswich, MA) at 37°C for 15 min. DNAsed RNA samples were mixed with Qiagen's RPE buffer, and standard RNeasy column cleanup was performed (Qiagen). One microgram of DNAsed RNA was then reverse transcribed to cDNA using Oligo dT primers and the Superscript III First-Strand Synthesis System (Life Technologies) alongside control reactions containing all components except for reverse transcriptase. The 20 μl reactions were incubated at 65°C for 5 min, followed by 50°C for 50 min and 85°C for 5 min. Next, cDNAs were diluted to 400 μl in DNAse-free water for use in qRT-PCR analysis for DUX4 (F—GGCCCGGTGAGAGACTCCACA, R—CCAGGAGATGTAACTCTAATCCAGGTTTGC), GAPDH (F—GTGAAGGTCGGAGTCAAC, R—TGAGGTCAATGAAGGGGTC) or BFP (F—ATCATGGCCGTCAAGCAGAA, R—TCTCGTTGGGGTCTTTGCTC), using the Roche Fast Start Universal SYBR Mastermix with ROX (Roche, Basel, Switzerland). The qRT-PCRs cycled on an ABI-7900HT machine at 50°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. qRT-PCR analyses on the same cDNAs that amplified DUX4-activated CCNA1 and MBD3L2 transcripts were performed using pre-validated TaqMan probes (Hs00544743_m1 and Hs00171105_m1, Applied Biosystems, Waltham, MA) and multiplexed with the RNAseP Taqman Endogenous Control Reference Assay (catalog #4401631, Applied Biosystems) using the ABI Absolute Mastermix System (Life Technologies). TaqMan qRT-PCRs were run on an ABI-7900HT machine at 95°C for 10 min, then for 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Samples for cell differentiation and RNA preparation experiments were collected from biological triplicates, and technical triplicates of all qRT-PCRs were analyzed.
RNA sequencing
Total RNA from BFP+ and BFP– cells from two different FSHD myoblast lines was prepped in triplicate as above, and RNA integrity was checked using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA) and quantified using a Trinean DropSense96 spectrophotometer (Caliper Life Sciences, PerkinElmer, Hopkinton, MA). RNA-seq libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA) on a Perkin Elmer Sciclone NGSx Workstation. Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies) and additional library QC, blending of pooled indexed libraries and cluster optimization were performed using Life Technologies’ Invitrogen Qubit® 2.0 Fluorometer. RNA-seq libraries were pooled (19-plex) and clustered onto three lanes of rapid-run flow cells using an Illumina cBot. Sequencing was performed using an Illumina HiSeq 2500 in rapid-run mode, employing a paired-end, 50 base read length (PE50) sequencing strategy. Image analysis and base calling were performed using RTA v1.18.61 Software (Illumina), files were demultiplexed and generated in FASTQ format using BCL2FASTQ Conversion Software v.1.8.4 (Illumina), and reads containing sequence that did not pass Illumina's base call quality threshold were eliminated. Raw sequence reads are available for download from the NCBI sequence read archive (SRA) database (study accession SRP058319). Reads were aligned to UCSC's genome build HG19 using TopHat v2.0.12 running Bowtie 2.2.3 ‘under the hood’. FastQC (Babraham Bioinformatics, Cambridge, UK) and RNASeQC reports were generated. RNA sequencing and read alignments were performed by the Fred Hutchison Cancer Research Center's Genomics Resource Core. The number of reads aligning to specific genomic features was quantified using the DESeq2 bioconductor package (38) with the intersection-strict overlapping mode and R scripting language. Gene pathway enrichment analysis was performed using the Generally Applicable Gene-set Enrichment for Pathway Analysis (GAGE) package (25) and referencing the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene sets (39) that were constructed on the fly using the keg.gsets command in R. Previously published CHIP-seq data were used to identify DUX4 binding sites (13). FASTQ files were downloaded and aligned to the hg19 reference genome using bowtie2 (40). Aligned reads were analyzed using ChIPpeakAnno package (41) to call peaks and to identify the nearest genomic feature. RNA splicing analysis was performed using the exon-centric analysis pathway in the python program MISO (mixture of isoforms) (32). Comparisons were made between RNA-seq data gathered from reporter-negative and reporter-positive cell populations of each line in biological triplicates. Percent spliced in Ψ values for a specific splicing event were then averaged over the six comparisons to obtain an average Ψ for a particular splicing event. Splicing data were plotted using a standalone python script called Sashimi plot (42).
Supplementary Material
Funding
This work was supported by the National Institutes of Health (NIAMS AR064197-01A1), the Friends of FSH Research, the FSHD Canada Foundation, Kacy Murray and the Anderson Family Foundation.
Supplementary Material
Acknowledgements
We thank Miller Lab members Gregory J. Block, Ira Martopullo and Alisa Jion Kim for technical assistance and discussions. We also thank Ron Seifert from the University of Washington Garvey Imaging Core, Thane Mittelstaedt and Michele Black from the University of Washington Cell Analysis Facility, and Alyssa Dawson and Jeffrey Delrow from the Fred Hutchison Cancer Research Center Genomics Resource. Lastly, we especially thank the volunteers whose generous tissue donations were used to generate the cell lines used in this study, and Dr. Rabi Tawil for his work to establish the Fields Center FSHD bio-repository.
Conflict of Interest statement. None declared.
References
- 1.Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tawil R. (2008) Facioscapulohumeral muscular dystrophy. Neurotherapeutics, 5, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor D.A., Carroll J.E., Smith M.E., Johnson M.O., Johnston G.P., Brooke M.H. (1982) Facioscapulohumeral dystrophy associated with hearing loss and Coats syndrome. Ann. Neurol., 12, 395–398. [DOI] [PubMed] [Google Scholar]
- 4.Gurwin E.B., Fitzsimons R.B., Sehmi K.S., Bird A.C. (1985) Retinal telangiectasis in facioscapulohumeral muscular dystrophy with deafness. Arch. Ophthalmol., 103, 1695–1700. [DOI] [PubMed] [Google Scholar]
- 5.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet., 35, 315–317. [DOI] [PubMed] [Google Scholar]
- 6.de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M. (2009) Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat., 30, 1449–1459. [DOI] [PubMed] [Google Scholar]
- 7.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A. et al. (2009) Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet., 5, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R. et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. [DOI] [PubMed] [Google Scholar]
- 9.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet., 44, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D. et al. (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene, 236, 25–32. [DOI] [PubMed] [Google Scholar]
- 11.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R. et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R. et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F. et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J., 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O. et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord., 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 16.Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol., 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dandapat A., Bosnakovski D., Hartweck L.M., Arpke R.W., Baltgalvis K.A., Vang D., Baik J., Darabi R., Perlingeiro R.C., Hamra F.K. et al. (2014) Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep., 8, 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Block G.J., Petek L.M., Narayanan D., Amell A.M., Moore J.M., Rabaia N.A., Tyler A., van der Maarel S.M., Tawil R., Filippova G.N. et al. (2012) Asymmetric bidirectional transcription from the FSHD-causing D4Z4 array modulates DUX4 production. PloS one, 7, e35532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandri M., El Meslemani A.H., Sandri C., Schjerling P., Vissing K., Andersen J.L., Rossini K., Carraro U., Angelini C. (2001) Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment? J. Neuropathol. Exp. Neurol., 60, 302–312. [DOI] [PubMed] [Google Scholar]
- 21.Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. (2013) Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet., 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce Paterson R.S. (1972) Myosin synthesis in cultures of differentiating chicken embryo skeletal muscle. Dev. Biol., 29, 113–138. [DOI] [PubMed] [Google Scholar]
- 23.Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P. Jr et al. (2012) Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet., 21, 4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet., 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics, 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Q., Snider L., Jagannathan S., Tawil R., van der Maarel S.M., Tapscott S.J., Bradley R.K. (2015) A feedback loop between nonsense-mediated decay and the retrogene in facioscapulohumeral muscular dystrophy. eLife, 4, e04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philips A.V., Timchenko L.T., Cooper T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Pegoraro E., Menegazzo E., Gennarelli M., Hoop R.C., Angelini C., Hoffman E.P. (1995) Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum. Mol. Genet., 4, 599–606. [DOI] [PubMed] [Google Scholar]
- 29.Timchenko N.A., Iakova P., Cai Z.J., Smith J.R., Timchenko L.T. (2001) Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol., 21, 6927–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puppo F., Dionnet E., Gaillard M.C., Gaildrat P., Castro C., Vovan C., Bertaux K., Bernard R., Attarian S., Goto K. et al. (2015) Identification of variants in the 4q35 gene FAT1 in patients with a facioscapulohumeral dystrophy-like phenotype. Hum. Mutat., 36, 443–453. [DOI] [PubMed] [Google Scholar]
- 31.Caruso N., Herberth B., Bartoli M., Puppo F., Dumonceaux J., Zimmermann A., Denadai S., Lebosse M., Roche S., Geng L. et al. (2013) Deregulation of the protocadherin gene FAT1 alters muscle shapes: implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet., 9, e1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krom Y.D., Thijssen P.E., Young J.M., den Hamer B., Balog J., Yao Z., Maves L., Snider L., Knopp P., Zammit P.S. et al. (2013) Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet., 9, e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek A., Rahimov F., Jones P.L., Kunkel L.M. (2015) Emerging preclinical animal models for FSHD. Trends Mol. Med., 21, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppee F., Belayew A. (2011) The FSHD atrophic myotube phenotype is caused by DUX4 expression. PloS one, 6, e26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariot V., Roche S., Hourde C., Portilho D., Sacconi S., Puppo F., Duguez S., Rameau P., Caruso N., Delezoide A.L. et al. (2015) Correlation between low FAT1 expression and early affected muscle in FSHD. Ann. Neurol., DOI:10.1002/ana.24446. [DOI] [PubMed] [Google Scholar]
- 37.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol., 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. (2010) ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics, 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katz Y., Wang E.T., Silterra J., Schwartz S., Wong B., Thorvaldsdottir H., Robinson J.T., Mesirov J.P., Airoldi E.M., Burge C.B. (2015) Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics, 31, 2400–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.