Abstract
The 46,XX male disorder of sex development (DSD) is rarely observed in humans. Patients with DSD are all male with testicular tissue differentiation. The mechanism of sex determination and differentiation remains to be elucidated. In the present case report, an 46,XX inv (9) infertile male negative for the sex-determining region of the Y chromosome (SRY) gene was examined. This infertile male was systemically assessed by semen analysis, serum hormone testing and gonadal biopsy. Formalin-fixed and paraffin-embedded gonad tissues were assessed histochemically. The SRY gene was analyzed by fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR). The other 23 specific loci, including the azoospermia factor region on the Y chromosome and the sequence-targeted sites of the SRY-box 9 (SOX9) gene were analyzed by PCR. The genes RSPO1, DAX1, SOX3, ROCK, DMRT1, SPRY2 and FGF9 were also assessed using sequencing analysis. Affymetrix Cytogenetics Whole Genome 2.7 M Arrays were used for detecting the genomic DNA from the patient and the parents. The patient with the 46,XX inv (9) (p11q13) karyotype exhibited male primary, however, not secondary sexual characteristics. However, the patient's mother with the 46, XX inv (9) karyotype was unaffected. The testicular tissue dysplasia of the patient was confirmed by tissue biopsy and absence of the SRY gene, and the other 23 loci on the Y chromosome were confirmed by FISH and/or PCR. The RSPO1, DAX1, SOX3, ROCK, DMRT1, SPRY2 and FGF9 genes were sequenced and no mutations were detected. A duplication on the 3 M site in the upstream region of SOX9 was identified in the patient as well as in the mother. The patient with the 46,XX testicular DSD and SRY-negative status was found to be infertile. The duplication on the 3 M site in the upstream region of SOX9 was a polymorphism, which indicated that the change was not a cause of 46,XX male SDS. These clinical, molecular and cytogenetic findings suggested that other unidentified genetic or environmental factors are significant in the regulation of SDS.
Keywords: 46, XX testicular disorder of sex development, SRY-negative, SOX9, inv (9)
Introduction
The 46,XX male disorder of sex development (DSD) is a rare genetic condition (1). Patients with the 46,XX karyotype display various degrees of testicular tissue development. As far as the sexual phenotype is concerned, three clinical categories of sex-reversed 46,XX individuals have been identified: i) Classic XX males, infertility with normal male internal and external genitalia; ii) XX males with ambiguous genitalia, usually detected at birth by external genital ambiguities, including hypospadias, micropenis or hyperclitoridy; iii) XX true hermaphrodites, who exhibit internal or external genital ambiguities detected at birth (2–4).
At the molecular level, XX males can be classified by the sex-determining region Y gene (SRY) location on the Y chromosome, as SRY-positive or SRY-negative, which is important for encoding testis determining factor (TDF) (5). It is known that ~90% of these patients exhibit Y chromosomal material, including the SRY gene, which is usually translocated to the distal tip of the short arm of the X chromosome or autosomal chromosomes (6). The 46,XX males with SRY-positive status are clinically heterogeneous. The majority of the patients are normal prior to puberty and are diagnosed following puberty based on infertility. By contrast, 20% of 46,XX males positive for SRY exhibit an external genitalia abnormality at birth with the classical phenotype being hypospadias. In addition, certain patients are negative for SRY, and these patients always exhibit external genital ambiguities and contact the doctor for infertility and gynecomastia (7). The mechanism by which the development of testicular tissue in 46,XX males negative for SRY is regulated remains to be elucidated.
Of note, several genes have been identified to be associated with SRY-negative 46,XX male sex reversal patients. SOX9 and DAX1 have been suggested to function as early mediators downstream of the SRY gene in the male sex-determination pathway (8,9). SOX3 has been demonstrated to upregulate the expression of SOX9 via a similar mechanism to that of SRY and to be responsible for XX male sex reversal in humans through gain-of-function mutations mediated by genomic rearrangements around SOX3, possibly leading to the alteration of regulation (10). Disruption in the R-spondin1 (RSPO1) gene has been previously reported in a recessive syndrome characterized by XX sex reversal, palmoplantar hyperkeratosis and pre-disposition to squamous cell carcinoma (11). SPRY2 and FGF9 are also associated with 46,XX male sex reversal (12,13). Testes formation may be initiated by an alternative signaling pathway attributed to ROCK1 activation in the XX testes. On the basis of the inhibitory assay in vitro, it has been suggested that ROCK1 phosphorylates and activates SOX9 in Sertoli cells (14). The haploid dose of DMRT1 may lead to testicular dysplasia with XY male-to-female sex reversal (15,16). Seeherunvong et al (17) described a partial duplication of 22q in a case of 46,XX sex reversal, SRY-negative, and almost complete androphany of patient's exogenous genitals. Others have revealed that 46,XX SRY-negative males have a duplication in a regulatory region upstream of the SOX9 gene, which was overexpressed in these individuals (18–23).
The present case report described the clinical, fluorescence in situ hybridization (FISH) and molecular analyses of a 46,XX male DSD patient negative for SRY and aimed to investigate the association between the clinical characteristics and the chromosomal karyotype. The possible mechanisms to explain the etiology of the 46,XX sex reversal male negative for the SRY gene was also investigated.
Materials and methods
Case presentation
A 29-year-old male visited the outpatient clinic of the Center for Reproduction and Genetics (Suzhou Municipal Hospital, Nanjing Medical University Affiliated Suzhou Hospital, Suzhou, China) with a complaint of infertility. The patient reported that he had a surgical history of correction of congenital hypospadias at the age of 5 and presented with the development of mammary glands at 19 years of age. The parents are in a non-consanguineous marriage and the family members exhibited no clinical manifestations. The patient was found to be short in stature and the physical examination revealed no prominentia laryngea, armpit hair or beard, pale skin, a marginal increase of breast bilaterally and a surgical scar on the abdomen. The testicular volumes were small and the texture was hard. Rectal touch revealed a detectable prostate; however, the volume was low. Endocrinological data were indicated as follows: No sperm and spermatogenic cells according to semen examination; fructose in the seminal plasma was normal; seminal plasma α-glucosidase levels of 22.0 U/ml (normal range, 35.1–87.7 U/ml); seminal plasma acid phosphatase levels of 31.2 U/ml (normal range, 48.8–208.6 U/ml); a Serum T levels of 6.02 nmol/l (normal range, 9.4–37 nmol/l); estradiol levels of 0.11 nmol/l (normal range, 0.129–0.239 nmol/l); follicle-stimulating hormone levels of 51 IU/l (normal range, 1.5–11.5 IU/l), luteinizing hormone levels of 35.5 IU/l (normal range, 1.1–8.2 IU/l) and prolactin levels of 332.6 IU/l (normal range, 95.4–400 IU/l). No abnormality was identified during brain and adrenal computerized tomography examination. No uterus or ovary was detected, and other clinical indicators were normal.
All procedures used in the present study were performed according to the Declaration of Helsinki. The Ethics Committee of Jinling Hospital (Nanjing, China) approved the present study. Written informed consent was obtained from all participants.
Histological analysis
Formalin-fixed and paraffin-embedded gonad tissue from the affected individual was obtained from the Department of Pathology, Faculty of Medicine, Jinling Hospital (Nanjing, China) by punch biopsy. A 5 µm-thick serial section was cut from the tissue block and stained with hematoxylin and eosin (Beyotime Institute of Biotechnology, Inc., Haimen, China).
Cytogenetic analysis and FISH analysis
G banding karyotype analysis was performed on the peripheral blood lymphocytes, including 100 metaphase cells, from the patient by conventional operating techniques. The identical analysis was performed on the blood samples from the parents and the sister. The karyotype analysis was performed using FISH with the following two probes: Dual color centromere probe, DXZ1, with Spectrum Green and DYZ3 with Spectrum Orange (Vysis, Downers Grove, IL, USA; cat. no. 32-111051), and SRY with Orange (Vysis; cat. no. 30-190079). A total of 10 mitotic phases were analyzed, according to the manufacturer's instructions. Microscopic examination was performed using an Olympus BX51 microscope (Olympus, Tokyo, Japan), and analyzed by Cytovision 3.0 image analysis software (Leica Biosystems, Oberkochen, Germany). FISH analysis was also performed on samples from the patient's father.
Polymerase chain reaction (PCR) amplification and sequencing of the coding region of candidate genes
The genomic DNA was isolated from whole-blood leukocytes from the parents and paraffin-embedded tissue from the patient using phenol/chloroform extraction. The purpose of DNA extraction was to determine the presence or absence of SRY in the peripheral blood lymphocytes. Two primer pairs for SRY were designed using Primer 5.0 software (Premier Biosoft, Palo Alto, CA, USA) and the primers were synthesized by BGI (Beijing, China). Each PCR reaction (25 µl) contained 2 µl genomic DNA, 1 µl forward primer, 1 µl reward primer, 2.5 µl 10X buffer, 1.5 µl Mg2+, 2 µl deoxynucleotide triphosphates, 0.25 µl Taq DNA polymerase and 15 µl double-distilled water. PCR was performed under the following conditions: 95°C for 5 min followed by 35 cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 60 sec. PCR amplification products were assessed on 2% agarose gels. The products were then sequenced by BGI company. A total of 24 sequence-targeted sites (STS) were investigated to determine the presence or absence of Y chromosome material in the genome of the patient's blood. Firstly, sY14 (SRY), sY84, sY86, sY124, sY127, sY132, sY134, sY152, sY157, sY239, sY242, sY254 and sY255 were detected. Primers and amplification conditions were derived from the Genome Database (http://www.ncbi.nlm.nih.gov). To further confirm the presence or absence of the Y chromosome, 11 STS, including DYS19, DYS385, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439, were detected. At the same time, 15 autosomal sites, including D3S1358, D21S11, D18S51, D5S818, D13S317, D7S820, D16S539, D8S117P, TH01, CSFIPO, Penta E, Penta D, vWA, TPOX and FGA, were detected. The PCR products were analyzed on a DNA PowerPlex-Y sequencing system (Promega, Madison, WI, USA). The RSPO1, DAX1, SOX9, SOX3, ROCK, DMRT1, SPRY2 and FGF9 genes were amplified using custom-synthesized oligonucleotide primers.
The internal locus, including D17S794, D17S1350, D17S1351, D17S1352 and D17S1807 in SOX9, and D9S1858 in DMRT1, were detected. These loci were amplified, according to the information provided by the Genome Database, using fluorescently-labeled forward primers. The PCR products were analyzed on a DNA sequencer (ABI3500; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) and sequence analysis was performed by Genecore Company (Shanghai, China).
Single nucleotide polymorphism (SNP) array
The patient and the parents were analyzed using Affymetrix Cytogenetics 2.7 M (Affymetrix, Inc., Santa Clara, CA, USA). The genomic DNA was extracted from whole-blood leukocytes from the patient and the parents. The array experiment was performed according to the manufacturer's instructions. Briefly, the genomic DNA was denatured and neutralized, and subsequently amplified by PCR. The PCR products were purified, fragmented and end-labeled with biotin. The fragmented, labeled PCR products were subsequently hybridized to the arrays overnight. The copy number variation was analyzed by Affymetrix Chromosome Analysis Suite software (ChAS) v1.2.2 (Affymetrix, Inc.). The GRCh37 (Hg19) was used to determine the chromosome positions. The abovementioned experiments were performed with the assistance of Capital Bio Corporation (Beijing, China).
Results
Histological analysis
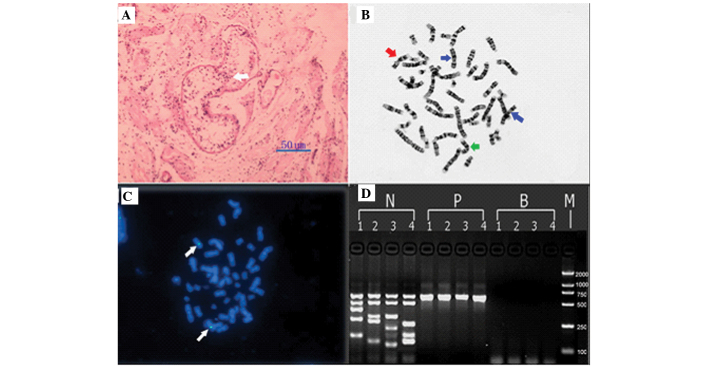
Assessment of a testicular biopsy of the patient demonstrated that the number of testicular seminiferous tubules significantly decreased in a few sertoli cells (Fig. 1A); however, this was not observed at all levels of spermatogenic cell.
Figure 1.
Patient results, including histological analysis, cytogenetic analysis, FISH analysis and PCR. (A) A percutaneous testicular biopsy demonstrating the convoluted seminiferous tubules were hypogenetic with interstitial fibrosis (hematoxylin and eosin staining; magnification, 50 µm). A few sertoli cells were observed in seminiferous tubules (white arrow). (B) Chromosome analysis demonstrating a pericentric inversion of the chromosome 9 (red arrow). The normal chromosome 9 is indicated by a green arrow and the normal chromosome X is indicated by a blue arrow. (C) FISH demonstrating that the orange signal of the SRY gene was absent in the patient, while two green signals were observed on the X chromosome (white arrows). (D) PCR analysis demonstrating the absence of azoospermia factor regions. Lanes: 1, ZFX/ZFY (690 bp), SRY (472 bp), sY254 (400 bp), sY127 (274 bp) and sY86 (155 bp); 2, ZFX/ZFY (690 bp), SRY (472 bp), sY134 (301 bp), sY84 (255 bp) and sY255 (126 bp); 3, ZFX/ZFY (690 bp), SRY (472 bp), sY157 (286 bp), sY239 (201 bp) and sY124 (109 bp); 4, ZFX/ZFY (690 bp), SRY (472 bp), sY242 (233 bp), sY132 (160 bp) and sY152 (126 bp). N, fertile male; P, patient; B, blank; M, DL2000 marker (2,000, 1,000, 750, 500, 250, 100 bp). FISH, fluorescent in situ hybridization; PCR, polymerase chain reaction.
Cytogenetic analysis and FISH analysis
Karyotype analysis confirmed the 46,XX inv (9) (p11q13) karyotype (Fig. 1B) and FISH analysis demonstrated negative expression of the SRY gene (Fig. 1C). It was determined that inv (9) (p11q13) was inherited from the patient's mother. No other abnormality was observed in the family members of the patient.
PCR amplification and sequencing of the coding region of candidate genes
The SRY gene was negative in peripheral blood leukocytes, as confirmed by FISH and PCR using two different primer pairs (Fig. 1C and D). STS detection revealed that the azoospermia factor (AZF)a, AZFb and AZFc regions were absent (Fig. 1D). Absence of PCR amplification products of the other 11 sites further confirmed the lack of Y chromosome sequences in the patient genomic DNA. The coding region and exon/intron boundaries of the RSPO1, DAX1, SOX9, SOX3, ROCK, DMRT1, SPRY2 and FGF9 genes were sequenced, and no mutation was detected.
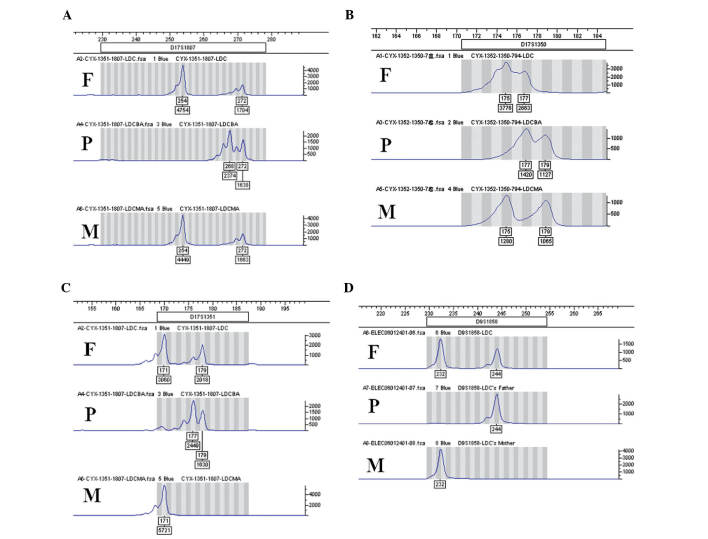
The five STS markers D17S794, D17S1352, D17S1807 (Fig. 2A), D17S1350 (Fig. 2B) and D17S1351 (Fig. 2C) in the SOX9 gene were normal. D9S1858 (Fig. 2D) was detected in the DMRT1 gene. All markers revealed no abnormality of dosage, in comparison with the results of the patient's parents.
Figure 2.
SOX9 and DMRT1 internal locus analysis. (A) Two kurtoses of 254 and 272 bp of D17S1807, (B) two kurtoses of 175 and 177 bp of D17S1350 and (C) two kurtoses of 171 and 179 bp of D17S1351 in the SOX9 gene. There were (D) two kurtoses of 232 and 244 bp of D9S1858 in the DMRT1 gene. No abnormal dose was detected in any STR site. P, patient; F, father; M, mother.
SNP array
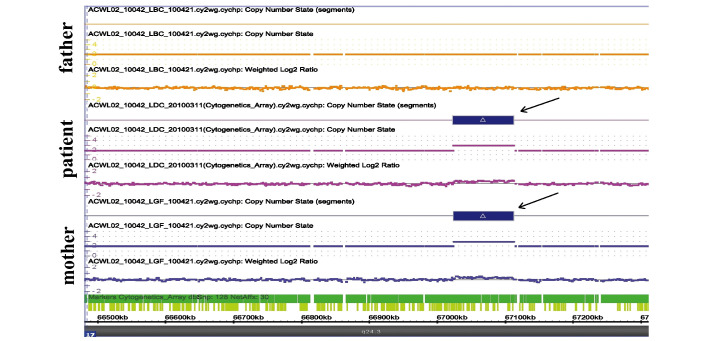
Following comparison of the results of the patient with those of the parents, it was identified that the patient exhibited a ~88-kb duplication in a region upstream of SOX9 at chromosome 17: 67,024,087–67,112,435 (Fig. 3). However, the duplicated region was also identified in the mother, which was a polymorphism (Fig. 3).
Figure 3.
Affymetrix cytogenetics whole genome 2.7 M arrays were used for detecting the genomic DNA from the patient and the parents. An 88-kb duplication was identified at the chromosome 17: 67,024,087-67,112,435 region in the patient and the mother (black arrows). This result indicated that the change was a polymorphism and not a cause of 46,XX male disorder of sex development.
Discussion
The 46,XX male DSD syndrome is a rare genetic condition, which occurs with an incidence of ~1:20,000 in newborn males (24,25). The 46,XX male DSD syndrome was first reported in 1964 by de la Chapelle (24). The patients are all male with testicular tissue differentiation. Normal male sexual differentiation predominantly depends on the Y chromosome, which contains TDF. In general, sex determination and differentiation of mammals begins with the expression of the SRY gene, which is located in the short arm of the Y chromosome, and is the major sex determination gene (26–28). The undifferentiated gonad is differentiated into the testicle via the SRY gene, which secretes testosterone and mullerian duet inhibiting substance (MIS) to launch a series of male sexual differentiation mechanisms. At the molecular level, XX males can be classified as SRY-positive and SRY-negative. In the present study, a case of 46,XX SRY-negative males was identified. The mechanism by which the induction of testicular tissue is performed in SRY-negative patients remains to be elucidated.
Several hypotheses have been suggested regarding testicular development in SRY-negative patients. Hiddengonadal mosaicism of SRY exists in patients. Domenice et al (29) indicated that no SRY was detected in the blood lymphocytes or skin tissue. Unfortunately, the present study was unable to detect the Y chromosome or the SRY gene in the blood samples. Certain autosomal or X-linked genes, which repress the male pathway (i.e. mutations in this pathway can result in de-repression of the male pathway in XX gonads) (1). For instance, a point mutation in the Wilm's tumor suppressor gene WT1 on the autosome can lead to sex reversal, as previously described (30). Altered expression of other sex-determining genes, such as the DAX1 gene, which are located downstream of SRY has been reported to be the cause of sex reversal (31). SRY is located in the short arm of the male Y chromosome, which is adjacent to the pseudoautosomal region. SRY negatively regulates the expression of the Z locus of the autosome, which may contain the male-determination gene. The reason that males with a deletion of SRY cannot express the male-determination gene is the degeneration of the Z locus. Therefore, a 46,XX male has normal male internal and external genitalia.
A previous study demonstrated that the overexpression of SOX9, which is located in 22q13, can trigger the development of testicles in the absence of SRY (17). The transcription factor SOX9 is the most important gene in the sex-determining gene high-mobility group box family, which is associated with SRY. SOX9 is the first gene expressed in precursor cells of Sertoli cells following SRY, and the normal expression of SOX9 is associated with testicular differentiation (20). Ectopic expression of SOX9 in the female gonad of XX mice, which lack SRY, caused complete female-to-male sex reversal (20,32). These data demonstrated that SOX9 can support the formation of functional Sertoli cells from precursor cells. As the primary molecular marker of SRY differentiation, MIS can induce precursor cells of Leydig cells in undifferentiated gonad to Leydig cells under the action of Sertoli cells secreting testosterone and promoting the normal development of the male reproductive tract (33). The genomic domain regulating the expression of SOX9 was located 500–600 kb upstream of SOX9 (1,21,23). It has been suggested that disruptions in certain regions upstream of SOX9 can lead to various diseases, which result from the removal of variable long-range tissue-specific regulatory elements, which alter the expression of SOX9 (34). Certain studies have suggested that duplication of the region containing SOX9 is able to trigger sex reversal. Huang et al (18) have reported a case of 46,XX SRY negative male sex reversal caused by duplication of SOX9. Benko et al (23) demonstrated that the copy number variations of the region containing SOX9 were the genetic basis for 46,XX DSDs of variable severity (ranging from mild to complete sex reversal).
In the present study, no change in the levels of SOX9 was detected by SOX9 internal locus analysis. In addition, deletion/duplication analysis was performed on the patient and a genomic-wide high-density Cyto2.7M™ Array kit (Affymetrix Inc.) was used to assess the parents. The patient exhibited a de novo partial duplication originating from the upstream of SOX9, involving ~88 kb in the chromosome 17: 67,024,087–67,112,435 region. An identical duplication was also detected in the mother. Therefore, it was suggested that the change was merely a polymorphism. The duplication located in the regulatory region upstream of SOX9 was not a direct cause of the 46,XX testicular disorder of sex development (18). Another explanation may be that besides SRY, there are other genes associated with male gonad differentiation. The clinical, molecular and cytogenetic findings of the present study suggested that other unidentified genetic or environmental factors are significant in the regulation of SDS. Further studies will aim at identifying the exact pathogenesis of the disease.
Acknowledgments
The authors would like to thank the patient and his parents for their kind participation and support. The authors are grateful to Dr Xiao-Qin Ye, Dr Yuan-Zhe Wu and Dr Hong-Lin Yin for clinical data and histopathological diagnosis. This study was supported by the Natural Science Foundation of Jiangsu Province (nos. BK2012601 and BK2011660), the Key Foundation of Jiangsu Science and Technology Bureau (no. BM2013058) and the Natural Science Foundation of China (no. 81170611).
References
- 1.Xiao B, Ji X, Xing Y, Chen YW, Tao J. A rare case of 46, XX SRY-negative male with approximately 74-kb duplication in a region upstream of SOX9. Eur J Med Genet. 2013;56:695–698. doi: 10.1016/j.ejmg.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Abbas NE, Toublanc JE, Boucekkine C, Toublanc M, Affara NA, Job JC, Fellous M. A possible common origin of 'Y-negative' human XX males and XX true hermaphrodites. Hum Genet. 1990;84:356–360. doi: 10.1007/BF00196234. [DOI] [PubMed] [Google Scholar]
- 3.Boucekkine C, Toublanc JE, Abbas N, Chaabouni S, Ouahid S, Semrouni M, Jaubert F, Toublanc M, McElreavey K, Vilain E. Clinical and anatomical spectrum in XX sex reversed patients. Relationship to the presence of Y specific DNA-sequences. Clin Endocrinol (Oxf) 1994;40:733–742. doi: 10.1111/j.1365-2265.1994.tb02506.x. [DOI] [PubMed] [Google Scholar]
- 4.McElreavey K, Rappaport R, Vilain E, Abbas N, Richaud F, Lortat-Jacob S, Berger R, Le Coniat M, Boucekkine C, Kucheria K. A minority of 46, XX true hermaphrodites are positive for the Y-DNA sequence including SRY. Hum Genet. 1992;90:121–125. doi: 10.1007/BF00210754. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson-Smith MA, Cooke A, Affara NA, Boyd E, Tolmie JL. Genotype-phenotype correlations in XX males and their bearing on current theories of sex determination. Hum Genet. 1990;84:198–202. doi: 10.1007/BF00208942. [DOI] [PubMed] [Google Scholar]
- 6.Ergun-Longmire B, Vinci G, Alonso L, Matthew S, Tansil S, Lin-Su K, McElreavey K, New MI. Clinical, hormonal and cytogenetic evaluation of 46,XX males and review of the literature. J Clin Endocrinol Metab. 2005;18:739–748. doi: 10.1515/jpem.2005.18.8.739. [DOI] [PubMed] [Google Scholar]
- 7.Vorona E, Zitzmann M, Gromoll J, Schuring AN, Nieschlag E. Clinical, endocrinological, and epigenetic features of the 46,XX male syndrome, compared with 47,XXY Klinefelter patients. J Clin Endocrinol Metab. 2007;92:3458–3465. doi: 10.1210/jc.2007-0447. [DOI] [PubMed] [Google Scholar]
- 8.Brennan J, Capel B. One tissue, two fates: Molecular genetic events that underlie testis versus ovary development. Nat Rev Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- 9.Meeks JJ, Weiss J, Jameson JL. Dax1 is required for testis determination. Nat Genet. 2003;34:32–33. doi: 10.1038/ng1141. [DOI] [PubMed] [Google Scholar]
- 10.Sutton E, Hughes J, White S, Sekido R, Tan J, Arboleda V, Rogers N, Knower K, Rowley L, Eyre H, et al. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest. 2011;121:328–341. doi: 10.1172/JCI42580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, Guerra L, Schedl A, Camerino G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 12.Chi L, Itäranta P, Zhang S, Vainio S. Sprouty2 is involved in male sex organogenesis by controlling fibroblast growth factor 9-induced mesonephric cell migration to the developing testis. Endocrinology. 2006;147:3777–3788. doi: 10.1210/en.2006-0299. [DOI] [PubMed] [Google Scholar]
- 13.Chiang HS, Wu YN, Wu CC, Hwang JL. Cytogenic and molecular analyses of 46, XX male syndrome with clinical comparison to other groups with testicular azoospermia of genetic origin. J Formos Med Assoc. 2013;112:72–78. doi: 10.1016/j.jfma.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno K, Kojima Y, Kamisawa H, Moritoki Y, Nishio H, Kohri K, Hayashi Y. Gene expression profile during testicular development in patients with SRY-negative 46,XX testicular disorder of sex development. Urology. 2013;82:1453.e1–e7. doi: 10.1016/j.urology.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 15.Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Evolution: Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- 16.Shan Z, Zabel B, Trautmann U, Hillig U, Ottolenghi C, Wang Y, Haaf T. FISH mapping of the sex-reversal region on human chromosome 9p in two XY females and in primates. Eur J Hum Genet. 2000;8:167–173. doi: 10.1038/sj.ejhg.5200431. [DOI] [PubMed] [Google Scholar]
- 17.Seeherunvong T, Perera EM, Bao Y, Benke PJ, Benigno A, Donahue RP, Berkovitz GD. 46, XX sex reversal with partial duplication of chromosome arm 22q. Am J Med Genet A. 2004;127:149–151. doi: 10.1002/ajmg.a.20630. [DOI] [PubMed] [Google Scholar]
- 18.Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–353. doi: 10.1002/(SICI)1096-8628(19991203)87:4<349::AID-AJMG13>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 19.Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- 20.Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 21.Cox JJ, Willatt L, Homfray T, Woods CG. A SOX9 duplication and familial 46, XX developmental testicular disorder. N Engl J Med. 2011;364:91–93. doi: 10.1056/NEJMc1010311. [DOI] [PubMed] [Google Scholar]
- 22.Vetro A, Ciccone R, Giorda R, Patricelli MG, Della Mina E, Forlino A, Zuffardi O. XX males SRY negative: A confirmed cause of infertility. J Med Genet. 2011;48:710–712. doi: 10.1136/jmedgenet-2011-100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benko S, Gordon CT, Mallet D, Sreenivasan R, Thauvin-Robinet C, Brendehaug A, Thomas S, Bruland O, David M, Nicolino M, et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet. 2011;48:825–830. doi: 10.1136/jmedgenet-2011-100255. [DOI] [PubMed] [Google Scholar]
- 24.de la Chapelle A. The etiology of maleness in XX men. Hum Genet. 1981;58:105–116. doi: 10.1007/BF00284157. [DOI] [PubMed] [Google Scholar]
- 25.Wachtel SS. XX sex reversal in the human Molecular genetics of sex determination. San Diego: Academic; 1994. p. 207. [Google Scholar]
- 26.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 27.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 28.Brennan J, Capel B. One tissue, two fates: Molecular genetic events that underlie testis versus ovary development. Nat Rev Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- 29.Domenice S, Nishi MY, Billerbeck AE, Carvalho FM, Frade EM, Latronico AC, Arnhold IJ, Mendonca BB. Molecular analysis of SRY gene in Brazilian 46,XX sex reversed patients: absence of SRY sequence in gonadal tissue. Med Sci Monit. 2001;7:238–241. [PubMed] [Google Scholar]
- 30.Ostrer H. Sex determination: Lessons from families and embryos. Clin Genet. 2001;59:207–215. doi: 10.1034/j.1399-0004.2001.590401.x. [DOI] [PubMed] [Google Scholar]
- 31.Sukumaran A, Desmangles JC, Gartner LA, Buchlis J. Duplication of dosage sensitive sex reversal area in a 46, XY patient with normal sex determining region of Y causing complete sex reversal. J Pediatr Endocrinol Metab. 2013;26:775–779. doi: 10.1515/jpem-2012-0354. [DOI] [PubMed] [Google Scholar]
- 32.Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 33.Habert R, Lejeune H, Saez JM. Origin, differentiation and regulation of fetal and adult Leydig cells. Mol Cell Endocrinol. 2001;179:47–74. doi: 10.1016/S0303-7207(01)00461-0. [DOI] [PubMed] [Google Scholar]
- 34.Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]