Abstract
Citrus greening (huanglongbing) is the most destructive citrus disease worldwide. The disease is associated with three species of ‘Candidatus Liberibacter’ among which ‘Ca. Liberibacter asiaticus’ has the widest distribution. ‘Ca. L. asiaticus’ is commonly transmitted by a phloem-feeding insect vector, the Asian citrus psyllid Diaphorina citri. A previous study showed that isolates of ‘Ca. L. asiaticus’ were clearly differentiated by variable number of tandem repeat (VNTR) profiles at four loci in the genome. In this study, the VNTR analysis was further validated by assessing the stability of these repeats after multiplication of the pathogen upon host-to-host transmission using a ‘Ca. L. asiaticus’ strain from Japan. The results showed that some tandem repeats showed detectable changes after insect transmission. To our knowledge, this is the first report to demonstrate that the repeat numbers VNTR 002 and 077 of ‘Ca. L. asiaticus’ change through psyllid transmission. VNTRs in the recipient plant were apparently unrelated to the growing phase of the vector. In contrast, changes in the number of tandem repeats increased with longer acquisition and inoculation access periods, whereas changes were not observed through psyllid transmission after relatively short acquisition and inoculation access periods, up to 20 and 19 days, respectively.
Introduction
Citrus greening (huanglongbing) is a devastating citrus disease globally. The disease was first reported in China in the early 20th century, and it was known as yellow shoot disease in this region [1]. By the 1920s, diseases similar to yellow shoot disease were recorded in Taiwan (likubin or drooping disease) [2] and India (citrus dieback) [3]. In the 1940s, the disease was initially recorded in Indonesia and was described as vein phloem degeneration [4]. Since then, citrus greening has been reported in Japan and South Africa [5, 6]. In the western hemisphere, citrus greening was first reported in Sao Paulo, Brazil [7], and then in Florida, USA [8]. After the initial findings, the disease has apparently spread all over the Caribbean, North and Central American countries in a relatively short period [9, 10, 11, 12, 13, 14, 15].
The causal agents are phloem-limited gram-negative bacteria. Thus far, three species of this organism have been identified; ‘Ca. Liberibacter africanus’ is mainly found in Africa, ‘Ca. Liberibacter americanus’ is found in Brazil [16], and ‘Ca. Liberibacter asiaticus’ is widely distributed in Asian countries as well as in the Americas. The pathogens are transmitted by the African citrus psyllid, Trioza erytreae (Del Guercio), in Africa [17] and the Asian citrus psyllid, Diaphorina citri Kuwayama, in Asia and the Americas [18]. The disease also spreads with contaminated plant material used for the propagation of nursery plants.
Methods for distinguishing bacterial isolates are important for epidemiological analyses to elucidate the genetic structure of microbial populations. Variable number of tandem repeat (VNTR) markers, also known as microsatellites or simple sequence repeats, are tandem repetitive DNA sequences with repeated motifs of 2–6 bp or more [19]. VNTRs in bacterial DNA have been used as markers for differentiating and subtyping strains of several bacterial species, including ‘Ca. L. asiaticus’ [20, 21, 22]. In a previous study, Katoh et al. [21] selected four highly polymorphic VNTR loci and these VNTR markers were used to estimate the genetic diversity and population structures of ‘Ca. L. asiaticus’ in Japan, Taiwan, India, Indonesia, Timor-Leste, Papua New Guinea, and Florida [21, 22]. Using these VNTR markers at four loci, Matos et al. [23] also performed an extended analysis of ‘Ca. L. asiaticus’ populations in several countries, including the Caribbean, Central America regions, and Mexico.
Matos et al. [23] also examined the stability of VNTRs in the four loci of the ‘Ca. L. asiaticus’ genome after transmission of the pathogen in citrus and psyllid hosts. It was shown that the profiles do not change upon passage of the pathogen in citrus and psyllid hosts as well as after it has remained within a host for over five years. However, the number of plant samples analyzed by Matos et al. [23] was smaller than that used in earlier studies [20, 21, 22], and did not include two important VNTRs: 002 and 077, which are designated as “cold Motif C and D” by Matos et al. [23]. In order to supplement the results of Matos et al. [23], we further analyzed changes in VNTRs through insect transmission and showed that some changes occurred even in the supposedly “cold Motifs”.
Materials and Methods
Psyllid and plant preparation
Wataru Ashihara, who worked in National Institute of Fruit Tree Science, received permission from the owner of the orchard to collect sample as well as the previous study [24]. A Diaphorina citri population originating from the Amami-ôshima Island of Kagoshima Prefecture, Japan was maintained on potted trees of orange jessamine, Murraya paniculata (L.) Jack (Rutaceae). To detect psyllid infection with ‘Ca. L. asiaticus’, PCR was conducted with OI1/OI2c primer [25, 26] and DNA extracted from feeding plant during periodic intervals. Rough lemon seedlings (Citrus jambhiri Lush, approximately 40 cm in height), grafted with ‘Ca. L. asiaticus’-infected scions originating from the Ishigaki Island of Okinawa Prefecture, Japan were used as a source for pathogen acquisition by psyllids. Plants were maintained in a temperature-controlled greenhouse at 30 and 25°C during the day and night, respectively. Healthy seedlings of yuzu (Citrus junos Tanaka), tankan mandarin (Citrus tankan Hayata), unzoki (Citrus keraji Tanaka), and orange jessamine (M. maniculata), approximately 10 cm in height, were prepared as transmission recipient plants. To detect infection with ‘Ca. L. asiaticus’, PCR was also conducted with OI1/OI2c primer [25, 26] and DNA extracted from their plant during periodic intervals.
Psyllid transmission experiment
All experiments using live D. citri were performed in growth chambers at 25°C with a 16L:8D photoperiod at the Kuchinotsu Citrus Research Station, NIFTS (Otsu 954, Kuchinotsu, Minamishimabara, Nagasaki 859–2501, Japan). For adult acquisition–adult transmission experimental tests, 10-day-old post-emerging ‘Ca. L. asiaticus’- negative adults were placed on a leaf of an infected rough lemon tree within a plastic tube (9 cm in length and 3 cm in diameter) for an acquisition feeding. After an acquisition access period (AAP) of 55 days, each adult individual was transferred to a healthy yuzu seedling for an inoculation access period (IAP) of 20 days.
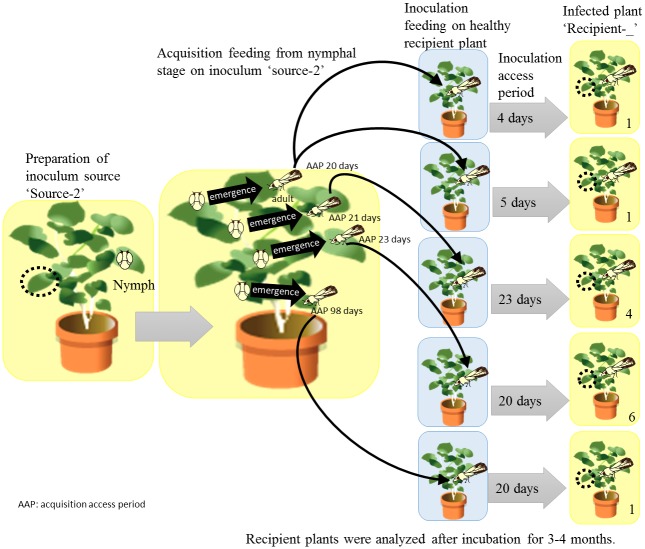
As for nymphal acquisition–adult transmission tests, healthy fifth instars maintained on ‘Ca. L. asiaticus’-negative M. paniculata were transferred to an infected rough lemon tree with a fine brush. These nymphs became adults during the acquisition feeding. After an AAP of 20–98 days, each adult individual was transferred to a healthy yuzu, tankan mandarin, unzoki, or orange jessamine seedling for an IAP of 4–23 days. The number of insect used for each IAP is dictate in Fig 1. Recipient plants were maintained for 3–4 months in a temperature-controlled greenhouse until DNA exrtraction. After inoculation feeding, all psyllids were collected and preserved at –50°C until DNA extraction. For all psyllid transmission experiment, single psyllid was used in IAP per plant. Twenty-seven of 144 inoculative psyllids died during IAPs (mortality rate = 18.8%) and the pathogen was successfully transmitted to 42 of 144 recipient plants by psyllids (transmission rate = 29.2%). Inoculative psyllid DNA samples from which sufficient copies of ‘Ca. L. asiaticus’ genome were detected by quantitative real-time PCR assays [24] were used for the subsequent analysis of VNTRs of the “Ca, L, asiaticus” genome in psyllids.
Fig 1. Schematic model of strategy of Table 2.
The black circle indicates the area used for DNA extraction. Numbers at the lower right, which are x n, indicate the number of samples in Table 2.
DNA extraction
Leaves were collected from the leaf midrib tissue of each leaf collected in citrus trees infected with ‘Ca. L. asiaticus’ was selected for DNA extraction. Total DNA was extracted with the DNeasy plant minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
In order to extract DNA from psyllids, the body was dissected into the anterior part (head and prothorax), which includes salivary glands, and the posterior part (mesothorax, metathorax, and abdomen) using a disposable razor. Total DNA was purified from each part of a single psyllid using the DNeasy Blood and Tissue Kit (Qiagen, Tokyo, Japan) and a plastic homogenizer pestle (As One, Tokyo, Japan) according to the manufacturer’s instructions, and eluted in 100 μL of AE buffer provided in the kit. PCR amplification was performed using each part of a single psyllid as template.
Primers and PCR
Four loci within the ‘Ca. L. asiaticus’ genome containing TACAGAA, CAGT, AGACACA, and TTTG motifs [20, 21, 27] were examined in this study. These motifs had been previously designated as 001, 002, 005, and 077, respectively, [21]. The fragments containing these loci were amplified by PCR using the primer sets listed in Table 1, as previously reported [21]. Additionally, another forward primer was designed in order to be easy to count the motif of VNTR ‘001’ by using a program available on the Primer3 website (http://frodo.wi.mit.edu/primer3/) (Table 1).
Table 1. Primers for characterization of variable tandem repeats in four loci of the genome of 'Ca. Liberibacter asiaticus' strains.
| Motif | Sequence | Genome position | Primers' seqoence 5'-3' | Reference | Amplicon size(bp) |
|---|---|---|---|---|---|
| 001 | TACAGGA | 255591–255646 | (+)ggtgaattaggatggaaatgc | [21] | 1159 |
| (-)tgaagtagctctgcaatatctga | |||||
| (-)tgcctcttctagggcacaa | this paper | The primer was used for only sequence. | |||
| 002 | CAGT | 537729–537760 | (+)ttgataatatagaaagaggcgaagc | [21] | 510 |
| (-)tccatacccaaaagaaaagca | |||||
| 005 | AGACACA | 354493–354527 | (+)attgaaggacgaaaccgatg | [21] | 598 |
| (-)tcccaaggttttcaaattgc | |||||
| 077 | TTTG | 655277–655332 | (+)tgactgatggcaaaagatgg | [21] | 528 |
| (-)agacacgccaaacaaggaat |
PCR was performed using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) in a 20-μL reaction mixture containing 1 μL of DNA template, 0.1 μM of each primer, 200 μM dNTP mixture, 1× PCR buffer, and 2.5 units of Ex Taq DNA polymerase Hot Start Version (TaKaRa, Shiga, Japan). The thermal cycling conditions were as follows: initial denaturation at 92°C for 2 min; 35 cycles of denaturing at 92°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 1 min. Amplified PCR products were separated by electrophoresis in a 1.5% (wt/vol) agarose gel in Tris-boric acid EDTA buffer. Before direct sequencing, the PCR products were extracted from the gel slices using a QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s instructions.
Direct sequencing and data analysis
The nucleotide sequences of the DNA fragments were obtained by directly sequencing both strands of the purified PCR products using the dideoxynucleotide triphosphate (ddNTP) termination method [28] with ABI 3130 sequencer (Applied Biosystems, Foster City, CA, USA). The number of repetitions for each VNTR was manually counted from the sequence data.
Results
Change in VNTRs through psyllid transmission
Some changes were confirmed in the profiles of the tandem repeats with psyllid transmission (Table 2, Fig 1). Changes in VNTR ‘001’ occurred with psyllid transmission in two different citrus hosts (yuzu and unzoki). In addition, changes in VNTR ‘077’ were observed in psyllid transmission to orange jessamine (Murraya paniculata), a citrus relative (Table 2).
Table 2. Analysis of the stability of tandem repeats through nymphal acquisition–adult transmission experiments.
| VNTR | |||||||
|---|---|---|---|---|---|---|---|
| Sample ID | Organism d | 001 | 002 | 005 | 077 | Acquisition access period (days) | Inoculation access period (days) |
| Source-2 a | Rough lemon | 14 | 8 | 8 | 9 | ||
| Recipient-4 b | Yuzu | 14 | 8 | 8 | 9 | 20 | 5 |
| Recipient-5 | Yuzu | 14 | 8 | 8 | 9 | 20 | 4 |
| Recipient-6 | Unzoki | 13 c | 8 | 8 | 9 | 23 | 20 |
| Recipient-8 | Unzoki | 14 | 8 | 8 | 9 | 23 | 20 |
| Recipient-9 | Unzoki | 14 | 8 | 8 | 9 | 23 | 20 |
| Recipient-12 | Unzoki | 14 | 8 | 8 | 9 | 23 | 20 |
| Recipient-13 | Unzoki | 15 | 8 | 8 | 9 | 23 | 20 |
| Recipient-17 | Unzoki | 14 | 8 | 8 | 9 | 23 | 20 |
| Recipient-18 | Tankan mandarin | 14 | 8 | 8 | 9 | 98 | 20 |
| Recipient-19 | Yuzu | 14 | 8 | 8 | 9 | 21 | 23 |
| Recipient-20 | Yuzu | 15 | 8 | 8 | 9 | 21 | 23 |
| Recipient-21 | Yuzu | 14 | 8 | 8 | 9 | 21 | 23 |
| Recipient-22 | Orange jessamine | 14 | 8 | 8 | 8 | 21 | 23 |
a‘Ca. L. asiaticus’-infected greenhouse grown rough lemon tree used as inoculum source for nymphal acquisition–adult transmission of the pathogen.
bRecepor plants that became infected upon psyllid transmission of the bacterium from 'Source-2' plant.
cUnderline shows varied number of VNTR compared with that from 'Source-2' plant.
dRough lemon, Citrus jambhiri; yuzu, Citrus junos; unzoki, Citrus keraji; tankan mandarin, Citrus tankan; orange jessamine, Murraya paniculata.
Changes in VNTRs in the ‘Ca. L. asiaticus’ genome occurred while the bacterium was in the psyllid body (Table 3, Fig 2). These VNTR changes in the psyllid body might have contributed to the changes of VNTRs in the ‘Ca. L. asiaticus’ genome in the recipient plant in some cases. For example, the profile of VNTRs of ‘Ca. L. asiaticus’ in one yuzu recipient plant (sample ID: Recipient-Y17) perfectly matches the VNTRs of ‘Ca. L. asiaticus’ in the psyllid body parts (Table 3).
Table 3. Analysis of the stability of tandem repeats through adult acquisition–adult transmission experiments.
| VNTR | |||||||
|---|---|---|---|---|---|---|---|
| Sample ID | Organism g | 001 | 002 | 005 | 077 | Acquisition access period (days) | Inoculation access period (days) |
| Source-R1 a | Rough lemon | 14 | 8 | 8 | 9 | ||
| Vector-head b -1 e | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen c -1 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient d -Y1 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-4 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-4 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y4 | Yuzu | 14 | 9 f | 9 | 9 | 20 | |
| Vector-head-6 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-6 | Psyllid | 14 | 9 | 8 | 9 | 55 | |
| Recipient-Y6 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-8 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-8 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y8 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-12 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-12 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y12 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-13 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-13 | Psllid | 15 | 8 | 8 | 9 | 55 | |
| Recipient-Y13 | Yuzu | 15 | 8 | 8 | 9 | 20 | |
| Vector-head-15 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-15 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y15 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-16 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-16 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y16 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-17 | Psyllid | 15 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-17 | Psyllid | 15 | 8 | 8 | 9 | 55 | |
| Recipient-Y17 | Yuzu | 15 | 8 | 8 | 9 | 20 | |
| Vector-head-20 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-20 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y20 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-21 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-21 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y21 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-24 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-24 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y24 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-27 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-27 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y27 | Yuzu | 14 | 8 | 8 | 9 | 20 | |
| Vector-head-30 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-30 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y30 | Yuzu | 14 | 8 | 9 | 9 | 20 | |
| Vector-head-11 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Vecor-abdomen-11 | Psyllid | 14 | 8 | 8 | 9 | 55 | |
| Recipient-Y11 | Yuzu | ND | ND | ND | ND | 20 | |
| Vector-head-29 | Psyllid | ND | ND | ND | ND | 55 | |
| Vecor-abdomen-29 | Psyllid | ND | ND | ND | ND | 55 | |
| Recipient-Y29 | Yuzu | 16 | 8 | 8 | 9 | 20 | |
a‘Ca. L. asiaticus’-infected greenhouse grown rough lemon (Source-R1) used as an inoculum source for psyllid transmission of the pathogen.
bAnterior part (including head and prothorax) of each psyllid individual after acquisition feeding on source-R1 plant and inoculation feeding on recipient yuzu seedling.
cPosterior part (including mesothorax, metathorax, and abdomen) of each psyllid individual after acquisition feeding on source-R1 plant and inoculation feeding on recipient yuzu seedling.
dRecipient plants that became infected upon psyllid transmission of the bacterium from the source plant 'Source-R1'.
eLast number between the psyllid as vector and the recipient plant was one-to-one correspondence, respectively.
fUnderline shows varied number of VNTR compared with that from source plant ‘Source-R1’.
gPsyllid, Diaphorina citri; yuzu, Citrus junos.
Fig 2. Schematic model of strategy of Table 3.
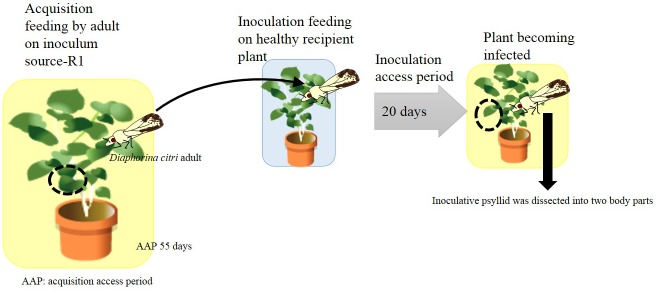
The black circle indicates the area used for DNA extraction. Numbers at the lower right, which are x n, indicate the number of samples in Table 3.
However, VNTRs of ‘Ca. L. asiaticus’ in the body parts of psyllids were not always related to those of ‘Ca. L. asiaticus’ in the recipient plants. For example, three VNTRs of ‘Ca. L. asiaticus’ in one a yuzu recipient plant (sample ID: Recipient-Y4) were different from those of the source plant, whereas all four VNTRs of ‘Ca. L. asiaticus’ in the body parts of the vector were unchanged. Such discrepancy was also observed in the transmission test using sample ID Vector-abdomen-6 (psyllid abdomen) and sample ID Recipient-Y6 (recipient), as well as in the test using sample ID Vector-head-13 (psyllid head) and sample ID Recipient-Y13 (recipient).
In Table 3, we show that VNTRs change after psyllid transmission with AAP and IAP of 55 and 20 days, respectively. Subsequently, we attempted psyllid transmission with a shorter AAP and IAP, and found that the four VNTRs did not change with shorter periods: AAP and IAP of 20 and 19 days, respectively (Table 4, Fig 3).
Table 4. Analysis of the stability of tandem repeats through nymphal acquisition–adult transmission of the pathogen from yuzu infected with ‘Ca. L. asiaticus’ for a short period.
| VNTR | |||||||
|---|---|---|---|---|---|---|---|
| Sample ID | Organism | 001 | 002 | 005 | 077 | Acquisition access period (days) | Inoculation access period (days) |
| Sorce-R2 a | Yuzu | 14 | 8 | 8 | 9 | 20 | 19 |
| Vector-head b -10 e | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen c -10 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient d -Y10 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-12 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-12 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y12 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-13 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-13 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y13 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-14 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-14 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y14 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-15 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-15 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y15 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-31 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-31 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y31 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-45 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-45 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y45 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
| Vector-head-48 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Vecor-abdomen-48 | Citrus psyllid | 14 | 8 | 8 | 9 | 20 | |
| Recipient-Y48 | Yuzu | 14 | 8 | 8 | 9 | 19 | |
a‘Ca. L. asiaticus’-infected greenhouse grown yuzu (Sorce-R2) used as an inoculum source for psyllid transmission of the pathogen.
bSame number between the psyllid as vector and the recipient plant was one-to-one correspondence, respectively.
cAnterior part (including head and prothorax) of each psyllid individual after acquisition feeding on source-R2 plant and inoculation feeding on recipient yuzu seedling.
dPosterior part (including mesothorax, metathorax, and abdomen) of each psyllid individual after acquisition feeding on source-R2 plant and inoculation feeding on recipient yuzu seedling.
eRecipient plants that became infected upon psyllid transmission of the bacterium from the source Sorce-R2 plant.
Fig 3. Schematic model of strategy of Table 4.
The black circle indicates the area used for DNA extraction. Numbers at the lower right, which are x n, indicate the number of samples in Table 4.
Discussion
This study suggested that psyllid transmission contributes to changes in the four VNTRs of ‘Ca. L. asiaticus’, and that both AAPs and IAPs by psyllids are involved. It was also suggested that longer acquisition/inoculation access would result in more changes in the four VNTRs, as significant differences in the frequency of alteration of tandem repeats among the four VNTRs were observed using direct sequencing. In this study, the most frequently varied tandem number among the four VNTRs was ‘001’, which changed in six samples in this study, and the VNTR ‘005’ locus was the second most frequently changed VNTR. In contrast, changes in VNTRs ‘002’ and ‘077’ were rarely seen in this study. These differences agree with Nei's measure of genetic diversity for each VNTR that was calculated among Japanese ‘Ca. L. asiaticus’ strains [21]. However, variability analysis of VNTRs with graft transmission is still insufficient because of the lack of specimens.
Matos et al. [23] indicated that the numbers of tandem repeats in the four loci tested displayed highly distinguishable “signature profiles” for the two Florida-type ‘Ca. L. asiaticus’ haplotype groups. However, the strain used in this study did not look like either of those haplotypes. The whole genome sequence of the strain used in this study also contained unique features [29]. The two haplotypes enumerated by Matos et al. [23] might show different trends when Japanese ‘Ca. L. asiaticus’ isolates are used for genetic analysis.
To our knowledge, this is the first report of a change in the tandem repeat VNTR ‘077’ after psyllid transmission. The results of this study suggested that the four VNTRs often change after psyllid transmission.
Folimonova et al. [30] indicated that real-time quantitative PCR assays of nonsymptomatic tissue located next to asymptomatic area for different hosts showed that this tissue sometimes did and sometimes did not contain detectable amounts of the bacterium [30]. Inoue et al. [24] suggested that longer AAPs result in higher percentages of ‘Ca. L. asiaticus’-positive psyllids, and that adult psyllids, after acquisition feeding in the nymph stage, enhanced proliferation and efficient transmission of the bacterium [24]. Growth efficiency might also affect stability of the four VNTRs in the bacterium.
The results of this study contribute to the understanding of relationships among different ‘Ca. L. asiaticus’ strains that are isolated from different fields and orchards, and might be used to examine effective transmission methods (by vegetative propagation or by psyllid transmission).
Acknowledgments
We also acknowledge Ms Hatomi for her meticulous technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
Funding was provided by National Agriculture and Food Research Organization (http://www.naro.affrc.go.jp/fruit/) and Center for Bioscience Research and Education, Utsunomiya University, http://c-bio.mine.utsunomiya-u.ac.jp/.
References
- 1. Zhao X. Citrus yellow shoot disease (Huanglongbing)—A review. Proc Int Soc Citriculture. 1981;1: 466–469. [Google Scholar]
- 2. Otake A. Bibliography of citrus greening disease and its vectors attached with indices and a critical review on the ecology of the vectors and their control. Japanese International Cooperative Agency, Tokyo: 1990; 161 p. [Google Scholar]
- 3. Capoor SP. Decline of citrus trees in India. Bull Natl Inst Sci India. 1963;24: 48–64. [Google Scholar]
- 4. Tirtawidjaja S, Hadewidjaja T, Lasheen AM. Citrus vein phloem degeneration virus, a possible cause of citrus chlorosis in Java. Proc Am Soc Hon Sci. 1965;86: 23–243. [Google Scholar]
- 5. Miyakawa T, Tsuno K. Occurrence of citrus greening disease in the southern islands of Japan. Ann Phytopathol Soc Jpn. 1989;55: 667–670. [Google Scholar]
- 6. Oberholzer PCJ, von Standen DFA, Basson WJ. Greening disease of sweet orange in South Africa In: Price WC, editor. Proceedings of the 3rd Conference of the International Organization of Citrus Virology; Gainesville, Florida. Gainesville: University of Florida Press; 1965. p. 213–19. [Google Scholar]
- 7. Coletta-Filho HD, Targon MLPN, Takita MA, De Negri JD, Pompeu JJ, Marcado MA. First report of the causal agent of Huanglongbing (“Candidatus Liberibacter asiaticus”) in Brazil. Plant Dis. 2004;88: 1382. [DOI] [PubMed] [Google Scholar]
- 8.Halbert SE. The discovery of huanglongbing in Florida. Proceedings of the 2nd International Citrus Canker and Huanglongbing Research Workshop; 2005 Nov; Orlando Florida, USA, 2005. p. 50.
- 9. Martínez Y, Llauger R, Batista L, Luis M, Iglesia A, Collazo C, et al. First report of ‘Candidatus Liberibacter asiaticus’ associated with Huanglongbing in Cuba. Plant Pathol. 2008;58: 389. [Google Scholar]
- 10. Matos L, Hilf ME, Camejo J. First report of ‘Candidatus Liberibacter asiaticus’ associated with citrus Huanglongbing in the Dominican Republic. Plant Dis. 2009;93: 668. [DOI] [PubMed] [Google Scholar]
- 11. Manjunath KL, Ramadugu C, Majil VM, Williams S, Irey M, Lee RF. First report of the citrus Huanglongbing associated bacterium ‘Candidatus Liberibacter asiaticus’ from sweet orange, Mexican lime, and Asian citrus psyllid in Belize. Plant Dis. 2010;94: 781. [DOI] [PubMed] [Google Scholar]
- 12. Estébeth de Jensen C, Vitroreli A, Roman F. Citrus greening in commercial orchards in Puerto Rico. Phytopathology. 2010;100: S34. [Google Scholar]
- 13.ORISA (Organismo Internacional Regional de Sanidad Agropecuaria) (2011) Taller sensibilization sobre centrification en la produccion material vegetativo. Santo Domingo, Republica Dominicana. Septembre 2011.
- 14.Trujillo-Arriaga J (2010) Situación actual, regulación y manejo del HLB en México. 2 Taller internacional sobre el Huanglongbing y el psílido asiático de los cítricos. Merida, Yucatan, Mexico.
- 15. Brown SE, Oberheim AP, Barret A, McLaughlin WA. First Report of 'Candidatus Liberibacter asiaticus' associated with Huanglongbing in the weeds Cleome rutidosperma, Pisonia aculeate, and Trichostigma octandrum in Jamaica. New Dis Rep. 2011;24: 25. [Google Scholar]
- 16. Bové JM. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 2006;88: 7–37. [Google Scholar]
- 17. Aubert B. Trioza erytreae Del Guercio and Diaphorina citri Kuwayama (Homoptera: Psylloidea), the two vectors of citrus greening disease: biological aspects and possible control strategies. Fruits. 1987;42: 149–162. [Google Scholar]
- 18. Halbert SE, Manjunath KL. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assessment of risk in Florida. Fla Entomol. 2004;87: 330–353. [Google Scholar]
- 19. van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1989;62: 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, Deng X, Sun X, Jones D, Irey M, et al. Guangdong and Florida populations of ‘Candidatus Liberibacter asiaticus’ distinguished by a genomic locus with short tandem repeats. Phytopathology. 2010;100: 567–572. 10.1094/PHYTO-100-6-0567 [DOI] [PubMed] [Google Scholar]
- 21. Katoh H, Subandiyah S, Tomimura K, Okuda M, Su HJ, Iwanami T. Differentiation of “Candidatus Liberibacter asiaticus” isolates by variable-number tandem-repeat analysis. Appl Environ Microbiol. 2011;77: 1910–1917. 10.1128/AEM.01571-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katoh H, Davis R, Smith MW, Weinert M, Iwanami T. Differentiation of Indian, East Timorese, Papuan and Floridian ‘Candidatus Liberibacter asiaticus’ isolates on the basis of simple sequence repeat and single nucleotide polymorphism profiles at 25 loci. Ann Appl Biol. 2012;160: 291–297. [Google Scholar]
- 23. Matos LA, Hilf ME, Chen J, Folimonova SY. Validation of ‘variable number of tandem repeat’-based approach for examination of ‘Candidatus Liberibacter asiaticus’ diversity and its applications for the analysis of the pathogen populations in the areas of recent introduction. PLoS ONE. 2013;8(11): e78994 10.1371/journal.pone.0078994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inoue H, Ohnishi J, Ito T, Tomimura K, Miyata S, Iwanami T, et al. Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann Appl Biol. 2009;155: 29–36. [Google Scholar]
- 25. Jagoueix S., Bove JM, & Garnier M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int J Syst bacteriol 1994:44: 379–386. [DOI] [PubMed] [Google Scholar]
- 26. Jagoueix S., Bové J. M., & Garnier M. (1996). PCR detection of the two ‘Candidatus’ liberobacter species associated with greening disease of citrus.Mol Cell Probes. 1996: 10: 43–50. [DOI] [PubMed] [Google Scholar]
- 27. Islam Md-S, Glynn JM, Bai Y, Duan YP, Coletta-Filho H D, et al. Multilocus microsatellite analysis of ‘Candidatus Liberibacter asiaticus’ associated with citrus Huanglongbing worldwide. BMC Microbiol. 2012;12: 39 10.1186/1471-2180-12-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanger F, Nickien S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad U. S. A. 1997;74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh H, Miyata S, Inoue H, Iwanami T. Unique features of a Japanese ‘Candidatus Liberibacter asiaticus’ strain revealed by whole genome sequencing. PLOS ONE. 2014;9(9): e106109 10.1371/journal.pone.0106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Folimonova SY, Robertson CJ, Garnsey SM, Gowda S, Dawson WO. Examination of the responses of different genotypes of citrus to Huanglongbing (citrus greening) under different conditions. Phytopathology. 2009;99: 1346–1354. 10.1094/PHYTO-99-12-1346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.