Abstract
Renal carcinoma is the most common type of kidney cancer in adults and is responsible for ~90–95% of the cases of kidney cancer. Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes, and is expressed at low levels in multiple malignancies. In the present study, a series of experiments were designed and performed to investigate whether the expression of TET1 is clinically correlated with clinical outcomes in renal carcinoma, and to examine the associations between TET1 expression level and the proliferation and migration in renal carcinoma cells. As a result, TET1 was observed to exhibit markedly low expression levels in 54 tumor tissue samples from 54 patients with renal carcinoma. Furthermore, statistical analysis revealed a clinical correlation between low expression levels of TET1 and the prognosis of patients with renal carcinoma. When TET1 was overexpressed in renal carcinoma cells, the viability and invasive abilities of the cells were decreased, and the rate of apoptosis was increased. In conclusion, the results demonstrated that TET1 is involved in tumor inhibition in renal carcinoma by promoting cell apoptosis and inhibiting cell proliferation and invasion, which may be exploited as a novel therapeutic target in the treatment of renal carcinoma.
Keywords: renal carcinoma, ten-eleven translocation methylcytosine dioxygenase 1, prognosis, proliferation, cell apoptosis, invasion
Introduction
Renal carcinoma is the most common type of kidney cancer in adults and is responsible for ~90–95% of the cases of kidney cancer (1). As a result of advances in chemotherapy, the long-term survival rates for patients with renal carcinoma, exhibiting no detectable metastases at diagnosis, have significantly improved. However, for patients that present with metastasis or exhibit disease recurrence, the long-term survival rates are low (2,3). There is an ongoing requirement for novel therapeutic strategies and a more thorough understanding of the mechanisms, which are involved in the development of renal carcinoma and the metastatic process, particularly to the lungs (4).
Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes (TET1/2/3), which, in humans, is encoded by the TET1 gene that possesses a zinc-binding CXXC domain containing eight conserved cysteine residues, which bind zinc (5). TET1 is identified as a 5-methylcytosine (5-mC) hydroxylase, which catalyzes the conversion of the modified DNA base, 5-mC, into 5-hydroxy-methylcytosine (5-hmC) in vitro or in vivo (6,7). Notably, substantial downregulation of all three TET genes has been reported in various types of solid tumor (8–12), and TET1 has been demonstrated to be an essential tumor suppressor in prostate and breast cancer (13,14). Until now, there have been no reports regarding the expression levels of TET1 and its association with clinical outcome in renal carcinoma.
Tumor tissue from patients with renal carcinoma has been found to exhibit a significantly higher incidence of hypermethylation in several genes, compared with corresponding normal tissue (15). Methylation of the promoter region of the P14ARF gene has been associated with repression of its protein expression and a poor prognosis in patients with renal carcinoma (16).
The present study aimed to demonstrate the significance of TET1 in the prognosis of renal cancer, and to investigate the roles of TET1 in the proliferative and migratory abilities of renal carcinoma cells as well as its pro-apoptotic effect.
Materials and methods
Tumor samples
The tumor samples were obtained from the tumor tissues of patients diagnosed with renal carcinoma, which had been preserved in the Cancer Tissue Bank between 2007 and 2013 at Changzhou First People's Hospital (Jiangsu, China). Among the 54 selected tumor samples, 35 were from male patients and 19 were from female patients. The average age of the patients was 36 years. Informed consent for the experimental use of surgical samples was obtained from all patients. All the tissue specimens were sampled from the tumors at the time of surgery, snap frozen and stored at −80°C until retrieval for experiments. The clinical specimens were divided into two groups; a low tumor grade group and a high tumor grade tumor group, graded according to the Price grading system. Normal renal tissue was used as a control, which was also obtained from the Cancer Tissue Bank in Changzhou First People's Hospital.
Cell culture, plasmid construction and transfection
The ACHN and 293T human renal carcinoma cell lines were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cultures were grown in Dulbecco's modified Eagle's Medium (DMEM), supplemented with 10% fetal calf serum (FCS; Life Technologies, Gaithersburg, MD, USA), penicillin (80 U/ml) and streptomycin (100 U/ml), at 37°C in a 5% CO2 atmosphere. All transfections were performed using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA). The TET1 gene cloned in vector pCMV6-XL5 was purchased from OriGene Technologies (Rockville, MD, USA) and the siRNA against TET1 was synthesized by GenePharma Co., Ltd. (Shanghai, China). The sequence of TET1 siRNA was 5′-GCTCGCGAGCTATAGAAGAAT-3′. The ACHN cells were transfected with the indicated plasmids prior to the subsequent western blot and flow cytometric analyses (BD Accuri™ C6 flow cytometer; BD Biosciences, Ann Arbor, MI, USA).
Immunohistochemistry and western blotting
The paraformaldehyde-fixed (10%) and paraffin-embedded tissues were sectioned at 3 µm. The paraffin sections were dried at 58°C for 2–4 h, then dewaxed with xylene and hydrated with alcohol. Antigen retrieval solution (0.01 mol/l sodium citrate, pH 6.0) was added after the sections were washed with phosphate-buffered saline (PBS). Subsequently, they were boiled at a constant temperature of 95°C for 15 min. To inhibit endogenous peroxidase activity, the sections were incubated with H2O2 (3%) at room temperature for 10 min. Following washing with PBS, 10% fetal calf serum was added, which was followed by a 30 min incubation at 37°C and washing with PBS three times. The expression levels and subcellular distributions of TET1 in the renal carcinoma tissue sections were detected using primary antibody (rabbit monoclonal anti-TET1; cat. no. ab157004; diluted 1:500), biotinylated secondary antibody (goat polyclonal anti-rabbit IgG; cat. no. ab150077; diluted 1:1,000) (Abcam Inc., Cambridge, MA, USA) and ABC reagent (Vector Laboratories, Inc., Burlingame, CA, USA), with 3,3′diaminobenzidine tetrahydrochloride (Sigma-Aldrich, St. Louis, MO, USA) as the substrate. The expression levels were measured by counting the number of cells with detectable immunoreactivity under an optical microscope (Olympus IX71; Olympus, Tokyo, Japan). The total proteins were extracted from the tissues or cells using lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China), containing phenylmethylsulfonyl fluoride, and were subsequently quantified using the Bradford method (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The proteins (20 µg) were separated on 10% SDS-PAGE gels (Sangon Biotech Co., Ltd., Shanghai, China), transferred onto polyvinilydine fluoride or nitrocellulose membranes (EMD Millipore, Billerica, MA, USA) and were visualized using a BCIP/NBT kit (Gibco Life Technologies, Carlsbad, CA, USA). The band densities were quantified by gray analysis using ImageJ version 1.41o (National Institutes of Health, Bethesda, MD, USA). The primary antibodies used were monoclonal anti-TET1 (dilution, 1:1,000) and anti-actin (dilution 1:100; cat. no. ab8227; Abcam Inc.).
Viability and apoptosis detection
The rate of cell proliferation was assessed using flow cytometry. Briefly, the cells (1×106) were stained with propidium iodide (PI) and were monitored using flow cytometry. The proliferation index was calculated using the following formula: Proliferation index = (S + G2/M) / (S + G2/M + G0/G1). The levels of apoptosis were monitored according to PI and annexin V binding, according to the manufacturer's instructions (FITC Annexin V Apoptosis Detection Kit I; BD Biosciences).
Cell proliferation assay
The cell counting kit-8 (CCK-8) assay was performed with a commercial kit (Beyotime Institute of Biotechnology) to analyze proliferation. The cells (1×105) were seeded in 96-well plates, and cultivated at 37°C for 1, 2, 3 and 4 days, respectively. After 10 µl CCK-8 reagent was added into each well, the plate was incubated at 37°C for 1 h, and the absorption was read at 450 nm. For each cell line, five wells were used to evaluate cell proliferation, which was performed in triplicate in independent groups.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted from the cells or tissues using TRIzol reagent (Invitrogen Life Technologies) and reverse transcribed using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). RT-qPCR was performed using an ABI 7500 fast sequence detection system (Applied Biosystems, Foster City, CA, USA) with a SYBR green fluorescent label. The reaction mixtures contained 10 µl 2X Power SYBR Green PCR Master mix (Applied Biosystems), 3–5 pmol of each primer (Sangon Biotech Co., Ltd.) and 0.25 µl of the RT reaction product. The samples were run in triplicate in optically clear 96-well plates (Corning Incorporated, Corning, New York, NY, USA). The thermocycling parameters were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, and a detection step at 72°C for 30 sec. The actin transcript was used in each sample as an internal reference standard to control for variations in mRNA content and quantity. Each RNA sample was run in triplicate. The primer sequences used were as follows: TET1, forward 5′-ACTCAACTCTCAGAAGCCCC-3′ and reverse 5′-GGTGGCTCTCCCTGTAAGTT-3′; actin, forward 5′-CGCGGATCCATGGATGATGATATCGCCGCG-3′ and reverse 5′-CCGCTCGAGGAAGCATTTGCGGTGGACGAT-3′.
Migration assay
The cells were trypsinized in 3 ml 0.05% trypsin, washed in PBS, and resuspended in FCS-free DMEM, supplemented with 0.1% bovine serum albumin. The migration assays were performed in a 24-well plate. The cell suspensions (5×105 cells/ml; 100 µl) were added into triplicate wells in the upper chambers of Transwells. The lower chambers of the Transwells (Corning Incorporated) were filled with 600 µl 15% FCS-containing DMEM. Following incubation at 37°C for 15 h, the cells that had migrated through the filter into the lower wells were fixed with 90% ethanol, stained with 3-(4,5-dimethylthiazol-2-)-2,5-diphenyltetrazolium bromide (5 mg/ml, 20 µl). The average numbers of cells, which had migrated was determined by calculating the numbers in four randomly-selected fields under a Olympus IX71 microscope (magnification, ×400), with the number of cells expressed as a percentage of the sum of the cells in the upper and lower wells.
Statistical analysis
The association between the expression levels of TET1 and the clinical characteristics were assessed using χ2 tests. Univariate survival analysis was performed using Kaplan-Meier's method and analyzed using a log-rank test to assess survival differences between the groups. P<0.05 was considered to indicate a statistically significant difference. Analyses were performed using SPSS 10.0 statistical software for Windows (SPSS, Inc., Chicago, IL, USA).
Results
Expression levels of TET1 are low in renal carcinoma, particularly in high grade tumors
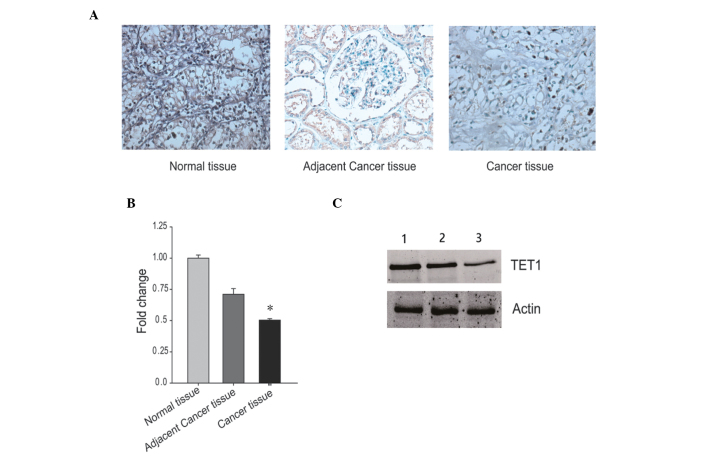
Renal carcinoma samples were collected from patients diagnosed with low tumor grade and from patients with high tumor grade, as defined by the Price grading system. The expression of TET1 was determined in the two different groups of renal carcinoma using immunohistochemistry. Immunoreactivity for TET1 in the high tumor grade tissue samples was lower, compared with that in the low tumor grade tissue samples. Notably, the two tumor groups exhibited low immunoreactivity for TET1, compared with the control tissues (Fig. 1A). In agreement with the immunohistochemical results, the mRNA expression levels of TET1 were significantly lower in the high tumor grade tissues, compared with the low tumor grade tissues (Fig. 1B; P<0.05). In addition, the results of the western blotting revealed that the protein expression levels of TET1 were lower in the high tumor grade tissues, compared with the low grade tissues (Fig. 1C), which was consistent with the results from the immunohistochemistry and RT-PCR analyses. These analyses demonstrated that the expression levels of TET1 were lower in the renal carcinoma samples than in the control samples, particularly in the group containing tissues from patients diagnosed with high tumor grades. This suggested that the expression levels of TET1 may be involved in renal carcinogenesis.
Figure 1.
Expression levels of TET1 in renal carcinoma tissues. (A) TET1 was detected in renal carcinoma tissues using immunohistochemical staining. The tumor tissue samples were obtained from patients with either low or high grade renal carcinoma. Normal renal tissues were used as a control. (B) mRNA expression levels of TET1 were measured in the renal carcinoma and control tissue samples samples using reverse transcription-quantitative polymerase chain reaction. The mRNA expression levels of actin were used as an internal control *P<0.05, compared with normal tissue and adjacent tissue. (C) Protein expression levels of TET1 were determined in the renal carcinoma and control tissue samples. Lane 1 was from normal renal tissue, Lane 2 was from low tumor grade renal carcinoma and Lane 3 was from high tumor grade renal carcinoma. The protein expression levels of actin were used as an internal control. TET1, ten-eleven translocation methylcytosine dioxygenase 1.
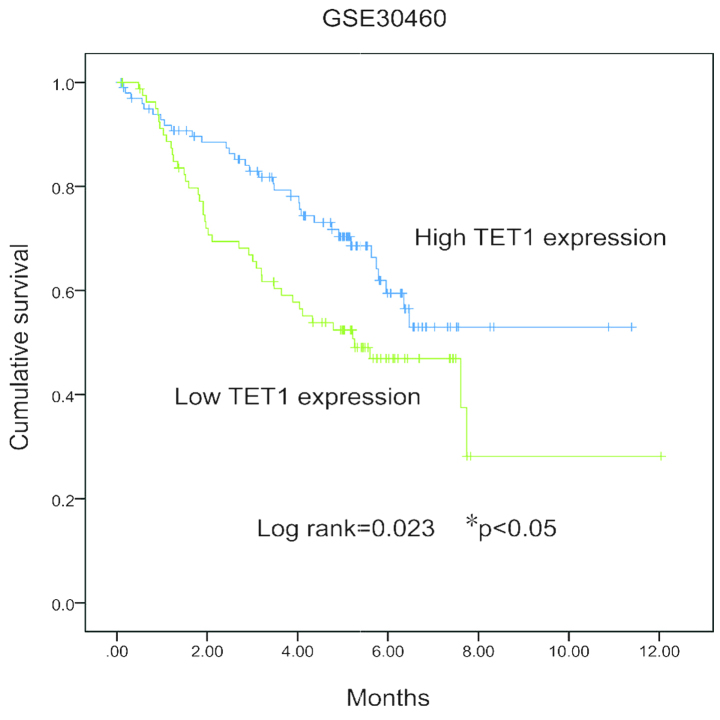
Low expression levels of TET1 are associated with poor prognosis in renal carcinoma
The statistical analyses demonstrated that the expression level of TET1 correlated negatively with the grade of the tumor. Univariate survival analysis revealed significant associations (P<0.05) between the patient prognosis, the tumor size and the expression levels of TET1. However, no significant associations were observed among genders or ages at diagnosis. The survival probability was reduced in patients with renal carcinoma tissues exhibiting lower expression levels of TET1 (Fig. 2).
Figure 2.
Kaplan-Meier curves of survival durations in patients with renal carcinoma, according to the expression levels of TET1. *P<0.05, compared with the high TET1 expression group.
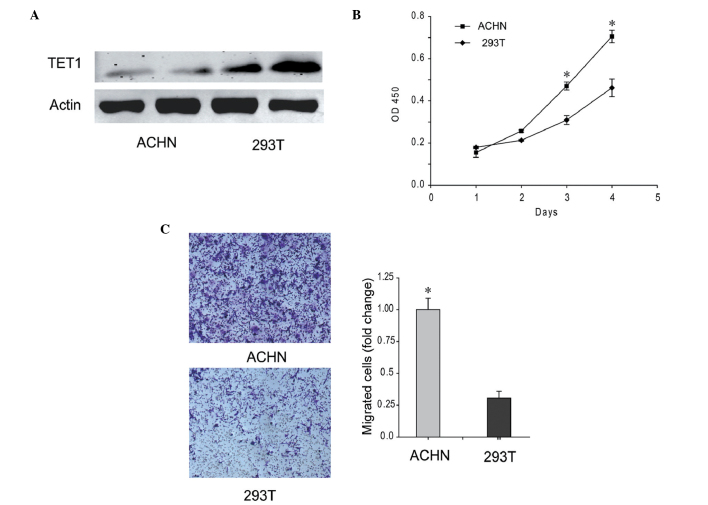
ACHN renal carcinoma cell line exhibits low levels of TET1 and high proliferation and migration abilities
The expression of TET1 was low in the ACHN renal carcinoma cell line, which exhibited the ability to metastasize to the lungs. As shown in Fig. 3A, the protein expression level of TET1 in the ACHN cells was decreased, and was ~33.67% of the level detected in the 293T cells. In addition, the proliferation and migration abilities of the ACHN cells were increased, compared with 293T cells(Fig. 3B and C). These results indicated that low expression levels of TET1 improved the proliferation and migration abilities of the ACHN cells.
Figure 3.
Low expression levels of TET1 were detected in the ACHN renal carcinoma cell line, which was accompanied by increased proliferation and migration abilities. (A) Protein levels of TET1 in the ACHN and 293T cells. ACHN is a renal carcinoma cell line and 293T is an embryonic kidney-derived cell line. (B) Proliferation abilities of the ACHN and 293T cells. (C) Metastatic abilities of ACHN and 293T cells were determined using a migration assay. Representative images and quantification are shown. *P<0.05, compared with 293T cells. TET1, ten-eleven translocation methylcytosine dioxygenase 1; OD, optical density.
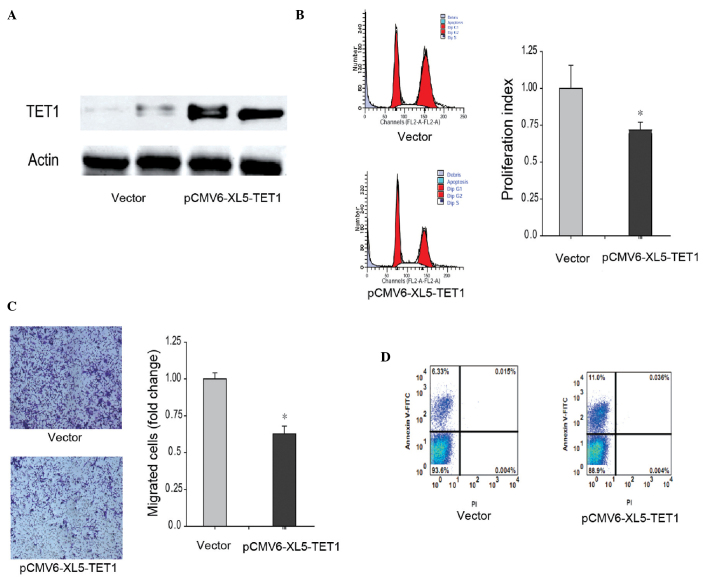
Overexpression of TET1 in ACHN cells reduces proliferation and migration abilities, and increases apoptosis
The present study also investigated the role of TET1 in renal carcinoma cells. Exogenous TET1 was expressed at high levels in the ACHN renal carcinoma cell line and was 217.34% of the level observed in the control cells (Fig. 4A). The proliferation index of the ACHN cells overexpressing TET1 was markedly decreased (P<0.05) compared with ACHN expressing background levels of TET1 (Fig. 4B). Additionally, the metastatic ability was also reduced when TET1 was overexpressed in the ACHN cells (Fig. 4C). The results of the flow cytometric analysis demonstrated that 12.5% of the ACHN cells containing exogenous TET1 were at the early stage of apoptosis, whereas only 1.28% of ACHN cells expressing background levels of TET1 were at the early stage (Fig. 4D). Taken together, these results suggested that the overexpression of TET1 inhibited renal carcinoma.
Figure 4.
Overexpressed TET1 inhibits ACHN cell proliferation and metastasis, and improves apoptosis. (A) TET1 was overexpressed in the ACHN renal carcinoma cell line. The ACHN cells were transfected with pCMV6-XL5-TET1 and screened using puromycin to generate a stable cell line. The protein levels in the whole cell extracts were analyzed using western blot analysis. (B) Proliferation rate of the TET1-overexpressed ACHN cells was determined using flow cytometric analysis. The PI was calculated as follows: PI= (S + G2/M) / (S + G2/M + G0/G1). (C) Metastatic ability of the TET1-overexpressed ACHN cells was determined using a Transwell assay. (D) Apoptotic rate of the TET1 overexpressed ACHN cells was analyzed using flow cytometry subsequent to labeling with PI and annexin V. (*P<0.05, compared with the vector group). TET1, ten-eleven translocation methylcytosine dioxygenase 1; FITC, fluorescein isothiocyanate.
Discussion
Of the patients with renal carcinoma, the mortality rate from pulmonary metastasis is >30% within 5 years after diagnosis (3). However, the cause of renal carcinoma remains to be fully elucidated. Downregulation of the expression of TET has been observed in several types of cancer, including human breast cancer, gastric cancer and prostate cancer (10,12,17,18). In the present study, the expression levels of TET1 were found to be lower in renal carcinoma (Figs. 1 and 3A). Until now, understanding of the function and mechanism of TET1 in renal cancer has been limited.
TET1 is identified as a 5-mC hydroxylase, which cata-lyzes the conversion of the modified DNA base, 5-mC, into 5-hmC, in vitro or in vivo (6,7). Methylation at the C5 position of the cytosine bases is an epigenetic modification of the mammalian genome, which is important in transcriptional regulation (19). Hypermethylation of CpG islands within the promoter and 5′ regions of genes is an important epigenetic mechanism for suppressing gene expression (20). TET1 exerts a role in DNA demethylation, by catalyzing the conversion of the modified DNA base, 5-mC, into 5-hmC in an iron- and α-ketoglutarate-dependent manner (7,21). The conversion of 5-mC into 5-hmC has been suggested as the initial step of active DNA demethylation in mammals (21).
Genetic alternations in the status of DNA methylation, termed epigenetic alterations, are the most common molecular alterations in human neoplasia (22). Previous evidence suggests that epigenetic alterations, including methylation and histone modification, of genes involved in cell cycle regulation and apoptosis may contribute to the pathogenesis of renal carcinoma (23). Patients with renal carcinoma exhibit a significantly higher (P<0.05) incidence of hypermethylation in several genes, compared with corresponding normal tissues (15). The present study demonstrated that lower expression levels of TET1 in tumor samples from patients with high tumor grading were associated with increased tumor size, metastasis and survival artes (Fig. 2). It appeared that TET1 may exert its antitumor role by demethylating the hypermethylated CpG islands within the promoter, and by activating gene expression, particularly of those genes involved in cell cycle regulation and apoptosis. The present study focused on the function of TET1 in renal cancer, however, the epigenetic regulatory mechanism of TET1 in renal cancer require investigation in the future.
In the present study, the ACHN cell line was selected for the functional investigation of TET1 in renal carcinoma, which demonstrated low expression levels of TET1 and high proliferation and migration abilities (Fig. 3). The overexpression of TET1 reduced the proliferation and migration of ACHN cells (Fig. 4B and C). In, addition, 12.5% of the cells overexpressing TET1 were positively-labeled with annexin V and negatively-labeled by PI, which suggested that cells overexpressing TET1 undergo early apoptosis (Fig. 4D). In conclusion, TET1 may exert its tumor inhibitory effects by downregulating the proliferation and migration ability of renal carcinoma cells and inducing the cells to undergo early apoptosis. These findings suggested that TET1 exerted its effects through the demethylation of hypermethylated CpG islands within the promoter and activating gene expression, particularly of those involved in cell cycle regulation and apoptosis, in an epigenetic manner.
Taken together, the findings of the present study demonstrated that lower expression levels of TET1 were associated with a high tumor grade, poor prognosis and other clinicopathological features in renal carcinoma, and that heterogenous TET1 exerts a tumor inhibitory role in renal tumor cells.
References
- 1.Naito S, Yamamoto N, Takayama T, Muramoto M, Shinohara N, Nishiyama K, Takahashi A, Maruyama R, Saika T, Hoshi S, et al. Prognosis of Japanese metastatic renal cell carcinoma patients in the cytokine era: A cooperative group report of 1463 patients. Eur Urol. 2010;57:317–326. doi: 10.1016/j.eururo.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Wang LL. Biology of osteogenic sarcoma. Cancer J. 2005;11:294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Shor AC, Keschman EA, Lee FY, Muro-Cacho C, Letson GD, Trent JC, Pledger WJ, Jove R. Dasatinib inhibits migration and invasion in diverse human sarcoma cell lines and induces apoptosis in bone sarcoma cells dependent on SRC kinase for survival. Cancer Res. 2007;67:2800–2808. doi: 10.1158/0008-5472.CAN-06-3469. [DOI] [PubMed] [Google Scholar]
- 4.Assouad J, Petkova B, Berna P, Dujon A, Foucault C, Riquet M. Renal cell carcinoma lung metastases surgery: Pathologic findings and prognostic factors. Ann Thorac Surg. 2007;84:1114–1120. doi: 10.1016/j.athoracsur.2007.04.118. [DOI] [PubMed] [Google Scholar]
- 5.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t (10;11) (q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 6.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu HL, Ma Y, Lu LG, Hou P, Li BJ, Jin WL, Cui DX. TET1 Exerts its tumor suppressor function by interacting with p53-EZH2 pathway in gastric cancer. J Biomed Nanotechnol. 2014;10:1217–1230. doi: 10.1166/jbn.2014.1861. [DOI] [PubMed] [Google Scholar]
- 13.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou P, Ji M, Yang B, Chen Z, Qiu J, Shi X, Lu Z. Quantitative analysis of promoter hypermethylation in multiple genes in osteosarcoma. Cancer. 2006;106:1602–1609. doi: 10.1002/cncr.21762. [DOI] [PubMed] [Google Scholar]
- 16.Oh JH, Kim HS, Kim HH, Kim WH, Lee SH. Aberrant methylation of p14ARF gene correlates with poor survival in osteosarcoma. Clin Orthop Relat Res. 2006;442:216–222. doi: 10.1097/01.blo.0000188063.56091.69. [DOI] [PubMed] [Google Scholar]
- 17.Frycz BA, Murawa D, Borejsza-Wysocki M, Marciniak R, Murawa P, Drews M, Kołodziejczak A, Tomela K, Jagodziński PP. Decreased expression of ten-eleven translocation 1 protein is associated with some clinicopathological features in gastric cancer. Biomed Pharmacother. 2014;68:209–212. doi: 10.1016/j.biopha.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov M, Barragan I, Ingelman-Sundberg M. Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci. 2014;35:384–396. doi: 10.1016/j.tips.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 21.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P, Ji M, Yang B, Chen Z, Qiu J, Shi X, Lu Z. Quantitative analysis of promoter hypermethylation in multiple genes in osteosarcoma. Cancer. 2006;106:1602–1609. doi: 10.1002/cncr.21762. [DOI] [PubMed] [Google Scholar]
- 23.Rao-Bindal K, Kleinerman ES. Epigenetic regulation of apoptosis and cell cycle in osteosarcoma. Sarcoma. 2011;2011:679457. doi: 10.1155/2011/679457. [DOI] [PMC free article] [PubMed] [Google Scholar]